Translate this page into:
Genome-wide identification and expression analysis of CC-NB-ARC-LRR (NB-ARC) disease-resistant family members from soybean (Glycine max L.) reveal their response to biotic stress
⁎Corresponding author. hmigdadi@ksu.edu.sa (Hussein H. Migdadi),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
Using disease-resistant genes is the most effective strategy for protecting crops and ensuring agricultural production, or and protection against infections of different pathogens. Under biotic and abiotic stresses, NB-ARC proteins play a critical role in regulating several critical plant metabolic processes and pathways.
Methods
NB-ARC identification and characterization in soybean are still in their infancy, even though R genes have been characterized by various major crop plants. NB-ARC encoding (R) genes in the soybean genome were identified and characterized in silico.
Results
The 103 NB-ARC genes were computationally identified in the soybean genome, randomly distributed on all soybean chromosomes except 5, 10, and 17. Phylogenetic analysis classified the NB-ARC proteins into nine primary groups. However, synteny analysis results of NB-ARC genes of soybean found the best orthologous hit in the A. thaliana representing sequence conservation up to 80%. Soybean NB-ARC genes displayed a plurality of introns between one to seven among the family members. Although their genomic regions have different sizes, a relatively conserved genetic structure was observed within phylogenetic tree groups. Twenty different domains were kept in a group-specific manner, together with the presence of the NB-ARC signatory. Moreover, the transcriptome based-data expression analysis suggested that NB-ARC genes in between non-pathogens and pathogens after the inoculation of Fusarium oxysporum (biotic stress) in the soybean transcriptome, supporting the conjecture of NB-ARC genes have disease resistance functions in the soybean genome and revealing the potential involvement of these genes in the conserved pathways of the biotic-stress-response.
Conclusion
This genome-wide in silico/ computational analysis will be used for accelerating NB-ARC members used for functional characterization, especially under biotic and abiotic stresses.
Keywords
Soybean
Genome-wide association
Biotic stress
Disease resistance
NB-ARC
1 Introduction
The present of wide variety of potential pathogens, such as fungi, viruses, bacteria, and nematodes, dynamically growing plants may encounter various biotic infections under natural conditions. Through the coevolution of plants and pathogens, plants have developed a range of advanced defense mechanisms that have enabled them to perceive various pathogens and defend against pathogenic infections (Muthamilarasan and Prasad, 2013). Plants have developed a sensory mechanism for detecting biotic stress that triggers systematic, localized disease resistance responses (Marone et al., 2013). When an elicitor, either a microbe-associated molecular pattern or damage-associated molecular pattern, is involved, a disease resistance response occurs (Boller and Felix, 2009). The virulence and virulence genes of pathogens are well known and are triggered directly or indirectly by plant disease resistance mechanisms, such as the hypersensitive response (HR) and gained systemic resistance (Künstler et al., 2016). Resistance to plant diseases driven by interaction with disease resistance (R) and avirulence (AVR) gene was first identified almost half a century ago as the “gene-for-gene model” (Flor, 1971). Moreover, the application of genome-wide association study (GWAS) and post-GWAS studies combined with transcriptome data helps to figure out candidate genes potentially regulating the various traits under certain conditions (Chen et al., 2021).
Most R-gene proteins contain the nuclear binding site (NBS) domain and the leucine-rich region (LRR), which are activated by pathogen-gene elicitors and send a systemic message to trigger plant defense reactions (Gao et al., 2013). R genes can be classified into at least five classes based on their structures (Li et al., 2016). The first R-gene family codes, a transmembrane reporter with extracellular regions and the Cladosporium fulvum (Cf) family, established the resistance in the tomato leaf (Li et al., 2016). The third category combines the above qualities of the receptor-like protein kinase shown by Xa21, which confers resistance to a bacterial blight disease in rice (Kim, 2018). A transmembrane domain and a cytoplasmic coil-coil (CC) domain (24) are included in the fourth class, and the fifth R cluster is NBS-LRR, where the bulk of R genes were present (Jorgensen and Emerson, 2009). An extracellular LRR and transmembrane region (TM), as well as cytoplasmic ser-thre-kinesin, make up the fifth class of genes. This suggests that there is an evolutionary relationship between distinct classes of plant disease resistance genes, based on the structure of Xa21 (Song et al., 1997). Finally, RPW8 helps to provide resistance to Arabidopsis against powdery mildew disorders (Jorgensen and Emerson, 2009). The NBS-LRR class is considered a cytoplasmic gene with a distinct N-terminal domain. To date, the application of disease resistance genes in crop plants is the most significant strategy to overcome the biotic stress issues against different pathogens. Up till now, GWAS have been used to identify the disease resistance genes (Sanseverino et al., 2012) in different crop plants i.e., Arabidopsis (Yu et al., 2014), potato (Lozano et al., 2012), wheat (Gu et al., 2015), rice (Singh et al., 2015), barley (Wang et al., 