Translate this page into:
Vaccine candidates for cellulitis from Staphylococcus aureus and Streptococcus pyogenes – In silico approach
⁎Corresponding author at: Biochematics Laboratory, Department of Bioinformatics, Bharathiar University, Coimbatore 641 046, Tamil Nadu, India. jeyam@buc.edu.in (Jeyam Muthusamy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Staphylococcus aureus and Streptococcus pyogenes are the causative agents of cellulitis, a bacterial skin infection. Cellulitis primarily affects the skin, and it later spreads to the other parts of the body, especially through the lymph nodes and bloodstream. Without proper medication, it becomes life-threatening. The main symptoms include inflammation, tenderness, tight and swollen skin, and fever. Vaccines using epitopes as antigens trigger quick immune responses; in addition, they are cost effective. These antigens are derived from bacterial proteins. In this study, virulence factors from membrane proteins such as sak, tst, isdA, clfB, and can fromS. aureus (Fig. 1) and SPy, scpA, and hylp1 from S. pyogenes (Fig. 2) were selected for predicting the vaccine epitopes. VaxiJen server was used to evaluate the antigenicity of the selected proteins; BCPred, AAPPred, and ABCpred were used for B-cell epitope prediction, while ProPred and ProPred I were used for T-cell epitope prediction. MHCPred was used for the selected alleles, DRB1*0101 and DRB1*0401. Pepitope server was used for epitope mapping of the selected peptides. Epitopes that are common among those from BCPred, AAPPred, and ABCpred were selected: LKYGPKFDK (tst) and MTFDDKNGK (cna) from S. aureus and YTNSDKGGS (SPy), FKIEPDTTV (SPy), MTPSERLDL (scpA), VKTDDQQDK (scpA), and LKFKPAATV (hylp1) from S. pyogenes. Among them, YTNSDKGGS, MTPSERLDL, and MTFDDKNGK, were identified as suitable vaccine candidates for eliciting immune responses. These results of this study can be used to create a peptide-based vaccine for preventing cellulitis.
Keywords
Cellulitis
S. aureus
S. pyogenes
Epitope
Immune response
1 Introduction
Skin is the largest organ in the body with multiple functions; its breakdown leads toseveral diseases (Tortora and Grabowski, 1993). The global burden of diseases estimated in 2013associated 1.79% of skin disorders withsignificant morbidity and mortality; the numbers were especially high for cellulitis (Karimkhani et al. 2017). Considering the disability-adjusted life years, skin illnesses contributed to 1.79% of the worldwide burden of disease, among the 306 diseases (DALY’s). Skin and soft tissue infections are majorly caused by bacteria, with skin inflammation (Karimkhani et al. 2017).
Cellulitis is a form of bacterial skin infection that can damage the dermis. It is caused by Staphylococcus aureus and Streptococcus pyogenes, which cause inflammatory immune responses (Dennis and Bryant, 2016). These bacteria are present on the skin, and when the skin in damaged, they enter in through the damaged regions resulting in infection of the skin. The disease-causing bacteria are more virulent during the winter months in cold countries (Olafsdottir et al. 2015). The main symptoms of such infections include skin redness, skin colonizing, skin maceration, and redness of the skin within a few days after infection. The entrance point may not be obvious, and the breach can include modification in the superficial skin or could manifest as more intrusive infections depending on the type of bacteria. More than 95% of infections occur below the knee (Vary and Connor, 2014). Diabetic patients and elderly people are majorly affected by this disease (Karimkhani et al. 2017). S. aureus is overrepresented in the skin microbiome, and the local microbiome and immunity influence the relationship between the microbiome and cellulitis (Christensen and Bruggemann, 2014).
“Virulence” is an organism’s capacity to infect its host and produce sickness. Virulence factors are components that help the bacteria establish an infection in the host at the cellular level. These elements either have a secretory, membrane-related, or cytosolic origin (Aditya Kumar et al., 2017). Virulence factors have contributed to the discovery of significant virulence traits in microbes, considerably advancing our understanding of microbial pathogenesis (Arturo and Pirofski, 2009).
When the pathogens enter into the break in the skin, they cause deep dermal and subcutaneous infection and cutaneous barrier disruption. Low surface pH, low temperature of the skin surface, and the occurrence of common microbes on the skin surface reduce colonization by pathogens (Raff and Kroshinsky, 2016). Heavy neutrophil infiltration around the blood vessels and dermal edema lymphatic dilation are important histological features of this disease (Burian et al., 2021). The primary strategy to combat cellulitis is vaccination. Peptide-based vaccines are novel and promising for the treatment of mild to chronic cellulitis (Wang and Walfield, 2005).
The UK National Health Service (NHS) conducted an analysis for cellulites diseases using the secure Anonymized information linkage databank, which compiles population-scale, individual-level anonymized linked data from a variety of sources, including 80% of primary care general practices in Wales (Humphreys et al., 2023) The number of general practice visits and in-patient stays were tracked for all patients linked to the pertinent codes in primary care settings for the last twenty years. The Welsh and UK NHS would experience significant financial savings through initiatives to assist patients and healthcare providers in identifying early indicators and dangers of cellulitis, enhance the accuracy of the initial diagnosis, avoid cellulitis recurrence, and improve evidence-based treatment pathways (Humphreys et al., 2023). In response to these discoveries, Wales has created the ground-breaking National Lymphoedema Cellulitis Improvement Program, which offers a proactive approach to cellulitis management (Humphreys et al., 2023).
