Translate this page into:
Phytochemicals of Withania coagulans (Stocks) Dunal against androgen receptor: An in silico insight
⁎Corresponding author. subinadhikari2018@gmail.com (Jhashanath Adhikari Subin)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
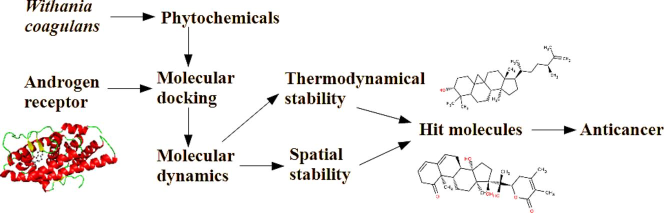
Abstract
Objectives
The androgen receptor is considered a magic target due to its involvement in multiple pathways leading to several maladies and its accessibility for small molecules to bind to it and modulate its normal functioning. The phytochemicals from the plant, Withania coagulans (Stocks) Dunal (common name: Rishyagandha) with traditional practice in healing different diseases were investigated in this regard. The higher safety profile and cost-effective features of plant-based resources in the design and development of drug like candidates seems to be worthy of exploration in the realm of alternate therapeutic options.
Methods
Several computational methods involving flexible receptor molecular docking calculations and molecular dynamics simulations were carried out. Spatially and thermodynamically stable adducts with best docking scores were identified using various parameters extracted from the molecular dynamics trajectory to find the hit candidates capable of inhibiting or disrupting the normal functioning of the target protein leading to the cure of the disease. The server based ADMET prediction was performed to comprehend the druglikeness, bioavailability, and toxicity profile of different phytochemicals.
Results
The molecule, 24-Methyl-9,19-cyclolanost-25-en-3-ol (PubChem CID: 185036) showed the binding score of −10.726 kcal/mol against androgen receptor (PDB ID: 1E3G) better than that of the native ligand, 17-β-hydroxy-17-methylestra-4,9,11-trien-3-one (PubChem CID: 261000) with −10.268 kcal/mol and selected anticancer drugs (best value of -10.080 kcal/mol for darolutamide). Withacoagulin I (PubChem CID: 71720665) displayed the binding score of −10.108 kcal/mol and was considered the second top molecule. The stability of the protein-phytochemical adducts as calculated from 200 ns long molecular dynamics simulations revealed good geometrical intactness from multiple spatial descriptors (root mean square deviation, radial pair distribution function, and hydrogen bond distribution) and sustained thermodynamical spontaneity from binding free energy changes of the protein-ligand complexes. The pharmacokinetics and pharmacodynamics showed acceptable druglike properties, good bioavailability and safety comparable with that of the reference drugs.
Conclusions
The hit molecules could be considered as possibly capable of inhibiting the target protein that would perform as anticancer agents and are recommended for further experimental trials for the verification of theoretical inferences.
Keywords
Anticancer
Docking scores
Free energy changes
Ligand RMSD
Molecular dynamics
MMPBSA
1 Introduction
Androgen receptor has been labeled as a magic drug target due to its involvement in various types of diseases and presence of different domains for small molecules to bind as agonists, antagonists or modulators (Li et al., 2019). It functions as a DNA-binding transcription factor regulating gene expression and controls cell behavior though protein levels (Pandini et al., 2005). It is triggered by hormones and synthetic molecules (testosterone, dihydrotestosterone, oxandrolone, and methyltestosterone) and deactivated by ralaniten, enzalutamide, and spironolactone to name a few (Elshan et al., 2019). A thorough review of the structure and its functions has been performed by Tan et al. (Tan et al., 2015) and several binding sites (N‐terminal domain, binding function‐3 site, activation function‐2 site, DNA‐binding domain, and ligand‐binding pocket) has been discussed (Li et al., 2019; Lallous et al., 2013). The ligand binding domain of the target protein as shown in Fig. 1, is responsible for the major catalytic activity since competition to bind to it has been the major mode of action (Ferraldeschi & de Bono, 2013) The imbalance in the cell differentiation and proliferation leads to prostate cancer, specially the up-regulation of proliferating genes and down-regulation of differentiating genes have been found to be the major causes (Helsen et al., 2014; Marques et al., 2011). Deprivation of androgen or blocking the activity of receptor are the therapeutic strategies that could treat the progression of cancerous cells. Any other molecule binding at this domain with better strength than the native ligand, 17-β-hydroxy-17-methyl-estra-4,9,11-trien-3-one (R1881) would likely suppress the function of the receptor (in case of an antagonist) leading to the treatment of the disease. The drug resistance due to mutations have resulted in ineffectiveness, posing a tough challenge for the development of stable drug leading to the cure of the disease (Watson et al., 2015; Joseph et al., 2013). Other different classes of small molecules that bind to it and remain stable would be desired for modulation of its activity. This strategy has been used initially in the virtual screening of molecules against androgen receptor by several authors (Tang et al., 2020; Pang et al., 2022). The relatively safer and cost-effective approach of employing plant-based therapeutics for fighting prostate cancer seems relevant and the initial computational screening was carried out in this pursuit in this work. Further optimizations were carried out by molecular dynamics simulations integrated with the results from bioassay. The research aiming to find a new type of molecule that could dock to the orthosteric site of androgen receptor and help in the treatment of prostate cancer is highly desirable and significant.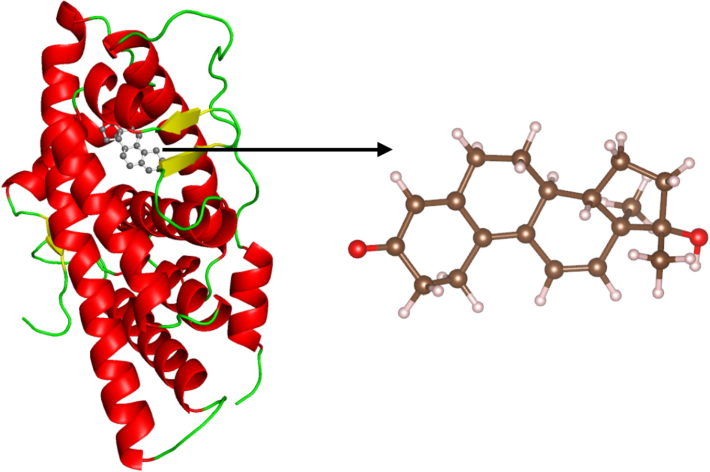
Structure of androgen receptor protein (ribbon representation) with native agonist ligand (R1881, ball-and-stick model) at the ligand binding pocket of the ligand binding domain (PDB ID: 1E3G).
The broad-spectrum therapeutic capabilities of Withania coagulans (Stocks) Dunal against various diseases in different medicinal systems have been reported (Gupta et al., 2022; Verma et al., 2021). The use of the plant against tumor or cancer related illness has been mentioned (Aljohny et al., 2022; Maqsood et al., 2018). In ancient eastern medical system, the plant has been extensively used against various diseases (Ahmad et al., 2023). The chemical components and its biological activities have been mentioned by multiple studies (Azmi et al., 2023; Rathore et al., 2018). The plant mainly contains withaferin A, withanolide A and withanone as active components and their anticancer potential has been studied (Grover et al., 2012; Lee & Choi, 2016). Withaferin A has been under preclinical trials against cancer (Hassannia et al., 2020). Withaferin A and withanone have shown differential activity in normal and human cancerous cells (Vaishnavi et al., 2012). Based on the premises on the capability of the phytochemicals from Withania coagulans (Stocks) Dunal to fight against cancer, the research with the chosen receptor in terms of computational techniques seems justifiable.
In this regard, in order to find a cost-effective molecule from plant-based resources and to understand the molecular-level details of the inherent binding mechanism against androgen receptor, the current work has been carried out by computational tools. The use of various free web-servers and programs would help to identify hit candidates that could possibly be extended to in vitro and in vivo trials. The determination of the orientations of the phytochemicals in the active pocket and calculation of geometrical and thermodynamical stability of the protein–ligand adducts at physiological condition would help to identify the candidates with acceptable inhibitory strength. The objective of the research is the in silico assessments of the phytochemicals of Withania coagulans (Stocks) Dunal for its capability to form stable adducts with the androgen receptor and recommend for further experimental trials.
2 Materials and methods
2.1 Ligand database and receptor protein structure preparation
A method similar to that reported in the literature was used in the database preparation (Adhikari Subin and Shrestha, 2024). Since no new method was developed or modified, the fine details as mentioned in the procedure and resources section of the supplementary information were followed.
2.2 Molecular docking, molecular dynamics simulations, and binding free energy change estimation
The possible binding orientations of the ligand at the active site of the protein was calculated by soft docking process with protein flexibility from the DockThor server (Guedes et al., 2021). The synopsis of the computational calculations and the control parameters have been mentioned in the supplementary information.
2.3 ADMET prediction
The suitability of phytochemicals as druglike candidates was determined through a quick server-based ADMET prediction (admetSAR 3.0, accessed November 04, 2024) (Gu et al. 2024). The bioavailability and toxicity were predicted and compared to those of the reference drugs.