2013), and Brachypodium distachyon (Tan and Wu, 2012). The R-gene class NBS-LRR comprises three domain-N terminal vectors, a nucleotide-binding site (NBS), and LRR (Chisholm et al., 2006). NBS protein domains are categories into five different conserved patterns (Panwar et al., 2011). The first pattern is called the P-loop, significant for binding protein domain and help for R gene product activities (Wan et al., 2012). The second conserved motif is known as Kinase 2. It has four hydrophobic amino acid residues and aspartic acid with a negative charge. LRR regions mediate protein–protein interactions, but they can play an essential role in gene-for-gene identification of pathogen-specific genes (Wan et al., 2012). The NBS-LRR group can be subdivided into two distinct groups based on the configuration of the N terminal. One type contains a coiled-coil (CC) motif for N terminals capable of participating in protein–protein interactions (Maekawa et al., 2011). The second form of NBS-LRR lacks the CC, whereas the N-terminus region has a TIR domain that shares homological features with a protein like Drosophila Toll Interleukin-1 mammals (TIR) (Pan et al., 2000). CNL and TNL comprise two families, usually found at the N-terminus of the R-protein, and differentiate themselves in a domain structure (Marone et al., 2013). Only TNL genes were present in monocots plants, while CNL genes were present in both dicots and monocots, making them appropriate for studying growth processes in plant species (Meyers et al., 1999).
Even though the research on R proteins imparting resistance to a variety of illnesses is limited to soybean. However, NB-ARC genes were found to co-segregate with the Rpg1-b locus, which confers resistance to biotic stress disease (Ashfield et al., 2003). Furthermore, it was also suggested that Toll/Interleukin-1 Receptor homology (TIR-NBS-LRR) was found to inhibit nodulation in soybean (Zhu et al., 2010). It could be that R genes control microbe entry into soybean plants because nodulation is a symbiotic rather than a pathogen-host interaction. However, the NBS-LRR genes that were present across the soybean genome may be recognized as pathogens and confer resistance (Kang et al., 2012).
Few NBS-LRR genes have some homologous with those of A. thaliana, but most NBS-LRR genes have noticeable variations compared to Arabidopsis NB-ARC (R) genes. For this purpose, in the current exploratory research, GWA investigations can be used to decode biological processes governing characteristics by employing candidate gene lists gained from GWAS analysis. Hence, the current study was done with systematic computational analysis in the soybean by defining a CC-NB-LRR function model wherein the LRR and CC domains co-regulate the NB domain's signaling behavior in a recognition-specific manner. Corresponding NB-ARC genes confirm the disease resistance against Fusarium oxysporum in the soybean. In this study, all NB-ARC disease-resistant genes in the soybean genome were collected, accompanied by homologous comparisons and phylogenetic analysis using NB-ARC proteins sequence. The classification of NB-ARC soybean types provides conclusive tools and essential data for continued functional exploration and finally shows their functions in the battle against biotic stress, i.e., fungus. The aim of this study is to investigate novel R-genes present throughout the genome of soybean, makes more convenient to understand the functioning of this specific domain. This will also help for accelerating NB-ARC members used for functional characterization, especially under biotic and abiotic stresses. To our best knowledge, the data regarding CC terminal in soybean was not reported before and concerted the effort to classify NB-ARC genes and their role in suppressing disease control.
2 Materials and methods
2.1 NB-ARC genes identification
The complete genome assembly of soybean and tabular form of protein sequences were downloaded from NCBI and verified from the phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#! info? alias = Org_Gmax/). A total of 436 disease resistance genes were chosen from the soybean genome derived from phytozome. Based on the phytozome database, 103 protein sequences were got from all NB-ARC genes resistant to the soybean genome (https://phytozome.jgi.doe.gov/pz/portal.html#! info? alias = Org_Gmax/) after duplication screening. Arabidopsis genes have been identified and listed. Sequences of Arabidopsis thialana NB-ARC genes were used for the comparative phylogenetic study (http://niblrrs.ucdavis.edu/ index.php) (Meyers et al., 2003). NBS-LRR disease resistance in a soybean proteome sequence file using CLC sequence viewer (v7.6.1) was used to find conserved protein sequences for NB-ARC (Knudsen et al., 2011). The local alignment search tool (blastP) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM = blastp&PAGE_TYPE = BlastSearch&LINK_LOC = blasthome) at the NCBI web server further verified the putative NB-ARC protein sequence. The NCBI database was also investigated and marked NBS-LRR gene information, including accession numbers (GI), chromosome numbers, genomic details, and protein size. All known NB-ARC genomic nucleotide sequences against specific protein sequences have also been recovered from NCBI. ExPASy bioinformatics systems resource platform was used to measure molecular weight and isoelectric point (IP) (http://web.expasy.org/compute_pi/) (Gasteiger et al., 2005). The conserved domain was confirmed from NCBI, and each gene's architecture is given in (Supplementary Table 1) presenting the location of the domain of interest in each gene.
