Translate this page into:
Protein carbonylation is a mediator in larvicidal mechanisms of Tabernaemontana cymosa ethanolic extract
⁎Corresponding authors at: Biology Department, Faculty of the Exact and Natural Science, Campus of San Pablo, University of Cartagena, Colombia (D.M. Cuadro). Pharmacy Department, Faculty of the Pharmaceutical Sciences, Campus Zaragocilla, University of Cartagena, Colombia (E.R. Cavallo). erodriguezc1@unicartagena.edu.co (Erika Rodríguez-Cavallo), dmendezc@unicartagena.edu.co (Darío Méndez-Cuadro)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Aedes aegypti mosquito is considered the most efficient vector for the spread of dengue, chikungunya, and zika viruses around the world and it is commonly controlled with synthetic insecticides. Despite their effectiveness, they have created problems like insecticide resistance become urgent to find novel insecticides or alternative methods for controlling it. Here, plants extracts, offer great promise as source of phytochemicals toxic against adult and immature mosquito stages. However, extracts are complexes mixture of compounds acting along at the same time on different biological targets and representing a real challenge when it is necessary to identify the mechanisms of action. Considering proteins are the main molecular targets of drugs, we used methods of redox proteomics to characterize pro-oxidant effects for larvicides, Ethanolic Extract of Tabernaemontana cymosa seeds (EETC) and the organophosphate temephos (Abate) on Aedes aegypti larvae midgut proteome. Initially, global oxidative damage caused by EETC and temephos was measured by carbonyl index quantitation in dot blot assay. After, combining Western blots with mass spectrometry, larvae proteins involved at the energy metabolism were identified as target of the pro-oxidant action. Results indicate that protein carbonylation plays an important role into the larvicide mechanisms of EETC and temephos. Finally, by phytochemical test were identified alkaloids, flavonoids and saponins as the major constituents.
Keywords
Protein carbonylation
Carbonyl index
Aedes aegypti
Midgut proteins
Temephos
Tabernaemontana cymosa
Pro-oxidant effect
1 Introduction
Mosquitos from the genus Aedes, specifically A. aegypti and A. albopictus, are responsible for the transmission of the arboviruses that cause dengue, chikungunya, and Zika; illnesses that have rapidly expanded across the globe in recent years (Patterson et al., 2016). The A. aegypti mosquito has traditionally been considered to be a much more efficient vector for the spread of these diseases due to several factors. It has evolved to live its entire life cycle from larvae to adult in close proximity with its human hosts, strongly preferring to feed on humans even in the presence of other mammals and will bite several individuals in the course of a single blood meal. This behavior can rapidly transmit a virus to multiple hosts in a short time frame, efficiently propagating disease (Patterson et al., 2016).
The use of synthetic insecticides is the most common approach to control mosquito vector and attenuate outbreak menaces. Even though their effectiveness, they have created many problems like insecticide resistance, pollution and toxic side effects on human beings (Mohankumar et al., 2016). In consequence, become urgent to find novel insecticides or alternative methods for controlling mosquito vectors. Here, plants offer great promise as source of phytochemicals toxic against adult and immature mosquito stages because they are in constant interaction and have coevolved with herbivores synthesizing complex molecules as defense (Toure et al., 2017).
In this sense, natural sources as plant extracts are complexes mixture of compounds acting along at the same time on different biological targets. Therefore, they represent a real challenge when it is necessary to identify the mechanisms that explain the biological action. Considering proteins represent the main molecular targets of marketed drugs, proteomics methods emerge as powerful tool to achieve a rigorous characterization of cellular targets and mechanism of bioactivity for phytochemicals and plant extracts (Salvador-Reyes and Luesch, 2015).
Using proteomics approaches it possible identify altered proteins as potential drug targets, and the global analysis of protein alterations can help understand its mechanism of action. By proteomics tools, can be also identified post-translational protein modifications as consequence of drug treatment, in special those that substantially affecting protein function and protein-protein interactions in vitro and in vivo. Through analysis of these protein changes in tissue and cultured cells before and after plant extracts treatment, is a possible elucidate the mechanism of action of natural remedies (Lao et al., 2014).
Into post-translational modifications, protein carbonylation is one of the more prominent oxidative alterations. It occurs without enzymatic catalysis, is irreversible, often leads to loss of function, increases the need for degradation of protein damaged and is more likely to happen in some proteins than others (Madian et al., 2011). More recently, enough evidence has been accumulated for the role of protein carbonylation in arthropod borne virus infections. Thus for example, the oxidative stress caused by pyrethroid insecticide permethrin plays a critical role in their toxicity and mechanism of action (Wang et al., 2016); while incapacitating arthritis/arthralgia on patients infected with the arbovirus Mayaro is associated to oxidative stress cellular response elicited (Camini et al., 2017).
In this work, we used methods of redox proteomics to characterize pro-oxidant effects of two larvicides, ethanolic extract of Tabernaemontana cymosa seeds (EETC) and the commercial organophosphate temephos (Abate) on Aedes aegypti midgut proteome.