3 Results and discussion
3.1 Docking scores from molecular docking
The binding affinities of various poses were calculated for the phytochemicals, the native ligand, and the drugs against prostate cancer after protocol validation (Figure S1 in the supplementary information). The best ones (top 5 and the native ligand) are shown in Table 1 along with the PubChem CID and molecular formula. Their smiles strings are given in Table S1 in the supplementary information. Molecule A, 24-Methyl-9,19-cyclolanost-25-en-3-ol possessed the best value (−10.726 kcal/mol) and was even better than those for the native ligand, 17-beta-hydroxy-17-methylestra-4,9,11-trien-3-one (−10.268 kcal/mol) and all the drugs studied. Molecule B, withacoagulin I had better binding affinity (−10.108 kcal/mol) than all the drugs. Both the molecules showed better docking scores than AR antagonists, Darolutamide and Bicalutamide hinting at stronger interactions and the formation of relatively stable adducts. The calculation results of some more drugs for comparative purposes are given in Table S2 in the supplementary information. The molecules A and B could be considered to be bound relatively strongly at the active site of the protein followed by other phytochemicals. Luteolin, a flavone from Chinese herbal medicine has been found to be the best docked candidate against androgen receptor from a set of 182 test molecules as reported in the literature (Li et al., 2023). Also, quercetin, kaempferol, and naringenin were studied by computational methods against androgen receptor and other targets. This showed that different class of organic molecules also could possibly be employed in finding a hit candidate in terms of binding strength. The sustained stability of the protein–ligand adducts at the physiological conditions would provide adequate premises in inferring it to be the effective candidates and is discussed in molecular dynamics simulation section.
Code
Other names
PubChem CID
Molecular Formula
Docking score
Drugs and hormones
PubChem CID
Docking score
Molecule A
24-Methyl-9,19-cyclolanost-25-en-3-ol
185,036
C31H52O
−10.726
Darolutamide
67,171,867
−10.080
Molecule B
Withacoagulin I
71,720,665
C28H38O6
−10.108
Oxandrolone
5878
−10.064
Molecule C
Withanolide E
301,751
C28H38O7
−9.867
Bicalutamide
2375
−10.040
Molecule D
Withanolide F
44,562,998
C28H38O6
−9.785
Cyproterone acetate
9880
−10.034
Molecule E
Coagulin F
24,941,991
C28H36O5
−9.704
Dihydrotestosterone
10,635
−10.009
Molecule N
Methyltrienolone
261,000
C19H24O2
−10.268
Methyltestosterone
6010
−10.001