2.2 Analysis of NB-ARC conserved motif structure
The NBS is a region that starts at the P-loop, ends with the MHDV motif, and contains approximately 260 to 300 amino acids. The P-Loop upstream segment is the N-terminal motive, and the LRR domain is the downstream segment of the MHDV. The program's absence or existence of TIR, NBS, and LRR domains was also verified, but it was inaccessible to examine smaller or fragmented patterns, such as those in the (NB-ARC) domain (Bailey et al., 2006). Therefore, to discover conserved motifs in NB-ARC protein sequences, conserved motif analysis was performed using the online MEME SUITE tool (http://meme-suite.org/) (Bailey et al., 2009).
2.3 Phylogenetic analysis of NB_ARC genes
An evolutionary tree of 103 NB-ARC protein sequences was constructed using Molecular Evolutionary Genetics Analysis software (MEGA version 7.0) (Tamura et al., 2013). First, all protein sequences were subjected to alignment through the MUSCLE algorithm with default parameters such as gap opening penalty −2.9, gap extension penalty 0, hydrophobicity multiplier 1.2, and unweighted pair group method with arithmetic mean (UPGMA) clustering method was used. After that, using aligned data, evolutionary history was inferred using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992). Finally, 1000 bootstrap replications, partial gap deletion, and 95% site coverage cutoff value were used.
2.4 Chromosomal mapping, intron/exon distribution, and conserved domain analysis
Phytozome database was used to record the chromosome position of NB-ARC genes, while the chromosomal location of all non-redundant NB-ARC genes was found using Map Chart (v. 2.32), and the map was constructed according to scale NB-ARC location on the chromosome (Voorrips, 2004). Gene Structure Display Server (v2.0http:/gsds.cbi.pku.edu.cn/) was used to constructing the gene structure that shows the intron–exon distribution of NB-ARC genes (Hu et al., 2015). Genomic DNA and CDS sequences of all NB-ARC genes were used to build the genome structure map and the intron phases (S1-figure; S3-Table hit data). Conserved motif analysis was performed using the online (http://meme-suite.org/) MEME SUITE tool to discover conserved motifs in NB-ARC protein sequences (Bailey et al., 2009). Different parameters were assessed one by one for motif discovery to display the conserved domains through identified motifs. However, maximum numbers of motifs; 20, minimum motif width; 15, and maximum motif width; 50 were finally used. All protein sequences of NB-ARC genes were arranged according to their clustering in the phylogenetic tree. As a result, all discovered motifs were adjusted in front of their respective gene name. Whereas conserved domain analysis was performed by visiting the online NCBI conserved domain database (https://structure.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) with default parameters, and final visualization was got using TBtools software.
2.5 Synteny analysis of NB-ARC genes family of soybean and Arabidopsis
Synteny analysis of the NB-ARC genes family of soybean and Arabidopsis was performed to determine the homologous NB-ARC genes family. There are 126 protein sequences of soybean, and Arabidopsis specific for NB-ARC domain was determined using synteny analysis was performed using the Circoletto online tool (http://mkweb.bcgsc.ca/circos/intro/circular_approach/), which was practiced using the strict E-value of 1x10-50 and BLOSSUM matrix.
2.6 Insilco expression analysis of NB-ARC of Fusarium oxysporum as biotic stress
Transcriptome data was taken from the soybean transcriptome database (https://soybase.org/soyseq/) (Lanubile et al., 2015), where biotic stress has been given to the roots. The soybean partially resistant genotype was briefly treated with a conidial suspension of non-pathogenic and pathogenic (F036 and F040) F. oxysporum isolates. Expression data of NB-ARC genes were retrieved from transcriptome induced by biotic stress root tissues. The expression of NB_LRR class NB-ARC genes was investigated from roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum (Lanubile et al., 2015). F. oxysporum penetrates the roots and moves through the vascular system (Ortiz et al., 2014). In these NB-ARC gene expression data were used to construct heat map expression (http://www.heatmapper.ca/expression/) profiles (Sturn et al., 2002), their differential root expression was shown in biotic stress condition to validate the study.