Therefore, this study aimed to identify possible B-cell and T-cell epitopes from the pathogenic membrane proteins that could stimulate immune responses, both cellular and humoral (Hajighahramani et al. 2017).
2 Materials and methods
2.1 Selection of virulence proteins
Virulence factors for cellulitis were selected from the membrane proteins of S. aureus and S. pyogenes. Five proteins were selected from S. aureus – sak, tst, isdA, clfB, and cna (Fig. 1) and three proteins from S. pyogenes – SPy, scpA, and hylpL (Fig. 2). The antigenic properties of these proteins were predicted.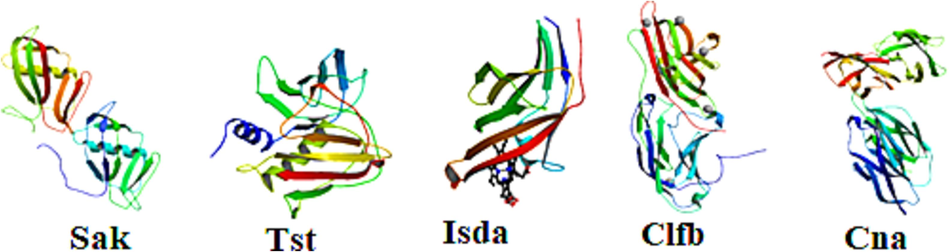
3D structures of selected S. aureus membrane proteins.

3D structures of selected S. pyogenes membrane proteins.
2.2 Retrieval of membrane protein sequences
The amino acid sequences of sak (P68802), tst (p06886), isdA (Q7A655), clfB (Q6GDH2), cna (Q53654), SPy (Q9A1S2), scpA (B6ETQ5), and hylpL (Q9A0M7) were retrieved from UniprotKB database.
2.3 Prediction of antigenicity of membrane proteins
The VaxiJen 2.0 server was used to determine the immunogenic characteristics of the recovered sequences for improving the prediction accuracy and reducing the false positives (Doytchinova and Flower, 2007). The sequences with a VaxiJen score of ≥ 0.7 were chosen.
2.4 B-cell epitope prediction
B-cell epitopes were predicted from sequences with VaxiJen score ≥ 0.7 using BCPred and AAPPred. This resulted in the identification of 20-mers using ABCpred, which resulted in 16-mer antigenic sequences (https://webs.iiitd.edu.in/raghava/propred1/) (Singh and Raghava, 2003).
2.5 T-cell epitope prediction
The chosen B-cell epitope antigens were used for T-cell epitope prediction using ProPred I based on the binding affinity with Major Histocompatibility Complex (MHC) classI, for 51 alleles (Singh and Raghava, 2003) and with MHC II, for 47 alleles (Singh and Raghava, 2001). Selected 9-mer epitopes with affinity to both MHC I and MHC II were selected. (https://webs.iiitd.edu.in/raghava/propred/).
2.6 MHCPred prediction
The prediction was made using MHCPred, based on MHC binding affinity. The IC50 values of the specific alleles, DRB1*0101 and DRB1*0401, such as predicted logIC50, predicted IC50, and confidence of prediction values were calculated. The sequences in FASTA format were evaluated using MHCPred (Guan et al., 2003) (https://www.ddgpharmfac.net/mhcpred/MHCPred/).
2.7 Target protein from protein data bank
The 3D structures of the selected 8 membrane proteins: sak (1C77), tst (2QIL), isdA (3QZO), clfB (4F24), and cna (1D2O) from S. aureus and SPy (3B2M), scpA (3EIF), and hylpL (3EKA) from S. pyogenes were retrieved from protein data bank (PDB).
2.8 Pepitope server
Epitopes were computationally predicted using the Pepitope server, and the epitope localization on the target protein was visualized. (Mayrose et al. 2007). To detect the epitope on the protein, epitope mapping was performed using PepSurf (Mayroseet al., 2006) and Mapitope (Bublil et al., 2007) (https://pepitope.tau.ac.il/).
3 Results
3.1 Virulence-associated membrane proteins and the characterization of their antigenic properties
sak, tst, isdA, clfB, cna, SPy, scpA, and hylpL were studied for their antigenic potential based on the criteria of VaxiJen score of ≥ 0.7 (Table1).