T. cymosa (Family Apocynaceae) is originally from Colombia, Venezuela, and Trinidad, and it is also distributed throughout the tropical and subtropical regions of the world. The biological activities of these plants, such as antimicrobial, antiparasitic, antitumoral, antifebrile, analgesic, and antiviral properties, have been widely studied, as well as their effects against the larvae and adults of A. aegypti (Gomez-Calderon et al., 2017).
Initially, quantitative dot blot assay was used to measure carbonyl index as biomarker of the global oxidative damage caused by EETC and temephos on Aedes aegypti midgut proteome. Secondly, combining Western blot analysis with tandem mass spectrometry were identified potential proteins carbonylated as target of the pro-oxidant action. Our results indicate that protein carbonylation plays an important role into the larvicide mechanisms of Tabernaemontana cymosa ethanolic extract.
2 Materials and methods
2.1 Collection and identification of plant sample
Tabernamontana cymosa seeds were collected in the municipally of Turbaco, in the department of Bolívar, located in the Colombian Caribean Coast. Voucher code JBGP6421 of taxonomic identification was made in the herbarium of the Botanical Garden “Guillermo Piñerez” of Cartagena (Colombia).
2.2 Preparation of plant extract
T. cymosa seeds were dried in the shade at the environmental temperatures (28–30 °C day time), and grounded in a mechanical mill. The ground was macerated with 96% ethanol using a relation 1:4 of plant material: solvent and gentle magnetic stirring during seven days at 25 °C. Extract obtained was filtered and concentrated under reduced pressure at 45 °C and the residues obtained divided into aliquots with DMSO/water at concentration of 1 µg/µL and stored at −20 °C until their use.
2.3 Phytochemical tests
Preliminary phytochemical evaluation of all the extracts was performed by following standard methods of Wagner and Bladt (1996) and Trease and Evans (Evans 2009) for alkaloids, coumarins, flavonoids, cardiotonic glycosides, saponins, triterpenes, sterols and quinones.
2.4 Larvicidal assay
The eggs of A. aegypti Rockefeller strain were obtained from the insectary of the Entomology Unit of Health Department Secretary of Atlántico, in Barranquilla, Colombia. Eggs were reared in water at 28 ± 2 °C and alternating photoperiods of 12-h. Appropriate amount of dog biscuits were added to enhance the growth until third and fourth instar larvae. The larvicidal bioassay was conducted following the WHO standard larvicidal bioassay method (WHO/OMS, 2005) with sligh modifications. Batches of 20 of third and fourth instar larvae were introduced to 200 mL cups and assayed by triplicates and divide into four groups, thus: 1. Control group (a blank of larva growth); 2. Solvent group (exposed to DMSO 1%); 3. Temephos group (exposed to commercial temephos at 0,5 ppm) and 4. TC group (exposed to T. cymosa extract). LC50, LC90, LC99.9 value and diagnostic concentration were calculated using a Probit mortality analysis with the statistical package Biostat statistical software®.
To study potential oxidative damage caused by temephos and T. cymosa ethanolic extract, larvae were exposed to a concentration equivalent to the LC50 value during 12 h.
2.5 Preparation of midgut protein extracts
In order to obtain the best yielding, four different solvents were assayed to extract the midgut proteins: ripa modified buffer (TRIS-HCl 50 mM pH 8, NaCl 50 mM y SDS 1%), PBS-buffer 100 mM pH 7,2 supplemented with triton X-100 at 1%, Urea 7M/Thiourea 2M/CHAPS 4% and DMSO/TFA (99.5%/0,5% v/v). All of them, were supplemented with protease inhibitor cocktail mini-complete (Roche). For this, 20 midguts dissected from larvae controls group were pooled and homogenized on ice using a pellet pestles cordless motor (Sigma) during five minutes in a 1.5 mL microtubes. Then, midgut homogenates were centrifuged for 10 min at 13,000 g at 4 °C and protein concentration in supernatants was determined by modified Bradford method (Bio-Rad Laboratories Inc., Munich, Germany). Proteins were aliquoted, derivatized with dinitrophenylhidrazine (DNPH) and stored to -80 °C until their use for measuring of carbonyl index and identification of proteins carbonylated.
2.6 Measurement of carbonyl index
Dot-blot inmune assay proposed by Wehr and Levine was used to measure oxidative damage caused by temephos and etanolic extract of T. cymosa on A. aegypti midgut proteins (Wehr and Levine, 2012). For this, a calibration curve was built using Bovine Serum Albumin (BSA) with different carbonyl index values. Here, BSA solution (1 mg/mL) was oxidized by treatment with FeSO4 10 mM/Ascorbic acid 25 mM during 2 h at 37 °C. A stock of BSA solution was reserved to determine basal carbonyl index value. Next, BSA basal and oxidized solutions were derivatized using DNPH 5 mM in 0.5 M phosphoric acid and incubated at room temperature for 10 min. Finally, BSA protein labeled solutions were alkalized with NaOH 6 M, incubated 10 min and read at 450 nm, according Mesquita’s procedure (Mesquita et al. 2014). Molar absorption coefficient of 22308 M−1 cm−1 of DNPH was used to calculate nmoles of carbonyl / mg of protein. BSA solutions were treated with TCA 10%, cleaned with chilled acetone, recovered in DMSO/TFA and quantified again by Bradford’s method. Stoichiometric mixing of BSA basal with BSA oxidized were realized to obtain the different points on calibration curve.