3 Results
3.1 Identification and distribution of NB-ARC in the soybean genome
We initially detected 250 non-redundant, disease-resistant NB_ARC genes from the whole soybean genome assembly, which putatively encoded NB-ARC. We screened the genes for the existence of the coding proteins of NB-ARC. The main domain was a basic requirement for including genes in the NB_LRR family in the full NB-ARC domain. Of the selection, 147 NB_ARC genes were deleted because of incomplete NB-ARC domains in their protein series. These pseudogenes may have lost their functional domain portion during evolution. The other 103 non-redundant NB-ARC proteins were rebuilt in ascendant order. The complete information of NB-ARC proteins sequence with different peptide lengths ranging from about 220 to 2199 amino acids with an average of 1016 amino acids (Table 1). The chromosome map revealed the uneven distribution of 103 NB-ARC genes on 17 out of 20 chromosomes (Fig. 1). The remaining chromosome numbers (5, 10, 17) were not mapped because of the scaffold regions. All the chromosomes share different positions of NB-ARC genes (Fig. 1). Interestingly, most NB-ARC genes were identified as clusters in the chromosome to form part of a single QTL within a cluster (Fig. 1).Fig. 2
Gene ID
New name
LOC ID
Chromosome
Start
End
Size
Protein
Accession #
Glyma.01G010500.1
GNBARC1
LOC100305458
1
1018932
1021736
934
NP_001235671.1
NC_016088.3
Glyma.01G010700.1
GNBARC2
LOC100805346
1
1029421
1033293
946
XP_003517650.1
NC_016088.3
Glyma01g01560
GNBARC3
LOC102662991
1
1157242
1160517
1091
XP_006572949.1
NC_016088.3
Glyma.01G013100.1
GNBARC4
LOC102666512
1
1234209
1237457
1082
XP_006573928.1
NC_016088.3
Glyma.01G035400.1
GNBARC5
LOC102660000
1
3693131
3696424
870
XP_006573066.2
NC_016088.3
Glyma.01G065800.1
GNBARC6
LOC100777175
1
3755286
3762981
897
XP_014627470.2
NC_016088.3
Glyma.01G171000.1
GNBARC7
LOC100788025
1
50833471
50836299
910
XP_003517205.1
NC_016088.3
Glyma.02G184300.1
GNBARC8
LOC100780033
2
32159928
32162528
866
XP_014623640.1
NC_016089.3
Glyma.02G026200.1
GNBARC9
LOC100775587
2
2338744
2341398
884
XP_014619948.1
NC_016089.3
Glyma.02G030700.1
GNBARC10
LOC100787353
2
2819837
2823095
859
XP_006574586.1
NC_016089.3
Glyma.03G034500.1
GNBARC11
LOC102668720
3
4054522
4057458
979
XP_014628961.1
NC_016090.3
Glyma.03G034800.1
GNBARC12
LOC106794107
3
4123259
4126987
1242
XP_003521994.1
NC_016090.3
Glyma.03G034900.1
GNBARC13
LOC100777231
3
4199841
4203584
1247
XP_014628962.1
NC_016090.3
Glyma.03G037000.1
GNBARC14
LOC100784015
3
4516156
4519896
1246
XP_003522002.1
NC_016090.3
Glyma.03G037100.1
GNBARC15
RPSHC18-BL1
3
4541962
4545711
1249
XP_006576451.1
NC_016090.3
Glyma.03G037300.1
GNBARC16
RPSHC18-BL2
3
4566919
4570596
1225
XP_006576452.1