S.No
Membrane proteins
Uniprot ID
VaxiJen score
Number of identified
B-cell epitopes using BCPred, AAPPred, and ABCpred
Number of accepted
B-cell epitopes from BCPred, AAPPred, and ABCpred
Staphylococcus aureus
1
Sak
P68802
0.7026
4 + 3 + 17
3 + 0 + 8
2
Tst
P06886
0.8659
5 + 6 + 27
3 + 4 + 21
3
isdA
Q2FZE9
0.7313
10 + 10 + 45
7 + 5 + 25
4
clfB
A0A0E1AQ60
1.1204
24 + 22 + 116
19 + 19 + 98
5
Can
A0A0E1AM77
0.7300
24 + 25 + 94
15 + 16 + 54
Streptococcus pyogenes
1
Spy
Q9A1S2
0.8431
9 + 9 + 41
8 + 8 + 26
2
scpA
P15926
0.5868
33 + 31 + 127
20 + 18 + 52
3
hylpL
Q9A0M7
0.8171
7 + 9 + 40
7 + 5 + 26
3.2 B-cell epitope prediction
The number of B-cell epitopes predicted using BCPred, AAPpred, and ABCpred for the selected proteins, sak, tst, isdA, clfB, and cna from S. aureus were 24, 38, 65, 162, and 143, but the accepted B-cell epitopes were 11, 28, 37, 136, and 85. The epitopes were narrowed down based on their VaxiJen antigenic score. For S. pyogenes, the number of epitopes identified from SPy, scpa, and hylpL were 59, 191, and 56 whereas the accepted B-cell epitopes were 42, 90, and 38 (Table 1). BCPred and AAPPred identified 20-amino acid B-cell epitopes, while ABCpred predicted epitopes with 16 amino acids (Supplementary Table 1).
3.3 T-cell Epitope prediction
To identify T cell epitopes, the chosen B-cell epitopes were evaluated using ProPred1 and Propred with the standard parameters. The VaxiJen score, MHCPred score, and the surface localization of the epitope in the theoretical model were used to identify epitopes. DRB1*0101 and DRB1*0401 are the MHC class I and II alleles, respectively, to which the common epitopes most frequentlybind. T-cell epitopes with the potential to interact with MHC class I and II molecules were predicted using ProPred1, for the MHC class I-binding regions of the antigens from 47 alleles and using ProPred, for the MHC class II-binding regions of the antigens from 51 alleles, from the B-cell VaxiJen antigenic epitopes. The binding regions identified using both the servers were used for further analysis of vaccine candidates. Finally, 9-mer epitopes predicted using both ProPred and ProPred 1 were shortlisted (Table 2) for the next step.
S.No
Protein name
B-cell epitopes
Simultaneous T-cell epitope
Number of selected alleles in MHC I + MHC II
Score for T-cell Epitope VaxiJen
Staphylococcus aureus
1
sak
KIEVTYYDKNKKKEETKSFP
YYDKNKKKE
5 + 3
0.7702
–
–
–
–
AKIEVTYYDKNKKKEE
YYDKNKKKE
1 + 3
0.7702
2
tst
GLYRSSDKTGGYWKITMNDGVHGKDSPLKYGPKFDKKQLA
YRSSDKTGG
LKYGPKFDK
3 + 11
2 + 112.37811.3422
VKVHGKDSPLKYGPKFDKKQTQIHGLYRSSDKTGGYWKIT
LKYGPKFDK
YRSSDKTGG2 + 11
3 + 111.3422
2.3781
DSPLKYGPKFDKKQLA
DGSISLIIFPSPYYSP
EGTYIHFQISGVTNTE