To measure carbonyl index in A. aegypti midgut proteins from controls and samples, these were derivatized with DNPH probe by incubation of proteins freshly solutions (1mg/mL) with derivatizing reagent (20 mM DNPH and 0.5% TFA in 92.5%). Next, the calibration curve of BSA and samples was obtained were spotted by triplicate on PVDF membrane (Immun-Blot® PVDF Membrane for Protein Blotting. Cat.#162-0177). Membranes were blocked for 1 h at room temperature with 10% non-fat dry milk in phosphate-buffered saline (PBS) and then incubated with rabbit polyclonal anti-DNPH antibody at 1:10.000 (Sigma) in PBS–Tween-20 0.05% and milk 10%, for 2 h at room temperature with gentle rocking. Next, membranes were washed and incubated with peroxidase-linked anti-rabbit IgG antibody at 1:10.000 (Amersham Biosciences; 1 h at room temperature). Chemiluminescence signals were developed using Western maxTM rabbit IgG detection kit from Amresco and captured in a ChemiDoc MP System (BioRad).
The intensity of each analyzed spot was obtained by optical densitometry using Image Lab software (Bio-Rad). In order to maintain equals conditions during quantitative analysis, an area of 43.86 mm2 was established for each protein spot and measured intensity were tabulate in a matrix table in Microsoft Excel version 2013. To establish difference among groups a variance analysis was performed in GraphPad PRISM 6.01 followed of Tukey Test. Dot blot assay was repeated one more time.
2.7 Identification of carbonylated proteins by EETC
To identify protein carbonylated in A. aegypti midgut from larvae exposed to larvicidal EETC and temephos a combined proteomic protocol of SDS-PAGE, Western blot and Tandem Mass Spectrometry was used. In this, DNPH-derivatized midgut proteins were electrophoresed by duplicate in 10% acrylamide/ bisacrylamide gels. One gel was transferred onto PVDF membrane in a semidry way during 7 min using a Trans blot turbo instrument (Biorad). Membrane was blocked, incubated with polyclonal anti-DNPH antibody and revealed by chemiluminescence as was mentioned above. In parallel, proteins separated in second gel were fixed in methanol 50%: phosphoric acid 2% solution and visualized by staining with Coomassie Blue Brilliant G-250. Images of immunoblotting and stained gel were obtained in a ChemiDoc apparatus to select oxidized proteins bands.
Proteins selected for analysis were in-gel reduced, alkylated and digested with trypsin according to Sechi and Chait (1998). Briefly, the samples were reduced with 10 mM dithioerytritol in 25 mM ammonium bicarbonate for 30 min at 56 °C and subsequently alkylated with 55 mM iodoacetamide in 25 mM ammonium bicarbonate for 15 min in the dark. Finally, samples were digested with 12.5 ng/µl sequencing grade Bovine Trypsin (Roche Molecular Biochemicals) in 25 mM ammonium bicarbonate (pH 8,5) overnight at 37 °C.
After digestion, the supernatant was collected and 1 µl was spotted onto a MALDI target plate and allowed to air-dry at room temperature. Then, 0.8 µl of a 3 mg/ml of α-cyano-4-hydroxy-cinnamic acid matrix (Sigma) in 50% acetonitrile 0.1% TFA were added to the dried peptide and allowed again to air-dry at room temperature.
MALDI-TOF MS analyses were performed in a 4800 Plus Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, MDS Sciex, Toronto, Canada) at the Proteomics Unit of Complutense University of Madrid. The MALDI-TOF/TOF operated in positive reflector mode with an accelerating voltage of 20000 V. All mass spectra were calibrated by default or internally using peptides from the auto digestion of trypsin when were appeared.
For protein identification searches for peptide mass fingerprints, tandem MS spectra and both combined were performed in the NCBInr database 160208 (79581714 sequences; 29080698065 residues) without taxonomy restriction, using MASCOT 2.3 (www.matrixscience.com) through the software Global Protein Server v 3.6 (ABSciex). Search parameters were carbamidomethyl Cystein as fixed modification and oxidized methionine as variable modification, Peptide mass tolerance, 50–100 ppm and 1 missed trypsin cleavage site.
In all protein identification, the protein scores were greater than the score fixed by mascot as significant with a p-value minor than 0.05.