NC_016090.3
Glyma.03G038800.1
GNBARC17
LOC100794060
3
4790805
4794512
1235
XP_014628972.1
NC_016090.3
Glyma.03G039300.1
GNBARC18
LOC100775612
3
4890834
4894580
1248
XP_003521990.1
NC_016090.3
Glyma.03G043000.1
GNBARC19
LOC106798139
3
4913423
4916401
992
XP_014629579.1
NC_016090.3
Glyma.03G043200.1
GNBARC20
LOC102659829
3
5457034
5460630
1198
XP_025983528.1
NC_016090.3
Glyma.03G043500.1
GNBARC21
LOC100811433
3
5503262
5521091
1242
XP_025983527.1
NC_016090.3
Glyma.03G043600.1
GNBARC22
LOC113001152
3
5517406
5520102
898
XP_025983530.1
NC_016090.3
Glyma.03G045700.1
GNBARC23
LOC100813244
3
5818186
5821920
1244
XP_003522031.1
NC_016090.3
Glyma.03G046500.1
GNBARC24
LOC100816969
3
5937187
5940662
1131
XP_006577442.3
NC_016090.3
Glyma.03G047000.1
GNBARC25
LOC100818566
3
5976716
5980375
1219
XP_025983532.1
NC_016090.3
Glyma.03G075200.1
GNBARC26
LOC100499652
3
18615820
18619545
1241
NP_001237787.1
NC_016090.3
Glyma.03G137200.1
GNBARC27
LOC100805394
3
35337199
35342255
858
XP_006576824.2
NC_016090.3
Glyma.04G137800.2
GNBARC28
LOC100781872
4
21176563
21179130
855
XP_006578494.1
NC_016091.3
Glyma.06G167200.1
GNBARC29
LOC100812584
6
13958598
13961228
876
XP_003526943.1
NC_038242.1
Glyma.06G311200.1
GNBARC30
LOC100801561
6
49978291
49981122
943
XP_003526348.1
NC_038242.1
Glyma.07G075700.1
GNBARC31
LOC102664581
7
6862792
6865801
220
XP_006583335.1
NC_038243.1
Glyma.08G259000.1
GNBARC32
LOC100801950
8
23381749
23384433
894
XP_006585845.1
NC_038244.1
Glyma.08G305400.1
GNBARC33
LOC100819372
8
42356991
42359831
946
XP_003530717.1
NC_038244.1
Glyma.08G317400.1
GNBARC34
LOC102661118
8
43699213
43701924
903
XP_014634887.1
NC_038244.1
Glyma.08G317700.1
GNBARC35
LOC100798933
8
43718126
43738036
900
XP_014634889.1
NC_038244.1
Glyma.08G319300.1
GNBARC36
LOC100802130
8
43849788
43852499
903
XP_006586093.1
NC_038244.1
Glyma.08G323200.1
GNBARC37
LOC102667193
8
44172401
44175121
906
XP_006586109.1
NC_038244.1
Glyma.08G328800.1
GNBARC38
LOC100782327
8
44662935
44670593
926
XP_003530797.1
NC_038244.1
Glyma.09G020500.1
GNBARC39
LOC102666390
9
1614752
1617772
1006
XP_006586820.1
NC_038245.1
Glyma.09G020700.1
GNBARC40
LOC102666690
9
1632002
1635025
1007
XP_006586822.1
NC_038245.1
Glyma.09G210400.1
GNBARC41
LOC100817624
9
43450724
43454330
948
XP_006587620.1
NC_038245.1
Glyma.09G210600.1
GNBARC42
LOC100805529
9
43458931
43461744
937
XP_003534302.1
NC_038245.1
Glyma.11G058900.2
GNBARC43
LOC100817860
11
4453650
4458075
835
XP_006590663.1
NC_038247.1
Glyma.11G072100.2
GNBARC44
LOC100788797
11
5378566
5381412
912
XP_003537613.1
NC_038247.1
Glyma.12G011700.1
GNBARC45
LOC100814688
12
853407
856196
929
XP_006591996.1
NC_038248.1
Glyma.12G218500.1
GNBARC46
LOC100799733
12
37820168
37823146
992
XP_006592896.1
NC_038248.1
Glyma.12G236500.4