PTPIELPLKVKVHGKD
YWKITMNDGSTYQSDL
NFFIVSPLLLATTATD
ESVMNKKLLMNFFIVSSMRIKNTDGSISLIIF
LKYGPKFDK
LIIFPSPYY
FQISGVTNT
YIHFQISGV
LPLKVKVHG
WKITMNDGS
FFIVSPLLL
FIVSPLLLA
LLMNFFIVSMRIKNTDGS2 + 11
9 + 21
6 + 18
5 + 8
2 + 10
3 + 5
12 + 29
5 + 18
1 + 10
3 + 381.3422
1.3917
0.8110
1.1966
1.1893
1.7053
0.269
1.26152.14782.3375
3
isdA
AKPNNVKPVQPKPAQPKTPT
VKPVQPKPAVQPKPAQPK
2 + 7
3 + 41.09471.3176
VEPGYKSLTTKVHIVVPQINKYQSEQRSSAMKKITMGTAS
VHIVVPQIN
YKSLTTKVH
YQSEQRSSA1 + 6
3 + 24
7 + 10.8454
0.7420
1.0402
LVYIGADSQQVNAATE
NSKYQSEQRSSAMKKI
QELATTVVNDNKKADT
KVHIVVPQINYNHRYT
EPGYKSLTTKVHIVVPSSTAPHYLCCCSARESYIGADSQQV
YQSEQRSSA
VVNDNKKAD
VHIVVPQIN
YKSLTTKVHYLCCCSARE5 + 7
7 + 1
2 + 11
1 + 6
3 + 24
3 + 30.8840
1.0402
1.0792
0.8454
0.74200.7326
4
clfB
ANSQVDNKTTNDANNIATNSGNTWVYIKGYQDKIEESSGK
VDNKTTNDAVYIKGYQDK
4 + 2
1 + 21.91520.7424
VPQEANSQVDNKTTNDANNI
VDNKTTNDA
4 + 2
1.9152
PNPNQYKVEFNTPDDQ
TDYVNTKDDVKATLTM
TVGIDSGTTVYPHQAG
TFKITVPKELNLNGVT
QAVNTSAPRMRAFSLANPENFEDVTNSVNITFYKVEFNTPD
YVNTKDDVK
VGIDSGTTV
FKITVPKEL
VNTSAPRMRVTNSVNITF2 + 4
5 + 2
8 + 8
11 + 14
2 + 2
8 + 31.1903
1.0564
1.0491
0.9177
1.50730.9582
5
cna
YVSKDITIKDQIQGGQQLDLVKMTFDDKNGKIQNGDTIKV
YVSKDITIK
IQNGDTIKV
MTFDDKNGK
5 + 9
9 + 12
5 + 80.99921.46721.8150
LPKYDEGKKIEYTVTEDHVK
PGSKITVDNTKNTIDVTIPQ
FAEFEVQGRNLTQTNTSDDK
NEKRYVSKDITIKDQIQGGQVKMTFDDKNGKIQNGDTIKVIEYTVTEDH
YVSKDITIK
ITVDNTKNT
VQGRNLTQT
YTVTEDHVK
MTFDDKNGK
1 + 2
5 + 2
3 + 4
5 + 1
5 + 9
6 + 81.1265
1.4125
0.8680
1.3175
0.9992
0.4141
QKEIEIKTDANGIANI
GKKIEYTVTEDHVKDY
ARDISSTNVTDLTVSP
GGKTTVKMTFDDKNGK
NGKIQNGDTIKVAWPT
EGTQKVKPTIYFKLYK
PTIYFKLYKQDDNQNT
TVKIEGYSKTVSLTVK
EINCNASSTAPHYLCC
APHYLCCCSARESSSPKMTFDDKNGKIQNGDTIKTDANGIA
IEYTVTEDH
YTVTEDHVK
ISSTNVTDL
MTFDDKNGK
IQNGDTIKV
VKPTIYFKL
YKQDDNQNT
YSKTVSLTV
IEGYSKTVS
INCNASSTA
YLCCCSARE
MTFDDKNGK
2 + 3
1 + 2
5 + 2
12 + 2
5 + 8
9 + 12
14 + 6
2 + 3
9 + 3
3 + 6
2 + 1
3 + 3
5 + 81.3925
1.1265
1.4125
0.7634
1.8150
1.4672
1.6698
1.6119
0.7988
0.9612
1.6269
0.73261.8150
Streptococcus pyogenes
6
SPy
YVVTEDDYKSEKYTTNVEVSMTKVTYTNSDKGGSNTKTAEPNTDFTFKIEPDTTVNEDGN
YVVTEDDYK
YTNSDKGGS
FKIEPDTTV
6 + 2
2 + 6
6 + 31.12172.24302.3066
YTNSDKGGSNTKTAEFDFSE
IPNTDFTFKIEPDTTVNEDGFGLTLKANQYYKASEKVMIE
YTNSDKGGS
FKIEPDTTV
FGLTLKANQLTLKANQYY2 + 6
6 + 3
2 + 10
8 + 12.2430
2.3066
1.33690.8411
TFKIEPDTTVNEDGNK
YVVTEDDYKSEKYTTN
VSPQDGAVKNIAGNST
TVVNGAKLTVTKNLDL
TTVHGETVVNGAKLTV
PMTKVTYTNSDKGGSN
PGVYYYKVTEEKIDKV
KVPIQFKNSLDSTTLT
DFEVPTGVAMTVAPYI
AVKNIAGNSTEQETSTSKDFNFGLTLKANQYY
FKIEPDTTV
YVVTEDDYK
VKNIAGNST
VVNGAKLTV
LTVTKNLDL
VVNGAKLTV
YTNSDKGGS
YYYKVTEEK
IQFKNSLDS
VPIQFKNSL
VAMTVAPYI
VKNIAGNSTFNFGLTLKA6 + 3
6 + 2
1 + 3
5 + 28
12 + 4
5 + 28
2 + 6
3 + 8
2 + 37
23 + 7
13 + 4
1 + 3
6 + 212.3066
1.1217
0.7509
0.8035
0.8892