The chemical structures of the top candidates and native ligand are shown in Fig. 2. The base skeletal structure is nearly the same in the molecules with variations in the functional groups. Molecule B, withanolide I have shown an IC50 of 8.8 μM against NF-κB signaling pathway which is responsible for cancer propagation among other functions (Xia et al., 2018; Taniguchi & Karin, 2018). The results of the experimental assays (Henrich et al. 2015; Yano et al. 2023; Ben Bakrim et al. 2018) provide good basis for selection of these phytochemicals for further analysis including in silico modeling. However, the biological activity for molecule A has not been reported by any experimental methods and could possibly be a new candidate worthy of exploration by different approaches.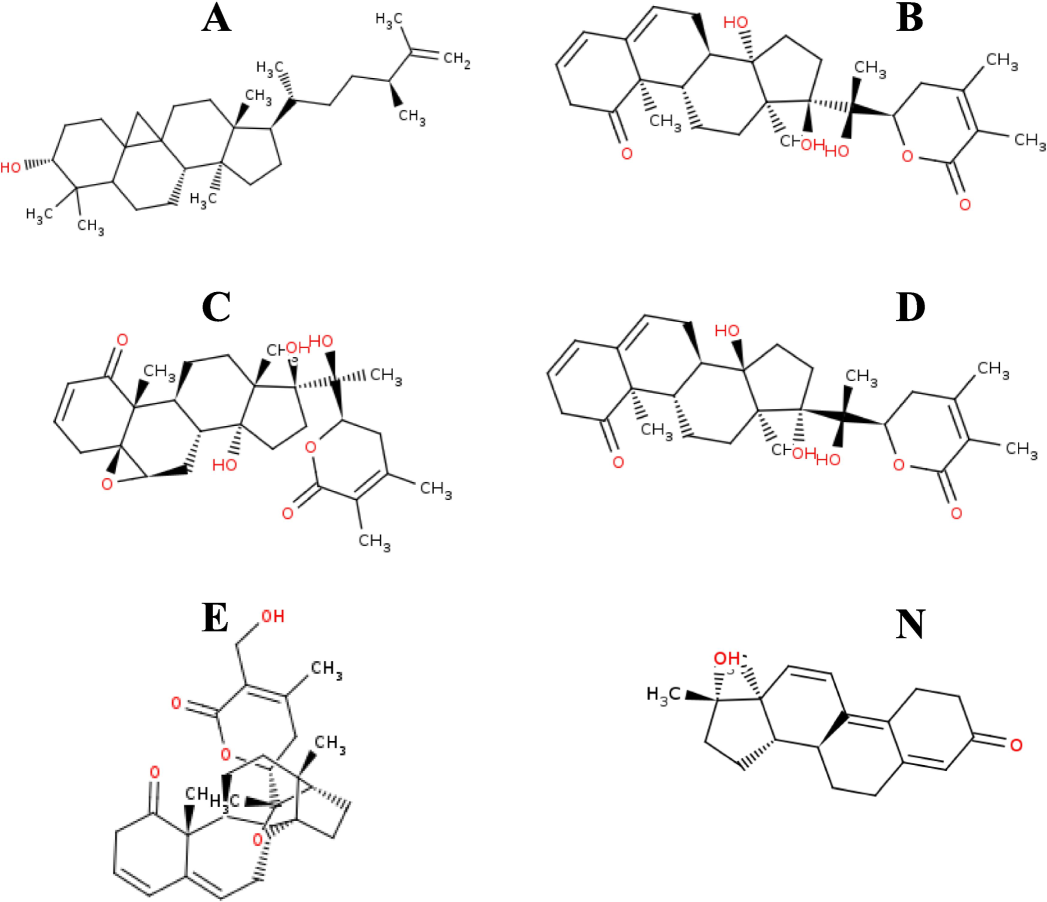
Chemical structures of selected phytochemicals and a native ligand (Molecule A, Molecule B, Molecule C, Molecule D, Molecule E, and Molecule N).
3.2 Stability of the adduct
3.2.1 Root mean square deviation (RMSD) of ligand
A quantitative measure of the deviation of ligand from its original location with time is given by Fig. 3 for various ligands in their adducts in terms of heavy atom RMSD of ligand relative to the protein backbone (least square fitting). The snapshots of the ligands showing the pose at the active site are shown at 1, 50, 100, 150, and 200 ns in the insets.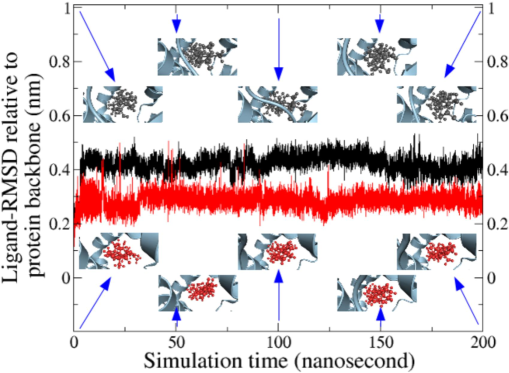
RMSD of ligands relative to protein backbone in different protein–ligand adducts during MDS; Snapshots at 1, 50, 100, 150, and 200 ns showing the location of ligands at the active site of protein are presented as small rectangular images (protein = greyish blue, molecule A = dark, and molecule B = red).
A quick visualization of the RMSD profile reveals exceptional stability of the ligand at the active pocket due to the lack of sharp and large rise or fall in the values. The smooth and flat nature of the curves for ligand A and ligand B (deviation below 5 Å) throughout the MDS hints that these two phytochemicals are strongly bound to the receptor without significant delocalization. The snapshots at various instances could help to augment the inference that the ligands remain in their docked location during the simulation run with minimal translational or rotational motion. The values obtained in this work is in par with those reported in the literature (Olubode et al. 2022) and justifies the inference of conservation of the pose and location of ligand at the catalytic site of the target protein.
The RMSD profile of other three ligands relative to the protein backbone is shown in Figure S2 in the supplementary information. The curve for molecule D is nearly flat (below 3 Å) with a slight bump at ca. 115 ns, whereas those for molecule C (deviation below 7 Å) and molecule E (deviation below 4 Å), there are multiple spikes. The curves are not smooth as that observed for other systems. In all the cases, the complexes seem to be equilibrated after 135 ns until the end. The adducts might be considered to possess moderate spatial stability that requires further experiments for a confirmation of the proposition.