3 Results
3.1 Plant extract and phytochemical tests
T. cymosa seeds were macerated with 96% ethanol during seven days until achieves an exhaustive extraction. At these conditions, a yield of 33.4% was obtained. Preliminary screening of ethanolic extract showed the presence of a variety of phytochemicals including alkaloids, coumarins, tannins, cardiotonic glycosides, flavonoids, saponins, quinones, triterpenes and sterols. The respective tests for the various phytochemical moieties and their results are presented in Table 1. D: Dragendorff reagent; M: Mayer reagent; W: Wagner reagent; B: Baljet reagent; Fe: Ferric chlorhyde; GS: Gelatine-salt reagent; K: Kedde reagent; R: Raymond-Marthoud reagent; KK: Keller-Kiliani reagent; Sh: shinoda reagent; Ci: Citrobóric acid reagent; FT: Foam test; LB: Liebermann-Buchard reagent; SW: Salkowski reagent; VS: Vainillina-sulfúric acid reagent; Bo: Bornträger reagent.
Extract
Alkaloids
Coumarins
Tannins
Cardiotonic Glycosides
Flavonoids
Saponins
Triterpenes and Sterols
Quinones
D
M
W
B
Fe
Gs
K
R
KK
Sh
Ci
FT
VS
LB
SW
Bo
T.cymosa
+
++
+
–
–
–
++
++
+
+++
++
+++
++
+++
++
++
3.2 Larvicidal assay
Larvicidal activity of EETC against 3rd late and 4th early instar larvae of A. aegypti was evaluated in the range of 5 to 200 µg/mL, following guidelines of WHO protocols (WHO/OMS, 2005). It exhibited a dose-response behavior as show in Fig. 1 with lethal concentrations LC50, LC90 and LC99.9 values of 37.58, 78.61 and 143.51 µg/mL, respectively.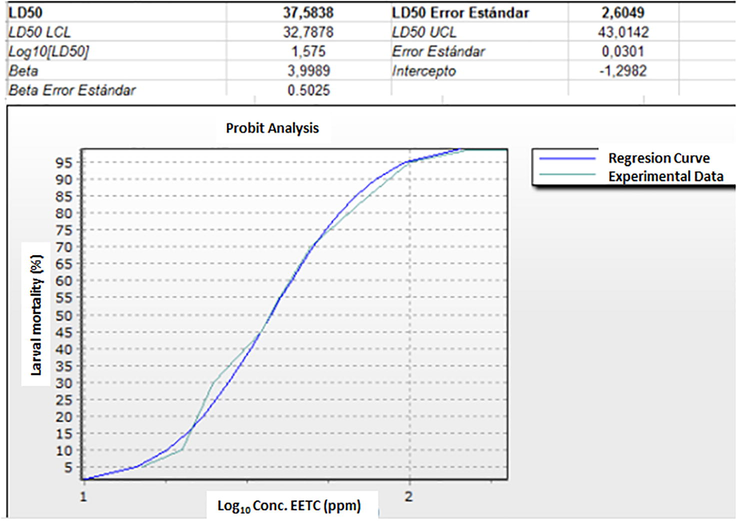
Curve dose-response for larvicidal effect of EETC on Aedes ageypti.
3.3 Midgut protein extracts obtained
Four different solvents were assayed to extract the Aedes aegypti midgut proteins and their yielding were compared. Thus, organic DMSO 99,5%/TFA 0,5% solvent was the most efficient with 21,6 ± 0,55 µg protein/midgut processed, followed by PBS-triton buffer and urea/thiourea/chaps with 13,11 ± 0,32 and 12,03 ± 0,42 µg protein/midgut processed, respectively. Finally, ripa buffer modified only yield 8,55 ± 0,35 µg protein/midgut processed. In addition to provide the best yielding, protein profiles obtained by SDS-PAGE showed a better performance of protein bands when DMSO 99,5%/TFA 0,5% was used as extraction solvent (Fig. 2).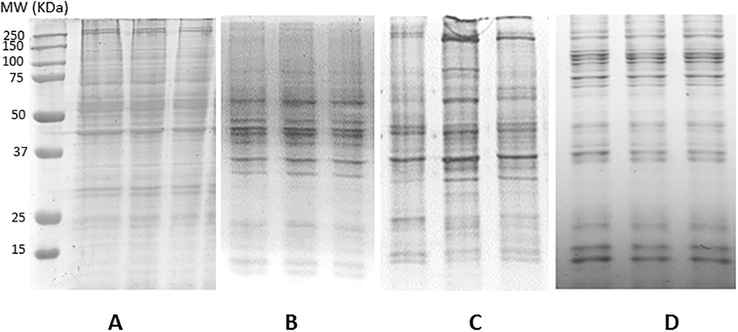
Electrophoregram of Aedes aegypti midgut proteins obtained with different solvents. (A) Ripa modified buffer (TRIS-HCl 50 mM pH 8, NaCl 50 mM y SDS 1%). (B) PBS-buffer 100 mM pH 7,2 suplemented with triton X-100 at 1%. (C) Urea 7M/Thiourea 2M/CHAPS 4%. (D) DMSO/TFA (99.5%/0,5% v/v). MW: Molecular Weight of protein markers.