0.8035
2.2430
1.1890
1.1565
0.7694
1.0109
0.75091.7373
7
scpA
DAKKASAATMYVTDKDNTSS
LQKQYETQYPDMTPSERLDL
AYANRGMKEDDFKDVKGKIA
KDQLDGDGLQFYALKNNFTA
TAMVKTDDQQDKEMPVLSTNRGDIDFKDKVANAKKAGAVGYVTDKDNTS
MTPSERLDL
YPDMTPSER
YANRGMKED
YALKNNFTA
VKTDDQQDKFKDKVANAK1 + 10
7 + 4
7 + 2
1 + 1
12 + 6
1 + 6
2 + 10.9313
1.1479
1.0347
0.8781
0.8630
1.83700.7177
TAMVKTDDQQDKEMPVLSTN
QYETQYPDMTPSERLDLAKKRTLEKRSSKRALATKASTRD
VKTDDQQDK
MTPSERLDL
YPDMTPSERLEKRSSKRA1 + 6
7 + 4
7 + 2
6 + 11.8370
1.1479
1.03471.7010
YETQYPDMTPSERLDL
TAMVKTDDQQDKEMPV
YIHRHANGEPYAAISP
GSSYYHEANSDAKDQL
SRTLEKRSSKRALATK
YVTDKDNTSSKVHLNN
ANNKYAKLSGTSMSAP
RGDIDFKDKVANAKKA
AYANRGMKEDDFKDVK
VQTDKVDGKHFALAPK
SRTLEKRSSKRALATK
YVTDKDNTSSKVHLNN
ANNKYAKLSGTSMSAP
RGDIDFKDKVANAKKA
AYANRGMKEDDFKDVK
VQTDKVDGKHFALAPK
KVVANGTYTYRVRYTP
ELYYQATVQTDKVDGKLVAHIFKTKRQKETKK
MTPSERLDL
YPDMTPSER
VKTDDQQDK
YIHRHANGE
YYHEANSDA
LEKRSSKRA
YVTDKDNTS
YAKLSGTSM
FKDKVANAK
YANRGMKED
VQTDKVDGK
LEKRSSKRA
YVTDKDNTS
YAKLSGTSM
FKDKVANAK
YANRGMKED
VQTDKVDGK
YTYRVRYTP
VQTDKVDGK
YQATVQTDKFKTKRQKET7 + 4
7 + 2
1 + 6
1 + 4
4 + 4
6 + 1
1 + 10
10 + 1
2 + 1
1 + 1
4 + 10
6 + 1
1 + 10
10 + 1
2 + 1
1 + 1
4 + 10
3 + 4
4 + 4
6 + 4
2 + 21.1479
1.0347
1.8370
1.2629
1.0072
1.7010
0.9313
0.7302
0.7177
0.8781
2.1472
1.7010
0.9313
0.7302
0.7177
0.8781
2.1472
1.1699
2.1472
0.85111.1338
8
hylpL
NITSGNENGSAMQLRGSEKANLKGGVMTGQLKFKPAATVA
MQLRGSEKA
LKFKPAATV
3 + 4
6 + 121.20061.7727
NGAGTAAQGIYINSTSGTTGMTGQLKFKPAATVAYSSSTG
IYINSTSGT
YINSTSGTT
LKFKPAATV
3 + 12
3 + 2
6 + 121.5134
1.72471.7727
GGVMTGQLKFKPAATV
AVNIDLSSTRGAGVVV
GNLKLKDPTANDHAAT
DYKGTTNAVNIAMRQP
AQGIYINSTSGTTGKL
GSAMQLRGSEKALGTLYNLLTNKPNIDGLATK
LKFKPAATV
VNIDLSSTR
IDLSSTRGA
LKLKDPTAN
YKGTTNAVN
IYINSTSGT
YINSTSGTT
MQLRGSEKAYNLLTNKPN6 + 12
2 + 12
6 + 1
2 + 5
3 + 4
3 + 12
3 + 2
3 + 4
2 + 71.7727
1.7435
1.5063
1.2230
0.8176
1.5134
1.7247
1.20060.9157
The epitopes identified using BCPred, AAPPred, and ABCpred (Table 2) were selected for screening using Pepitope.
The final epitopes predicted from S. aureus are LKYGPKFDK and MTFDDKNGK and from S. pyogenes are YTNSDKGGS, FKIEPDTTV, MTPSERLDL, VKTDDQQDK, and LKFKPAATV. Pepitope, which identifies the best epitope cluster, was used to find the epitope clusters on the protein’s outer membrane (Table 3).
S.No
Protein name
Epitope/s
Staphylococcus aureus
1.
tst
LKYGPKFDK
2.
cna
MTFDDKNGK
Streptococcus pyogenes
1.
Spy
YTNSDKGGSFKIEPDTTV
2.
scpA
MTPSERLDLVKTDDQQDK
3.
hylpL
LKFKPAATV
3.4 Pepitope cluster
Among the 58 epitopes identified from S. aureus, only the two best clusters, LKYGPKFDK and MTFDDKNGK, were present on the outer surface of the protein (Fig. 3).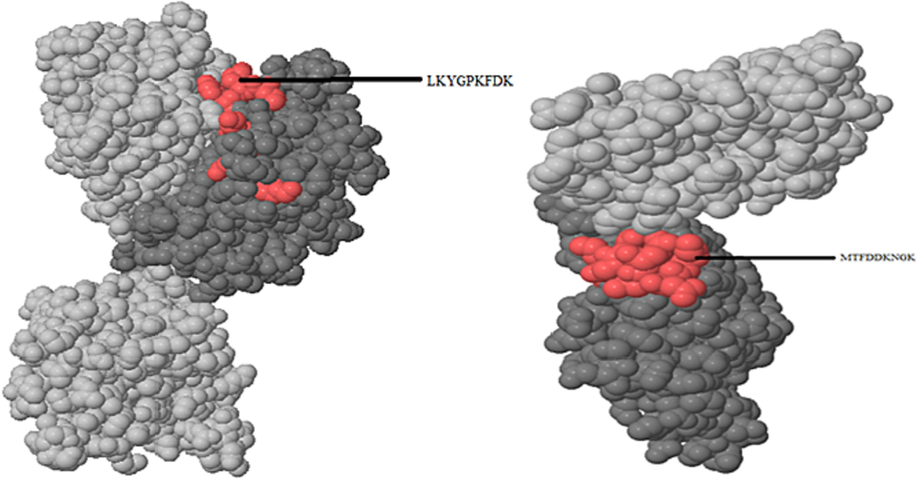
Epitopes of S. aureus predicted using Pepitope.