3.2.2 Radial pair distribution function
The radial pair distribution functions, g(r) between the center of mass of the ligand and the center of mass of the protein are shown in Fig. 4 for different adducts. Molecule A and molecule B have single peak each at ca. 0.934 nm and at ca. 1.180 nm, respectively, referring to the occupation of single location at the orthosteric site of the receptor during the simulation run. For other ligands, there are multiple peaks with different intensities and refer to the occupation of different locations in the protein during the production run. A significant and sustained variation in the position of ligand have yielded non-singular peaks for molecule C, molecule D, and molecule E. The plot is consistent with the ligand RMSD profile and the overall inference is that molecule A and molecule B could bind with the key amino acid residues for long duration residing at a single site leading to the inhibition of the enzyme. The literature on this parameter for androgen receptor was scanty or unavailable and comparative analysis could not be performed.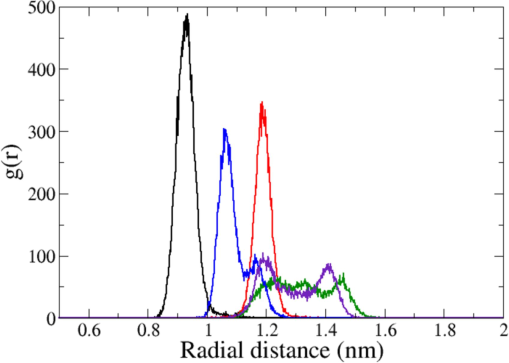
RPDF of ligands’ center of mass relative to protein’s center of mass in different protein–ligand adducts during MDS (molecule A = dark, molecule B = red, molecule C = green, molecule D = blue, and molecule E = indigo).
3.3 Stability of protein structure
The geometrical intactness of the protein structure upon ligand binding was determined in terms of several parameters (RMS fluctuation, gyradius of protein, solvent accessible surface area, and RMS deviation of protein backbone). The plots are shown in supplementary information as Figure S3-S6. If the unexposed region of the surface of the protein becomes wettable, then the rise in SASA would be observed and the protein is considered to be unstable. The stable SASA profile, as shown in Figure S3, infers to the lack of major conformational changes in the protein geometry upon ligand binding. The expansion or contraction of the protein is given by gyradius and a flat curve with minimal variation suggests rigidity of the receptor geometry. The maximal gyradius of 18.9 Å was calculated for the protein structure with smooth nature of curve, Figure S4. The ligand binding, therefore, do not change the receptor structure. The translational motion of protein backbone during the simulation relative to that at the beginning is given by protein backbone RMSD curve as presented in Figure S5. For all the cases, the flat curves with values less than 2 Å imply excellent stability of the system. The fluctuation of the alpha carbon atoms of the amino acid residues shows the dynamics of the main protein framework. The smaller value suggests better stability and are mostly absent in the sheets or helices. The value less than 3 Å was obtained for all the cases, Figure S6, and it being minimal with small variation infers good structural integrity of the adduct upon ligand binding. The hydrogen bond distribution in five different adducts showed single peak for most of the cases hinting at its major contribution to protein–ligand binding (Figure S7). The relevant variation of donor–acceptor distance (<4 Å) is shown in Figure S8. All the parameters point to a single unified proposition that the protein geometry remained intact during the simulation and the binding of the ligand was not affected or disrupted. The overall results suggest that the ligand and protein form a stable adduct and the ligand could modulate the function of androgen receptor.
3.4 Molecular-level analysis of the protein–ligand interactions
The amino acid residues participating in interaction with the ligand in two most stable adducts are shown in Fig. 5. The presence of hydrophobic interaction in the first adduct and both hydrophobic and hydrogen bond in the second adduct were observed. The amino acid residues involved in different interactions along with the distances are shown in Table 2.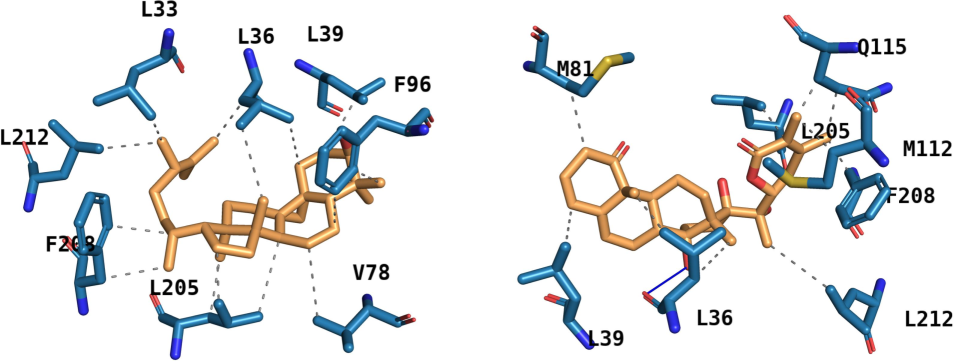
Different amino acid residues involved in bonding with the ligand in different protein–ligand adducts (left) complex of molecule A and (right) complex of molecule B.