3.4 Quantitation of prooxidant effects by measuring of carbonyl index
Oxidative damage of larvicides, temephos and EETC, on Aedes aegypti midgut proteome was evaluated by measuring their carbonyl indexes and compared against controls. Here, larvae exposed to the LC50 of the larvicides during 12 h were processed and the midguts excised, treated with DMSO / TFA, quantified proteins and derivatized with DNPH. The corresponding carbonyl indexes were calculated in nmol of carbonyls / mg of protein, using a calibration curve of BSA oxidized (Fig. 3) and the results obtained are listed in Table 2. A significant rising of carbonyl indexes only was observed in larvae exposed to temephos and EETC in comparison with the control growth group; being 1,65 and 1,42-fold up, respectively. Carbonyl index values for each group corresponding to averages and standard deviations for triplicate of dot blot analyses. Significant differences were found for larvae groups exposed to larvicides temephos and ETCC.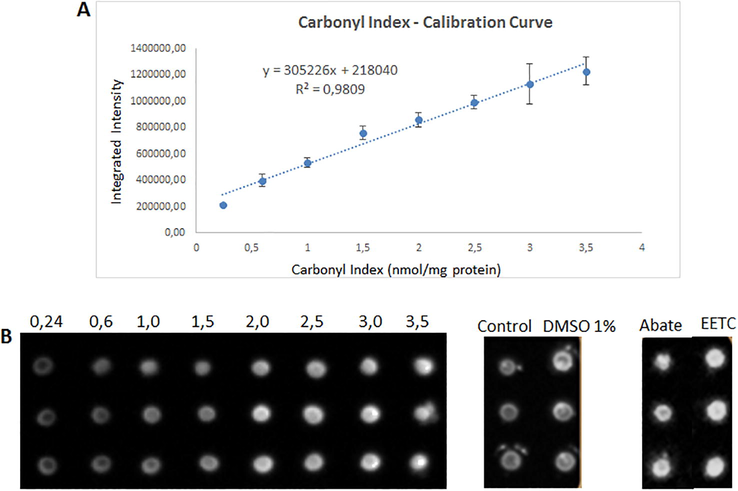
Calibration curve for quantitative dot blot. (A) Graph shows a typical result of regression analysis for dot blot membrane. (B) Dot blot pictures displaying BSA with different carbonyl indexes (from 0,24 to 3,5 nmol of carbonyl/mg of protein) and samples spotted on PVDF membrane by triplicate. Results of quantitation are listed in Table 1.
Test de Tuckey
Samples
Carbonyl Index (nmol/mg protein)
Means Dif.
Confidence
Significant dif.
Control Growth
0,78 ± 0,04
Solvent (DMSO)
0,92 ± 0,08
−2,763
−7,781 to 2,255
No
Temephos
1,29 ± 0,1
−9,863
−14,88 to 4,845
Yes
E.E T.C
1,11 ± 0,16
−6,363
−11,38 to 1,345
Yes
3.5 Identification of carbonylated proteins by larvicidal temephos and EETC
To identify potentially protein carbonylated as consequence of treatment with temephos and EETC a pattern of protein oxidized was obtained by Western blot (Fig. 4). Qualitative analysis of the oxyblots obtained exhibited more proteins bands carbonylated in proteome of larvae exposed to temephos and EETC. These results are agreement with the previously observed in the dot blot assay.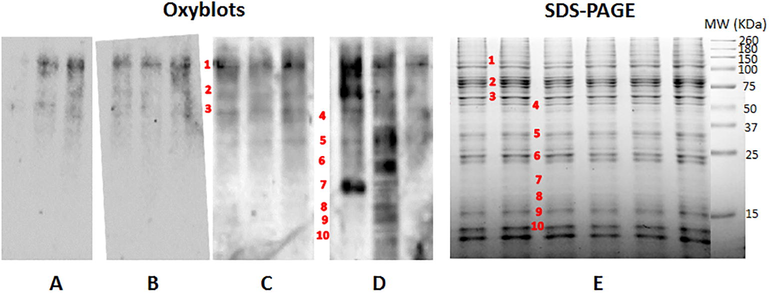
Carbonylation patterns and SDS-PAGE of Aedes aegypti midgut proteins. Comparative 1D OxyBlots from Aedes aegypti midgut proteins of studies groups for (A) Normal growth controls, and exposed to (B) DMSO solvent, (C) Temephos and (D) EETC. (E) Protein bands carbonylated in oxyblots were compared against SDS-PAGE and matched bands were numbered, excised, digested and analyzed by MS spectrometry.