GNBARC47
LOC102660573
12
39528938
39533282
1024
XP_006592961.1
NC_038248.1
Glyma.13G071900.1
GNBARC48
LOC100803330
13
17288420
17292130
1236
XP_003543829.1
NC_038249.1
Glyma.13G187900.1
GNBARC49
LOC100806158
13
29858322
29862900
1185
XP_014621132.1
NC_038249.1
Glyma.13G184800.1
GNBARC50
LOC100806158
13
29858322
29862900
1263
XP_014621131.1
NC_038249.1
Glyma.13G190300.1
GNBARC51
LOC102661203
13
29872256
29877088
1095
XP_014621743.1
NC_038249.1
Glyma.13G188300.1
GNBARC52
LOC100804921
13
30207288
30211729
1181
XP_025980833.1
NC_038249.1
Glyma.13G190400.1
GNBARC53
LOC100818432
13
30402232
30408831
2199
XP_006594359.1
NC_038249.1
Glyma.13G190800.1
GNBARC54
LOC100499655
13
30426359
30430201
1280
NP_001237835.1
NC_038249.1
Glyma.13G192100.2
GNBARC55
LOC100777280
13
30532501
30536199
1232
XP_006594365.1
NC_038249.1
Glyma.13G193100.1
GNBARC56
LOC100778337
13
30643477
30647103
1208
XP_003541580.2
NC_038249.1
Glyma.13G194100.1
GNBARC57
LOC547607
13
30726801
30730421
1206
XP_014621169.1
NC_038249.1
Glyma.13G194500.2
GNBARC58
LOC100781012
13
30763920
30767591
1223
XP_006594377.1
NC_038249.1
Glyma.13G195600.1
GNBARC59
LOC100783712
13
30914885
30918517
1132
XP_006594385.1
NC_038249.1
Glyma.14G199400.1
GNBARC60
LOC100787796
14
46435759
46438392
877
XP_014622020.1
NC_038250.1
Glyma.15G126900.1
GNBARC61
LOC100784635
15
10069700
10072719
1005
XP_006597652.2
NC_038251.1
Glyma.15G127100.1
GNBARC62
LOC102663592
15
10079713
10082742
1009
XP_006598349.1
NC_038251.1
Glyma.15G168500.1
GNBARC63
LOC100305356
15
15007753
15012351
979
NP_001237924.1
NC_038251.1
Glyma.15G186800.1
GNBARC64
LOC106796110
15
19312523
19315555
900
XP_025981675.1
NC_038251.1
Glyma.15G226100.1
GNBARC65
LOC100789590
15
41539364
41543065
1233
XP_014623006.1
NC_038251.1
Glyma.15G230700.1
GNBARC66
LOC100776964
15
43210480
43214280
1266
XP_006598091.1
NC_038251.1
Glyma.15G232800.1
GNBARC67
LOC547639
15
43744171
43747779
1191
XP_025981489.1
NC_038251.1
Glyma.15G233100.1
GNBARC68
LOC100784466
15
43796673
43800785
1370
XP_025981563.1
NC_038251.1
Glyma.15G233400.1
GNBARC69
LOC100792404
15
43919306
43922920
1193
XP_006598101.1
NC_038251.1
Glyma.16G079400.1
GNBARC70
LOC100777510
16
8197004
8200591
1195
XP_006599131.1
NC_038252.1
Glyma.18G078000.4
GNBARC71
LOC100779508
18
7420171
7443944
938
XP_003551452.2
NC_038254.1
Glyma.18G082100.1
GNBARC72
LOC100805006
18
7972983
7975742
919
XP_003551523.1
NC_038254.1
Glyma.18G082300.1
GNBARC73
LOC100809266
18
8018442
8021201
919
XP_003551528.1
NC_038254.1
Glyma.18G082400.1
GNBARC74
LOC100787897
18
8061135
8063909
913
XP_014625972.1
NC_038254.1
Glyma.18G083200.1
GNBARC75
LOC100784168
18
8189521
8192283
920
XP_003551547.1
NC_038254.1
Glyma.18G084400.1
GNBARC76
LOC100798997
18
8302552
8305323
923
XP_003551565.1