Five best epitopes of S. pyogenes were shortlisted from among the 64 epitopes. Among them, FKIEPDTTV, MTPSERLDL, and LKFKPAATV (Fig. 4) were present on the outer surface of the protein. The red regions are the epitopes present on the outer surface of the protein.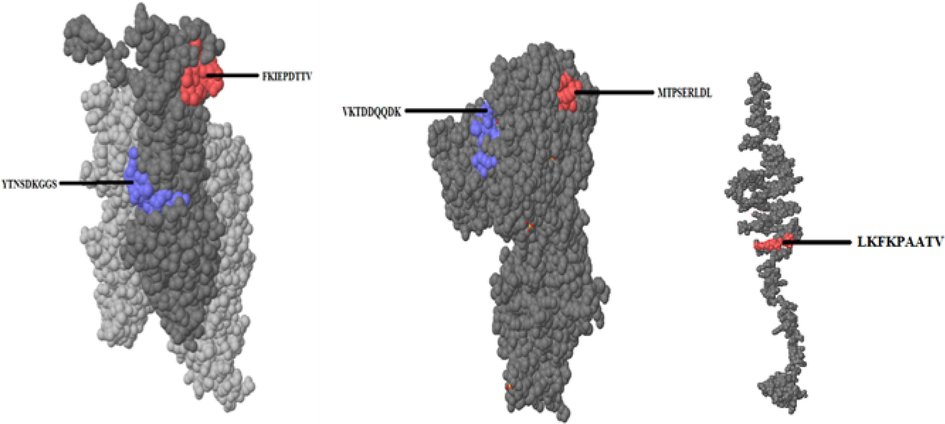
Epitopes of S. pyogenes predicted using Pepitope.
IC50 values of 0.01 to 5000 nM indicate that the sequence is eligible; < 5000 nM indicates low affinity, < 500 nM indicates intermediate affinity, and < 50 nMindicates high affinity. The epitopes were selected as vaccine candidates based on the IC50 values for both alleles, DRB1*0101 and DRB1*0401 (Table 4), the most-prevalent MHC alleles in mammals. Further experimental validation of the vaccine candidates is necessary. Among the two epitopes from S. aureus, MTFDDKNGK exhibited an intermediate binding affinity for DRB1*0101 with a IC50 value of 84.33, which is slightly higher than the threshold value of 50 nM; it had a low affinity for DRB1*0401. LKYGPKFDK exhibited low affinity for both alleles. In case of S. pyogenes epitopes, both YTNSDKGGS and MTPSERLDL had a high binding affinity for DRB1*0101; the former had an intermediate and the latter had a low affinity for DRB1*0401. FKIEPDTTV had intermediate binding affinity for DRB1*0101 and low binding affinity for DRB1*0401. Both VKTDDQQDK and LKFKPAATV showed low affinity for both alleles.
S.No
Protein name
T-cell epitopes
IC50values of DRB1*0101 (MHCPred)
IC50 values of DRB1*0401 (MHCPred)
Staphylococcus aureus
1.
tst
LKYGPKFDK
944.06
2697.74
2.
cna
MTFDDKNGK
84.33
1729.82
Streptococcus pyogenes
1.
SPy
YTNSDKGGSFKIEPDTTV
33.81268.53
597.04672.98
2.
scpA
MTPSERLDLVKTDDQQDK
29.85620.87
1945.36571.48
3.
hylpL
LKFKPAATV
706.32
3118.89
4 Discussion
The two most common bacteria that cause cellulitis are S. aureus and S. pyogenes (Bennett et al., 2010); they are frequently encountered and clinically challenging (Sullivan and Barra, 2018). Methicillin-resistant Staphylococcus aureus (MRSA) in the population increased skin and soft tissue infections from 1993 to 2005 (Pallinet al., 2008). In a recent studyincluding40 locations and 9 countries encompassing 7477 chronic oedema patients, 16% had experienced cellulitis in the previous 12 months with a prevalence of 37% (Burian et al., 2021).
To avoid the risk factors associated with cellulitis, it is essential to identify an effective vaccine; this strategy is widely used to tackle the incidence of diseases with high risk (Oscherwitz, 2016). A vaccine for S. aureus against bacteremia and pneumonia is available (Adhikari et al. 2012). This study aimed to predict a vaccine for cellulitis.
Both the humoral response involving B-cells and cell-mediated immunity involvingT-cells are important for adaptive immunity. Pathogens as a whole are not recognized by B- and T-cells; however, molecular components known as antigens are recognized (Paul et al., 2015). Epitopes as vaccine candidates have no undesirable effect; therefore, they are preferred over whole protein vaccines (Gallimore et al., 1998). The research of Correia et al. (2014) supports the epitope-focused vaccine design employed in this study.