Preparative duplicates of SDS-PAGE were matched against oxyblots to identify protein bands carbonylated (Fig. 4). Then, ten protein bands were excised, digested with trypsin and analyzed in a MALDI-TOF-TOF spectrometer. Bands from1 to 3 were commons either controls and treated with synthetic and natural larvicidals. Nevertheless, chemoluminiscense intensity emitted by the bands in groups treated with temephos and EETC was higher than controls. Bands 4 to 6 were common for temephos and EETC groups and the bands 7 to 10 were only observed in larvae treated with EETC. A total of 12 proteins were identified and classified according their function thus: 4 serinproteases, 2 mitochondrial ATPases, 2 actin isoforms, 1 fatty acid binding protein (FABP), 1 Integral component of the filamentous matrix of the centrosome and 2 proteins without function described. Proteins identified are listed in Table 3 along their accession number, mass (Da), score, submitted name, coverage and functional class (annotated in Uniprot-Swissprot database). Mascot data for each protein identified are listed in Supplementary Material S1. According oxyblots showed in Fig. 4, bands of carbonylated proteins were detected for different group samples established. Then, carbonylated proteins of Aedes aegypti larvae midgut from controls and treated samples were identified using a MALDI-TOF-TOF spectrometer. Thus, proteins identified from bands 1 to 3 were observed in all samples analyzed, while protein bands 4 to 6 were identified in larvae exposed to temephos and EETC. Other proteins were only identified from larvae exposed to EETC.
Band
Accession Number
Mass (Da)
Score
Submitted name
Coverage
Class*
Sample
1
gi|157119583
53,093
267
AAEL008708-PA [Aedes aegypti]
12%
Serine-type peptidase activity
All
gi|973173869
517,335
101
PREDICTED: pericentrin isoform X3 [Lepisosteus oculatus]
Integral component of the filamentous matrix of the centrosome
2
gi|157119815
68,528
225
AAEL008787-PA [Aedes aegypti]
29%
ATP binding. proton-transporting ATPase activity, rotational mechanism
gi|2454488
68,898
198
V-ATPase A-subunit [Aedes aegypti]
ATP synthase alpha subunit vacuolar
3
gi|157119583
53,093
146
AAEL008708-PA [Aedes aegypti]
8%
Serine-type peptidase activity
4
gi|576695773
43,780
92
Actin [Echinococcus granulosus]
34%
cell motility
Temephos + EETC
5
gi|195402199
54,241
398
ATPsyn-beta [Drosophila virilis]
40%
Mitochondrial membrane ATP synthase
gi|157136033
53,940
333
AAEL003393-PA [Aedes aegypti]
ATP binding. proton-transporting ATP synthase activity, rotational mechanism
6
gi|157113225
32,821
84
AAEL006384-PA [Aedes aegypti]
20%
Serine-type endopeptidase activity.
7
gi|157167794
27,516
119
AAEL011917-PA [Aedes aegypti]
40%
Serine-type enodpeptidase
EETC
8
gi|585185897
42,238
108
PREDICTED: beta-actin-like protein 2-like isoform X1 [Leptonychotes weddellii]
32%
ATP binding
9
gi|157109281
19,268
92
AAEL005259-PC [Aedes aegypti]
34%
Not Described
gi|157109285
21,865
90
AAEL005259-PA [Aedes aegypti]
Not Described
10
gi|157111905
14,908
294
AAEL005997-PA [Aedes aegypti]
80%
Lipid binding and transporter activity. Interacting selectively and non-covalently with a lipid
gi|58585202
15,140
158
fatty acid binding protein [Apis mellifera]
Lipid binding and transporter activity. Interacting selectively and non-covalently with a lipid
From these, actin, ATPsyn-beta and AAEL003393-PA were identified in protein bands carbonylated by temephos and EETC (bands 4 to 6, respectively). Actin is a cytoskeletal protein involved in various types of cell motility and is ubiquitously expressed in all eukaryotic cells. While the others, correspond to mitochondrial membrane ATP synthase and ATP binding protein, respectively. In the case of protein bands oxidized only by EETC, were identified a Serine-type endopeptidase named AAEL011917-PA, beta-actin-like protein 2-like isoform X1, a Lipid binding and transporter activity, and two proteins AAEL005259-PC and AAEL005259-PA without function known.
4 Discussion
Nowadays ascertaining the multiple target proteins is required for understanding the mechanism of action of natural products, as well as addressing potential adverse effects related to off-target actions (Chang et al., 2016). In this sense, different proteomic approaches have been proposed to improve the discovery process of effective compounds in plant extracts and elucidate their potentials mechanisms of action. Thus is possible characterizes protein functions, protein–protein interactions, and protein modification in tissues or animals, and use it to investigate signaling pathway perturbations in cells or the whole body (Lao et al., 2014). Herein, we applied methods of redox proteomics to identify potential target proteins involved in the larvicides mechanisms of action to EETC and the commercial organophosphate temephos.
First, we demonstrate that both larvicidal agents produce irreversible oxidative damage by significantly increasing the carbonyl indexes values of the midgut proteome larvae exposed to them. It value ascertains the grade of protein carbonylation, which is the most common biomarker used to measure oxidative modifications on proteins (Wehr and Levine, 2012). Carbonylated proteins cannot be repaired by cellular enzymes, they disrupts protein-stabilizing elements, such as salt bridges and proline kinks, tend to aggregate and, if not eliminated, result in cell death (Fedorova et al., 2013).