Type of interactions
Amino acid residues (distances, Å)
Complex of A
Hydrophobic
L33 (2.58), L36 (2.96, 3.72, 3.79), L39 (3.25), V78 (3.86), F96 (2.63, 3.60), L205 (2.60, 2.96, 3.91), F208 (3.09, 3.20), L212 (3.10)
Complex of B
Hydrophobic
L36 (3.00, 3.81), L39 (3.57), M81 (3.24), M112 (3.52), Q115 (2.15, 2.60), L205 (2.55), F208 (2.47), L212 (3.95)
Hydrogen bonding
L36 (2.52), L205 (2.16)
Different parts of the molecule seem to be interacting with the amino acid residues at the active pocket of the protein. Basically, leucine appears multiple times with different types of interactions in case of the two adducts. The first complex lacks hydrogen bonding in the initial docked pose whereas the second one has two residues at the distances of 2.52 Å and 2.16 Å. The short distance and the sustaining hydrogen bonds contribute to the overall stability of the adduct. For comparative purposes, the complexes formed by the antagonists, Darolutamide and Bicalutamide, were evaluated for stability in terms of the number of different types of interactions (Figure S9-S10). There were 8 hydrophobic, 1 hydrogen bond, and 1 π-stacking interactions holding the drug, Darolutamide at the orthosteric pocket of the protein. In case of protein-Bicalutamide complex, there were 6 hydrophobic interactions and 3 hydrogen bonds. These numbers were lower than those observed for the phytochemicals possibly resulting in weaker interactions. The lack of hydrogen bonding was reported for antagonists and weaker binding free energy was reported for antagonists and agonists/antagonists (Singam et al., 2019). Based on these premises, molecule A might be an antagonist and molecule B could perform in dual role. The results of the current study indicate that the hit candidate could modulate the functioning of the receptor (as competitive inhibitor) with better effectiveness than the native and other reference molecules.
3.5 Thermodynamical stability
The changes in binding free energy (ΔGBFE) and several components for the two complexes are given in Table 3 and that for other three complexes are shown in Table S3. Also, the variation in ΔGBFE for the equilibrated portion of the trajectory (last 20 ns with 200 frames) are shown in Figure S11. The negative value of ΔGBFE implied that the reactions were spontaneous in nature and the sustainability was observed until the end of the production run as depicted in the frame-wise variation. The energetics and the spatial analysis have both independently pointed out to the same inference about the adducts. The ligands are therefore bound sturdily to the androgen receptor and could inhibit its normal functioning as antagonists.
Components, kcal/mol
Complex of A
Complex of B
ΔEvdw
−64.89 ± 2.25
−64.81 ± 2.99
ΔEEL
−9.23 ± 2.29
−21.57 ± 5.38
ΔEPB
27.75 ± 3.08
48.37 ± 3.12
ΔENPOLAR
−5.40 ± 0.08
−4.97 ± 0.08
ΔGBFE
−51.77 ± 3.06
−42.98 ± 3.91
The recent literature has revealed several different kinds of chemical compounds studied as antagonist or agonist of androgen receptor (Abdurrahman et al., 2022; Selvaraj et al., 2021; Serçinoğlu et al., 2021; Singh et al., 2017). Finding an effective and safe one with a distinct mechanistic role seems to be challenging in structural analysis point of view and would be carried out as further work. This work has provided different set of molecules from plant-based resources with the possibility of modulating the functioning of androgen receptor. The druglikeness and safety assessments would be required for establishing these phytochemicals for therapeutic uses in human and will be discussed next.
3.6 Druglikeness and safety
The Lipinski’s rule of 5 was followed by all the top molecules (1 violation for 1 molecule only), native ligand, and four drugs (Darolutamide, Bicalutamide, Enobosarm and Abiraterone) showing good solubility and membrane permeability leading to optimal bioavailability (oral formulations) for effective therapeutics. The pharmacokinetics (ADME) and pharmacodynamics (T) showed comparable properties with those of the reference drugs and is mentioned in Table S4-S6.
The analysis of all the results showed that molecule A and molecule B formed stable adducts whereas molecule D formed moderately stable adduct with the androgen receptor. These ligands could cease the activation of receptor by binding to it on a competitive basis with androgens or bring conformational changes that could disrupt gene transcription. The androgen assisted cancer cell progression (prostate cancer) would then be prevented and ultimately the hit candidates could be carried forward to next stages in drug design and discovery process.
4 Conclusions
The phytochemicals present in Withania coagulans (Stocks) Dunal, Rishyagandha showed promising capability in binding with the androgen receptor at the orthosteric site and the geometrical as well as the thermodynamical stability of the adducts with top docking scores were excellent for molecules, 24-Methyl-9,19-cyclolanost-25-en-3-ol (PubChem CID: 185036) and Withacoagulin I (PubChem CID: 71720665), and good for Withanolide F (PubChem CID: 44562998). Major hydrophobic interactions and some hydrogen bonds were responsible for yielding the protein–ligand adduct with sustained stability owing to which the normal functioning of the protein could be disrupted or inhibited as antagonist. This corroborates with the traditional practices of the plant being used against cancer. The modelling results and ADMET predictions could be used in proposing the hit molecules from plant-based resources as analogously safe and druglike therapeutic agents against prostate cancer. A recommendation could be made for further in vitro (enzyme-based essays/cell-lines) and in vivo (mouse model) experimental trials for verification (efficacy and ADMET profile) and for application perspective of the theoretical outcomes.