Locating possible B- and T-cell epitopes using computational techniques, such as immunoinformatics reduces the time and lowers the cost involved in pathogen gene product investigation in the laboratory. (Davies and Flower, 2007). Mondal et al. (2019) showed that the virulence factor and outer membrane proteins are promising candidates for designing epitopic vaccines. Therefore, the virulence factor-based membrane proteins, such as sak, tst, isdA, clfB, and can from S. aureus and SPy, scpA, and hylp1 from S. pyogenes were selected using VaxiJen server with a VaxiJen antigenicity cutoff score of ≥ 0.7, with 70–89% prediction accuracy (Verjovski-Almeida et al., 2003). According to Tong et al., (2009), the majority of investigations use linear epitopes, similar to that predicted in this study, because these B-cell epitopes interact with B-cell receptors to form antigenic determinants on the surface of pathogens. Chen et al. (2007) suggested that utilizing the three tools, BCPred, AAPPred, and ABCpred will increase the accuracy of epitope prediction. B-cells provide long-term protection against pathogens and harmful molecules (Jespersen et al., 2019); therefore, 20-mer B-cell epitopes (Singh et al., 2013) were predicted using BCPred and AAPred, and 16 mer-epitopes (EL-Manzalawyet al., 2008) were predicted using ABCpred. The number of accepted B-cell epitopes were 11, 28, 37, 136, and 85 from sak, tst, isdA, clfB, and cna of S. aureus and 42, 90, and 38 fromSPy, scpa, and hylpL of S. pyogens.
A vaccine delivers antigenic substances that trigger immune reactions to provide an efficient degree of protection against a disease (Kreikemeyeret al., 2017). Our earlier research concentrated on identifying conserved epitopes in several S. pyogenes membrane proteins. Therefore, it is possible to develop conserved peptide-based streptococcal vaccines against human illnesses (Ebrahimi and Mohabatkar, 2018). Clinical application of these peptide-based vaccines against S. pyogenes requires further studies and experimental validation.
Numerous S. aureus strains are resistant to common clinical drugs, including vancomycin (Howdenet al., 2010), daptomycin (Fowler et al., 2006), mupirocin (Cadillaet al., 2011), and linezolid (Locke et al., 2009). There is ongoing research for developing novel molecules, especially vaccines, to fight these infections. An important strategy to combat S. aureus is creating potent monoclonal antibodies and vaccines. However, traditional inactivated vaccines and capsular polysaccharide vaccines are not the best options for providing immunoprotection. Therefore, the B-cell epitope, 272GYTEDEIV279 is promising for eliciting a potent immunoprotection against S. aureus infection.
Predicting T-cell epitopes requires the identification of MHC-binding peptides because T-cell recognition of antigenic peptides requires their association with MHC (Tomar and De, 2010). Short peptides of 9–11 amino acids can bind to MHC I; therefore, the 9-mer T-cell epitopes, which bind to MHC I and MHC II with high intensity were selected for predicting the affinity for the human MHC alleles, DRB1*0101 and DRB1*0401. The frequency of the two selected alleles, DRB1*0101 and DRB1*0401 within MHC class-II ranges between 20 and 50%. They are among the most common HLA-DR alleles; therefore, these two HLA molecules were chosen during MHCPred analysis (Panigada et al., 2002; Saha et al., 2017). It is essential for a peptide to bind to more than one HLA allele to be considered as a potential peptide for vaccine development. All five epitopes assessed in this study, two from S. aureus and three from S. pyogenes, were present on the surface, as indicated by the results of epitope mapping.
5 Conclusion
Cellulitis is a typical bacterial skin infection that causes pain, swelling, and redness in the infected region. We aimed to develop a new vaccine candidate from virulence-associated membrane proteins, through epitope identification, that is selective against the pathogens and invokes effective immune responses against this skin disease. Several steps of screening were performed to identify the appropriate vaccine candidates. The epitope, YTNSDKGGS, identified from fctA of S. pyogenes is promising as a prospective vaccine candidate, considering the binding affinity and high antigenic values. In addition, the epitopes, MTPSERLDL from scpA of S. pyogenes and MTFDDKNGK from cna of S. aureus, are present on the outer surface of their respective proteins. They exhibited better affinity for the two MHC alleles; therefore, they are promising as vaccine candidates as well. Future studies should focus on in vitro and in vivo evaluations focused onverifying the effectiveness of these vaccine candidates.
Funding
Researchers Supporting Project number (RSP2023R360) King Saud University, Riyadh, Saudi Arabia.
Ethical statement
This research work does not contain any human/animal sample.
Acknowledgement
Authors are grateful to the Researchers Supporting Project number (RSP2023R360) King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Novel structurally designed vaccine for S. aureus a-hemolysin: protection against bacteremia and pneumonia. PLoS One. 2012;7(6):e38567.
- [Google Scholar]
- Bacterial Virulence Factors: Secreted for Survival. Indian J. Microbiol.. 2017;57(1):1-10.
- [Google Scholar]
- Virulence factors and their mechanisms of action: the view from a damage- response framework. J. Water Health 2009:1-17.
- [Google Scholar]
- Rescue of Drosophila Melanogaster l(2)35Aa lethality is only mediated by polypeptide GalNAc-transferase pgant35A, but not by the evolutionary conserved human ortholog GalNAc-transferase-T11. Glycoconj. J.. 2010;27(4):435-444.
- [Google Scholar]
- Stepwise prediction of conformational discontinuous B-cell epitopes using the Mapitope algorithm. Proteins. 2007;68(l):294-304.
- [Google Scholar]
- Cellulitis in chronic oedema of the lower leg: an international cross- sectional study. Br. J. Dermatol.. 2021;2021:110-118.