Pro-oxidant effect had been observed for plant extracts in the gut of many insect herbivores. There, alkaline conditions prevalent in the gut promoting the autoxidation of different classes of phenolics compounds that have variable abilities to function as pro-oxidants (Vihakas et al., 2014). Therefore the mixture of alkaloid, flavonoids, saponins, triterpenes and sterols present in the EETC cause an irreversible oxidative damage on Aedes aegypti larval midgut proteome.
In particular, the monoterpene indole alkaloids are present as major secondary components in all parts of the plants of the genus Tabernaemontana and they have attracted the attention of the scientific community for new alkaloids derivatives and bioactivities (Marinho et al., 2016). These compounds had shown moderate to null antioxidant activity in vitro assays (Kasote et al., 2015) and by contrast they can affect insects at all levels of biological organization, because their action generally disturbs cellular and physiological processes by altering redox balance, hormonal regulation, neuronal signalization or reproduction in exposed individuals (Chowanski et al., 2016). Despite of these finding, the identification of molecular targets is still requiered to characterize pro-oxidants effects and their potential role in mechanisms of action larvicides for EETC.
Here, using proteomic redox approaches, we found that commercial organophosphate temephos and EETC producing carbonylation on the Aedes aegypti midgut proteins actin, subunit beta of ATP synthase and the protein ATP-binding AAEL003393-PA. All of them are components of two proton pumps, the F-ATPase and the V-ATPase (Nakanishi-Matsui et al., 2010). In insect midgut epithelium, ATP synthase/vacuolar H+-ATPase (V-ATPase) system, serves as a major energy source for secretion and absorption by playing a role as an H+/K+ transporter and it is downregulated in the larvae of Culex quinquefasciatus mosquitoes exposed to temephos (Games et al., 2016).
In consequence, it feasible considerer these protein carbonylations disrupt in anyway the interaction among ATP bombs components and affecting energy metabolism at the midgut of Aedes aegypti larvae. Now, taken account ATP proton pumps are highly conserved in eukaryotic cells, any larvicide able to disturbed it should be poorly selective, as was observed to EETC during the ictiotoxicity assay (Data not Shown).
Moreover effects on energy metabolism were stronger in larvae exposed to EETC than temephos, because the former induced additional protein carbonylation on Lipid binding binding protein AAEL005997-PA and FABP. In general, these proteins playing a central role as regulators of lipid metabolism, inflammation and energy homeostasis (Thumser et al., 2014). Furthermore, different insect FABPs have been investigated in several species and their physiological roles as fatty acid transporters to provide energy source for sustaining flight have been demonstrated (Huang et al., 2012). However, any reporting as potential target of natural and synthetic larvicidals was no found in our review for FABP.
The relationship between alterations of energy metabolism and protein carbonylation has been described as deleterious of cell life, recently. Thus for example, association among alterations of energy metabolism, signaling cell and protein carbonylation might represent a self-sustaining triangle of harmful events that trigger the degeneration and death of neurons and the development and progression of Alzheimer’s disease brain (Di Domenico et al., 2016).
On the other hand, serine endopeptidases AAEL006384-PA and AAEL011917-PA were observed only oxidized in larvae exposed to temephos and EETC, respectively. These endoproteases are the dominant class of proteolytic enzymes in many insect species and include peptidases such as trypsins, chymotrypsins and elastases (Park and Kwak, 2008). These enzymes were find down regulated too in the larvae of Culex quinquefasciatus mosquitoes exposed to temephos (Games et al., 2016). In particular, the A. aegypti genome contains 369 genes coding for serine peptidases, but only 5 (three trypsins and two chymotrypsins) are well characterized in the midgut of females of this insect, where playing pivotal roles in oogenesis, immunity, metamorphosis, modulation of embryonic development and nutrition. Moreover, they can provide energy and essential amino acids or be secreted into the lumen of the midgut in defense against pathogens (Park and Kwak, 2008). Hence, protein carbonylation on these serine endopeptidases could be others important targets of the larvicidal assayed.
Additionally, two hypothetical and conserved proteins were found carbonylated in larvae exposed to EETC but they do not have some functions described.
Finally, our findings indicate that protein carbonylation is an important biochemistry biomarker useful to study potential mechanisms of action for natural sources with larvicidal properties such as the Tabernaemontana cymosa ethanolic extract.
In conclusion, we propose to DMSO/TFA as an efficient solvent for the extraction of Aedes aegypti larvae midgut proteins. Temephos and Tabernaemontana cymosa etanolic extract are larvicides with oxidant effects on Aedes aegypti larvae midgut proteins. Both larvicides increasing the carbonylation of proteins belonging to the energy metabolism pathways.
Acknowledgements
The authors would like to thank Judith Rodriguez Cavallo for her technical assistance at the Analytical Chemistry and Biomedicine Labs. Lola Gutierrez and Maria Luisa Hernáez of the Complutense University (Madrid) for proteomic analysis and Jorge Mendez by check the spelling.