NC_038254.1
Glyma.18G086600.1
GNBARC77
LOC100799057
18
8527932
8530700
922
XP_014625795.1
NC_038254.1
Glyma.18G087800.1
GNBARC78
LOC100786451
18
8691630
8694350
906
XP_014625802.1
NC_038254.1
Glyma.18G088300.1
GNBARC79
LOC100806153
18
8750100
8752865
921
XP_014625825.1
NC_038254.1
Glyma.18G093400.1
GNBARC80
LOC100782760
18
9424633
9427344
903
XP_003553063.1
NC_038254.1
Glyma.18G093500.1
GNBARC81
LOC100784361
18
9492578
9495304
908
XP_006603185.1
NC_038254.1
Glyma.18G093600.1
GNBARC82
LOC100784890
18
9499458
9502375
912
XP_003553066.1
NC_038254.1
Glyma.18G093800.1
GNBARC83
LOC100785955
18
9535721
9538414
897
XP_003553068.1
NC_038254.1
Glyma.18G093900.1
GNBARC84
LOC102662760
18
9542826
9545235
769
XP_006603186.1
NC_038254.1
Glyma.18G105100.1
GNBARC85
LOC102663437
18
9585492
9587972
918
XP_014625904.1
NC_038254.1
Glyma.18G190900.1
GNBARC86
LOC102667903
18
46052489
46057089
925
XP_014625814.2
NC_038254.1
Glyma.18G269500.1
GNBARC87
LOC100805727
18
55321894
55325641
919
XP_006602948.1
NC_038254.1
Glyma.18G287000.1
GNBARC88
LOC100780593
18
56706975
56709665
896
XP_006603027.1
NC_038254.1
Glyma.18G287100.1
GNBARC89
LOC100499631
18
56710526
56713416
884
XP_006601748.1
NC_038254.1
Glyma.19G085600.1
GNBARC90
LOC100305368
19
30577076
30581436
909
NP_001238129.1
NC_038255.1
Glyma.19G134100.1
GNBARC91
LOC100777049
19
39510186
39512804
872
XP_006604334.1
NC_038255.1
Glyma.19G134200.1
GNBARC92
LOC100499628
19
39523292
39529108
694
NP_001237395.1
NC_038255.1
Glyma.19G135600.1
GNBARC93
LOC100305457
19
39674352
39676943
863
NP_001235657.1
NC_038255.1
Glyma.19G135800.1
GNBARC94
LOC100781317
19
39707093
39709726
877
XP_006604341.1
NC_038255.1
Glyma.19G136900.1
GNBARC95
LOC106797500
19
39833860
39836484
874
XP_014627443.1
NC_038255.1
Glyma.19G137200.1
GNBARC96
LOC100786131
19
39849808
39852429
873
XP_006604349.1
NC_038255.1
Glyma.19G139600.1
GNBARC97
LOC100795479
19
40084248
40086890
880
XP_003553414.2
NC_038255.1
Glyma.19G139700.1
GNBARC98
LOC100796004
19
40105944
40108505
853
XP_006604363.1
NC_038255.1
Glyma.20G042400.1
GNBARC99
LOC100787762
20
7632667
7635447
926
XP_014627876.1
NC_038256.1
Glyma.20G042700.1
GNBARC100
LOC100789363
20
7689993
7692791
932
XP_003556794.1
NC_038256.1
Glyma.20G046200.1
GNBARC101
LOC100801544
20
8605208
8608984
1258
XP_003556802.1
NC_038256.1
Glyma.20G193300.1
GNBARC102
LOC102663592
20
43217821
43222056
1411
XP_006606921.1
NC_038256.1
Glyma.20G195400.1
GNBARC103
LOC102663848
20
43372511
43375327
938
XP_014627824.1
NC_038256.1
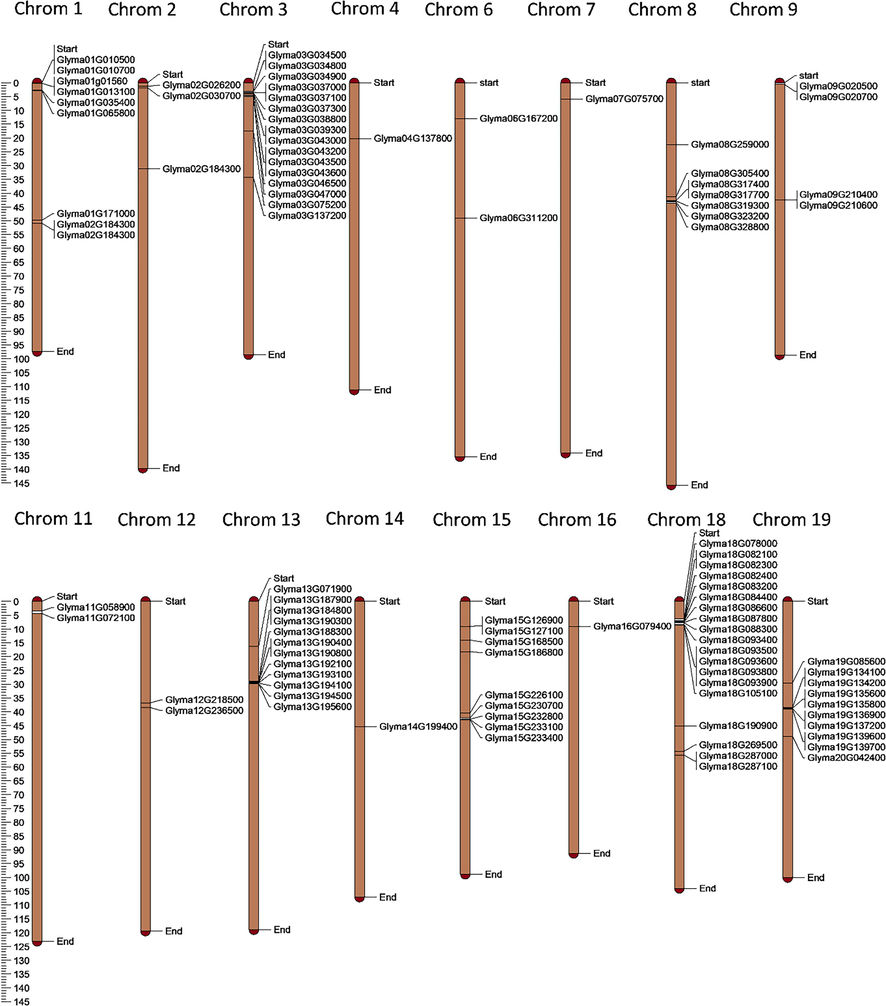
Distribution of 103 NB-ARC genes on soybean chromosomes. The numbers at the top of each bar represent the soybean chromosome numbers. The location of each gene is shown on the right-hand side of the respected chromosome.