- [Google Scholar]
- Association of high-level mupirocin resistance and multidrug-resistant methicillin resistant Staphylococcus aureus at an academic center in the Midwestern United States. N. Engl. J. Med.. 2011;49(1):95-100.
- [Google Scholar]
- Prediction of linear B-cell epitopes usingamino acid pair antigenicity scale. Amino Acids. 2007;33(3):423-428.
- [Google Scholar]
- Bacterial skin commensals and their role as host guardians. Benefic. Microbes. 2014;5(2):201-215.
- [Google Scholar]
- Proof of principle for epitope- focused vaccine design. Nature. 2014;507(7491):201-206.
- [Google Scholar]
- Harnessing bioinformatics to discover new vaccine. Drug Discov. Today. 2007;12(9–10):389-395.
- [Google Scholar]
- Dennis, L.S., Bryant, E.A., 2016. Impetigo, Erysipelas and Cellulitis. Basic Biology to Clinical Manifestation.
- VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf.. 2007;8:4.
- [Google Scholar]
- Prediction of T-cell epitopes for designing a reverse vaccine against streptococcal bacteria. Mol. Biol. Res. Commun.. 2018;7:35-41.
- [Google Scholar]
- Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit.. 2008;21(4):243-255.
- [Google Scholar]
- Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med.. 2006;355(7):653-665.
- [Google Scholar]
- Induction and exhaustion of lymphocytic choriomeningitis virus–specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class i-peptide complexes. J. Exp. Med.. 1998;187(9):1383-1393.
- [Google Scholar]
- MHCPred: a server for quantitative prediction of peptide-MHC binding. Nucleic Acids Res.. 2003;31(13):3621-3624.
- [Google Scholar]
- Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus. Infect. Genet. Evol.. 2017;48(1):83-94.
- [Google Scholar]
- Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin- intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. J. Clin. Microbiol.. 2010;23(1):99-139.
- [Google Scholar]
- Evaluating the cost of managing patients with cellulitis in Wales, UK: A 20-year population-scale study. Int. Wound J.. 2023;2023:1-12.
- [Google Scholar]
- Systematic identification of recognition motifs for the hub protein LC8. Life Sci. Alliance. 2019;2(4):1-16.
- [Google Scholar]
- Global skin disease morbidity and mortality an update from the global burden of diseases study 2013. JAMA Dermatol.. 2017;1(153):406-412.
- [Google Scholar]
- International journal of medical microbiology genomic organization, structure, regulation and pathogenic role of pilus constituents in major pathogenic Streptococci and Enterococci. Int. J. Med. Microbiol.. 2017;301(3):240-251.
- [Google Scholar]
- Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700) Antimicrob. Agents Chemother.. 2009;53(11):4580-4587.
- [Google Scholar]
- Epitope mapping using combinatorial phage-display libraries: a graph based algorithm. Nucleic Acids Res.. 2006;35(1):69-78.
- [Google Scholar]
- Pepitope: epitope mapping from affinity-selected peptides. Bioinformatics. 2007;23(23):3244-3246.
- [Google Scholar]
- Relationship of serum antileishmanial antibody with development of visceral leishmaniasis, post-kala-azar dermal leishmaniasis and visceral leishmaniasis relapse. Front. Microbiol.. 2019;10:22-68.
- [Google Scholar]
- Invasive infections due to Streptococcus pyogenes: seasonal variation of severity and clinicalcharacteristics, Iceland 1975 to 2012. European Centre for Disease Prevention and Control. 2015;19(17):5-14.
- [Google Scholar]
- The promise and challenge of epitope- focused vaccines. Hum. Vaccin. Immunother.. 2016;12(8):2113-2116.
- [Google Scholar]
- Increased US emergency department visits for skin and soft tissue infections, and changes in antibiotic choices, during the emergence of community-associated methicillin-resistant Staphylococcus aureus. Ann. Emerg. Med.. 2008;51(3):291-298.
- [Google Scholar]
- Identification of a promiscuous T-cell epitope in Mycobacterium tuberculosis Mce proteins. Infect. Immun.. 2002;70:79-85.
- [Google Scholar]
- A population response analysis approach to assign class II HLA epitope restrictions. J. Immunol.. 2015;194(12):6164-6176.
- [Google Scholar]
- In silico identification and characterization of common epitope-based peptide vaccine for Nipah and Hendra viruses. Asian Pac. J. Trop. Med.. 2017;10:529-538.
- [Google Scholar]
- ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17(12):1236-1237.
- [Google Scholar]
- ProPred1: prediction of promiscuous MHC Class-Ibinding sites. Bioinformatics. 2003;19(8):1009-1014.
- [Google Scholar]
- Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS One. 2013;8(5):1-8.
- [Google Scholar]
- Clinicopathological study on peripheral T-cell non-Hodgkin lymphoma with bone marrow involvement: a retrospective analysis from China. Int. J. Hematol.. 2009;90(3):303-310.
- [Google Scholar]
- Principles of Anatomy and Physiology (seventh ed.). HarperCollins College Publishers; 1993.
- Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nat. Genet.. 2003;35(2):148-157.
- [Google Scholar]
- Site-specific peptide vaccines for immunotherapy and im-munization against chronic diseases, cancer, infectious diseases, and for veterinary applications. Vaccine. 2005;23(17–18):2049-2056.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102917.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