CRediT authorship contribution statement
Ram Lal Swagat Shrestha: Writing – original draft, Visualization, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation. Jhashanath Adhikari Subin: Writing – review & editing, Validation, Supervision, Methodology, Formal analysis, Conceptualization.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Acknowledgement
Authors would like to thank Dr. Bishnu Prasad Marasini (Nepal Health Research Council, Government of Nepal) for helpful insights on the biological aspect of research. The administrative support from Binita Maharjan and Timila Shrestha is also cordially acknowledged.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Active antialopecia chemical identification of merremia peltata leaves and computational study toward androgen receptor using molecular docking and molecular dynamic simulation. Sci. World J.. 2022;2022:1-17.
- [CrossRef] [Google Scholar]
- Computational assessment of the phytochemicals of panax ginseng C.A. Meyer against Dopamine Receptor D1 for Early Huntington’s Disease Prophylactics. Cell Biochem. Biophys. 2024:1-11.
- [CrossRef] [Google Scholar]
- Biotechnological Intervention and Withanolide Production in Withania coagulans. Agronomy. 2023;13:1997.
- [CrossRef] [Google Scholar]
- In vitro anticancer and antibacterial potentials of selected medicinal plants and isolation and characterization of a natural compound from Withania coagulans. Zeitschrift Für Naturforschung C. 2022;77:263-270.
- [CrossRef] [Google Scholar]
- In Silico Characterization of Withania coagulans Bioactive Compounds as Potential Inhibitors of Hydroxymethylglutaryl (HMG-CoA) Reductase of Mus musculus. ACS Omega. 2023;8:5057-5071.
- [CrossRef] [Google Scholar]
- Bioactive metabolites from the leaves of Withania adpressa. Pharm. Biol.. 2018;56:505-510.
- [CrossRef] [Google Scholar]
- Molecules targeting the androgen receptor (AR) signaling axis beyond the AR‐Ligand binding domain. Med. Res. Rev.. 2019;39:910-960.
- [CrossRef] [Google Scholar]
- Agents That Target Androgen Synthesis in Castration-Resistant Prostate Cancer. The Cancer Journal. 2013;19:34-42.
- [CrossRef] [Google Scholar]
- Grover, A., Singh, R., Shandilya, A., Priyandoko, D., Agrawal, V., Bisaria, V.S., Wadhwa, R., Kaul, S.C., Sundar, D., 2012. Ashwagandha Derived Withanone Targets TPX2-Aurora A Complex: Computational and Experimental Evidence to its Anticancer Activity. PLoS ONE 7, e30890. doi: 10.1371/journal.pone.0030890.
- Gu, Y., Yu, Z., Wang, Y., Chen, L., Lou, C., Yang, C., Li, W., Liu, G., Tang, Y., 2024. admetSAR3.0: a comprehensive platform for exploration, prediction and optimization of chemical ADMET properties. Nucleic Acids Research 52, W432–W438. doi: 10.1093/nar/gkae298.
- Drug design and repurposing with DockThor-VS web server focusing on SARS-CoV-2 therapeutic targets and their non-synonym variants. Sci. Rep.. 2021;11:5543.
- [CrossRef] [Google Scholar]
- An overview on pharmaceutical properties and biotechnological advancement of Withania coagulans. ADV TRADIT MED (ADTM). 2022;22:673-683.
- [CrossRef] [Google Scholar]
- Withaferin A: From ayurvedic folk medicine to preclinical anti-cancer drug. Biochem. Pharmacol.. 2020;173:113602
- [CrossRef] [Google Scholar]
- Helsen, C., Van Den Broeck, T., Voet, A., Prekovic, S., Van Poppel, H., Joniau, S., Claessens, F., 2014. Androgen receptor antagonists for prostate cancer therapy. Endocrine-Related Cancer 21, T105–T118. doi: 10.1530/ERC-13-0545.
- Withanolide E sensitizes renal carcinoma cells to TRAIL-induced apoptosis by increasing cFLIP degradation. Cell Death Dis.. 2015;6:e1666-e.
- [CrossRef] [Google Scholar]
- A Clinically Relevant Androgen Receptor Mutation Confers Resistance to Second-Generation Antiandrogens Enzalutamide and ARN-509. Cancer Discov.. 2013;3:1020-1029.
- [CrossRef] [Google Scholar]
- Targeting Alternative Sites on the Androgen Receptor to Treat Castration-Resistant Prostate Cancer. IJMS. 2013;14:12496-12519.
- [CrossRef] [Google Scholar]
- Withaferin-A—A Natural Anticancer Agent with Pleitropic Mechanisms of Action. IJMS. 2016;17:290.