Funding
This work was funded by the Departamento Administrativo de Ciencia, Tecnología e Investigación—COLCIENCIAS [Administrative Department of Science, Technology and Research], the University Hospital of the Caribbean and The University of Cartagena. Grant 1107-545-31632. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Adriana Yepez and Javier Guarnizo were funded by Young Research Program of Colciencias 2015-2016. The proteomic analysis was performed in the Proteomics Unit at Complutense University of Madrid that belongs to ProteoRed, PRB2-ISCIII, supported by grant PT13/0001.
References
- Advances in identification and validation of protein targets of natural products without chemical modification. Nat. Prod. Rep.. 2016;33(5):719-730.
- [Google Scholar]
- A review of bioinsecticidal activity of solanaceae alkaloids. Toxins (Basel). 2016;8(3)
- [Google Scholar]
- The triangle of death in Alzheimer's disease brain: the aberrant cross-talk among energy metabolism, mammalian target of rapamycin signaling, and protein homeostasis revealed by redox proteomics. Antioxid Redox Signal. 2016;26(8):364-387.
- [Google Scholar]
- Trease and Evans Pharmacognosy. Saunders Elsevier; 2009.
- Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev.. 2013;33(2):79-97.
- [Google Scholar]
- Differential protein expression in the midgut of Culex quinquefasciatus mosquitoes induced by the insecticide temephos. Med. Vet. Entomol.. 2016;30(3):253-263.
- [Google Scholar]
- Antiviral effect of compounds derived from the seeds of Mammea americana and Tabernaemontana cymosa on Dengue and Chikungunya virus infections. BMC Complement Altern. Med.. 2017;17(1):57.
- [Google Scholar]
- Cloning and characterization of a midgut-specific fatty acid binding protein in Spodoptera litura. Arch. Insect Biochem. Physiol.. 2012;79(1):1-17.
- [Google Scholar]
- Significance of antioxidant potential of plants and its relevance to therapeutic applications. Int. J. Biol. Sci.. 2015;11(8):982-991.
- [Google Scholar]
- Application of proteomics to determine the mechanism of action of traditional Chinese medicine remedies. J. Ethnopharmacol.. 2014;155(1):1-8.
- [Google Scholar]
- Differential carbonylation of proteins as a function of in vivo oxidative stress. J. Proteome Res.. 2011;10(9):3959-3972.
- [Google Scholar]
- Brazilian Tabernaemontana genus: indole alkaloids and phytochemical activities. Fitoterapia. 2016;114:127-137.
- [Google Scholar]
- Simplified 2,4-dinitrophenylhydrazine spectrophotometric assay for quantification of carbonyls in oxidized proteins. Anal. Biochem.. 2014;458:69-71.
- [Google Scholar]
- Screening of methanolic plant extracts against larvae of aedes aegypti and anopheles stephensi in Mysore. J. Arthropod. Borne Dis.. 2016;10(3):303-314.
- [Google Scholar]
- The mechanism of rotating proton pumping ATPases. Biochim. Biophys. Acta. 2010;1797(8):1343-1352.
- [Google Scholar]
- Expression of Chironomus riparius serine-type endopeptidase gene under di-(2-ethylhexyl)-phthalate (DEHP) exposure. Comput. Biochem. Phys. B Biochem. Mol. Biol.. 2008;151(3):349-354.
- [Google Scholar]
- Dengue, Zika and Chikungunya: emerging Arboviruses in the New World. West J. Emerg. Med.. 2016;17(6):671-679.
- [Google Scholar]
- Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat. Prod. Rep.. 2015;32(3):478-503.
- [Google Scholar]
- Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal. Chem.. 1998;70(24):5150-5158.
- [Google Scholar]
- Fatty acid binding proteins: tissue-specific functions in health and disease. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17(2):124-129.
- [Google Scholar]
- Aedes aegypti Larvicidal Sesquiterpene Alkaloids from Maytenus oblongata. J. Nat. Prod.. 2017;80(2):384-390.
- [Google Scholar]
- Rapid estimation of the oxidative activities of individual phenolics in crude plant extracts. Phytochemistry. 2014;103:76-84.
- [Google Scholar]
- Plant Drug Analysis. A Thin Layer Chromatography Atlas (2nd ed.). Berlin, Heidelberg, New York: Springer-Verlag; 1996. ISBN 3-540-58676-8
- Permethrin-induced oxidative stress and toxicity and metabolism. A review. Environ. Res.. 2016;149:86-104.
- [Google Scholar]
- Quantitation of protein carbonylation by dot blot. Anal. Biochem.. 2012;423(2):241-245.
- [Google Scholar]
- WHO/OMS, 2005. Guidelines for laboratory and field testing of mosquito larvicides. Geneve, WORLD HEALTH ORGANIZATION, 41.
Appendix A
Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jksus.2018.04.019.
Appendix A
Supplementary data