- [CrossRef] [Google Scholar]
- Luteolin inhibits A549 cells proliferation and migration by down-regulating androgen receptors. Eur. J. Med. Res.. 2023;28:353.
- [CrossRef] [Google Scholar]
- A magic drug target: Androgen receptor. Med. Res. Rev.. 2019;39:1485-1514.
- [CrossRef] [Google Scholar]
- In vitro anticancer activities of Withania coagulans against HeLa, MCF-7, RD, RG2, and INS-1 cancer cells and phytochemical analysis. Integrative Medicine Research. 2018;7:184-191.
- [CrossRef] [Google Scholar]
- Marques, R.B., Dits, N.F., Erkens-Schulze, S., Van IJcken, W.F.J., Van Weerden, W.M., Jenster, G., 2011. Modulation of Androgen Receptor Signaling in Hormonal Therapy-Resistant Prostate Cancer Cell Lines. PLoS ONE 6, e23144. doi: 10.1371/journal.pone.0023144.
- Olubode, S.O., Bankole, M.O., Akinnusi, P.A., Adanlawo, O.S., Ojubola, K.I., Nwankwo, D.O., Edjebah, O.E., Adebesin, A.O., Ayodele, A.O., 2022. Molecular Modeling Studies of Natural Inhibitors of Androgen Signaling in Prostate Cancer. Cancer Inform 21, 117693512211185. doi: 10.1177/11769351221118556.
- Androgens Up-regulate the Insulin-like Growth Factor-I Receptor in Prostate Cancer Cells. Cancer Res.. 2005;65:1849-1857.
- [CrossRef] [Google Scholar]
- Discovery of novel antagonists targeting the DNA binding domain of androgen receptor by integrated docking-based virtual screening and bioassays. Acta Pharmacol. Sin.. 2022;43:229-239.
- [CrossRef] [Google Scholar]
- Rathore, M.S., Khatri, K., Kheni, J., Shekhawat, N.S., 2018. Biotechnological Advancement in an Important Medicinal Plant, Withania coagulans: An Overview and Recent Updates, in: Kumar, N. (Ed.), Biotechnological Approaches for Medicinal and Aromatic Plants. Springer Singapore, Singapore, pp. 445–465. doi: 10.1007/978-981-13-0535-1_20.
- Syringaresinol as a novel androgen receptor antagonist against wild and mutant androgen receptors for the treatment of castration-resistant prostate cancer: molecular docking, in-vitro and molecular dynamics study. J. Biomol. Struct. Dyn.. 2021;39:621-634.
- [CrossRef] [Google Scholar]
- In silico and in vitro assessment of androgen receptor antagonists. Comput. Biol. Chem.. 2021;92:107490
- [CrossRef] [Google Scholar]
- Structural Dynamics of Agonist and Antagonist Binding to the Androgen Receptor. J. Phys. Chem. B. 2019;123:7657-7666.
- [CrossRef] [Google Scholar]
- Structure Based docking studies towards exploring potential anti-androgen activity of selected phytochemicals against Prostate Cancer. Sci. Rep.. 2017;7:1955.
- [CrossRef] [Google Scholar]
- Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacologica Sinica. 2015;36(1):3-23.
- [CrossRef] [Google Scholar]
- Novel androgen receptor antagonist identified by structure-based virtual screening, structural optimization, and biological evaluation. Eur. J. Med. Chem.. 2020;192:112156
- [CrossRef] [Google Scholar]
- NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol.. 2018;18:309-324.
- [CrossRef] [Google Scholar]
- Vaishnavi, K., Saxena, N., Shah, N., Singh, R., Manjunath, K., Uthayakumar, M., Kanaujia, S.P., Kaul, S.C., Sekar, K., Wadhwa, R., 2012. Differential Activities of the Two Closely Related Withanolides, Withaferin A and Withanone: Bioinformatics and Experimental Evidences. PLoS ONE 7, e44419. doi: 10.1371/journal.pone.0044419.
- Verma, S., Lall, N., Meyer, D., 2021. A Wonder Plant Withania: Pharmacological and Chemical Perspectives, in: Mandal, S.C., Chakraborty, R., Sen, S. (Eds.), Evidence Based Validation of Traditional Medicines. Springer Singapore, Singapore, pp. 873–900. doi: 10.1007/978-981-15-8127-4_41.
- Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer. 2015;15:701-711.
- [CrossRef] [Google Scholar]
- OTT. 2018;11:2063-2073.
- [CrossRef]
- Golden berry leaf extract containing withanolides suppresses TNF-α and IL-17 induced IL-6 expression in HeLa Cells. Biosci. Biotech. Bioch.. 2023;87:972-980.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103558.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