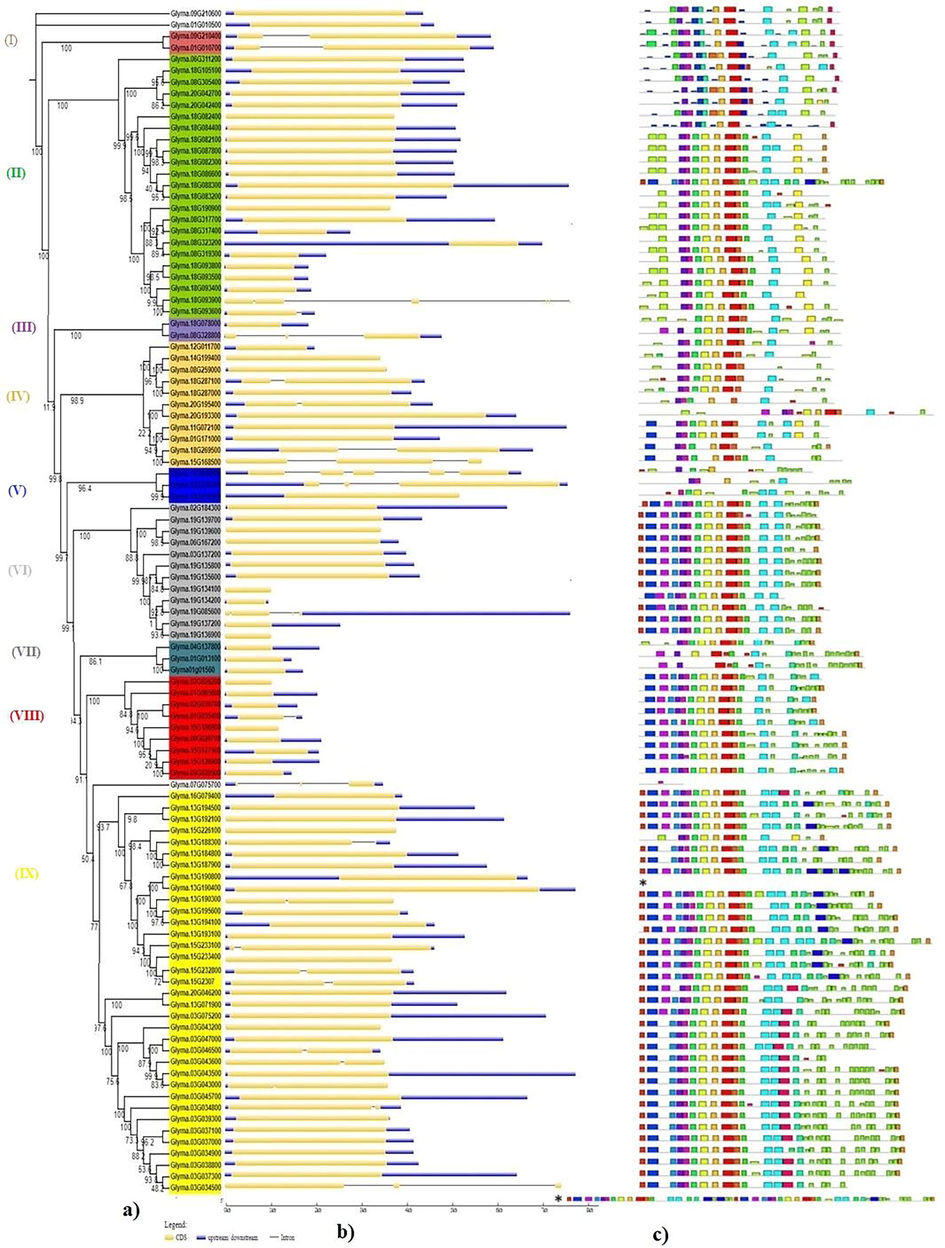
a. Phylogenetic tree-based classification of CC-NB-ARC-LRR (NB-ARC). An un-rooted phylogenetic tree was created based on the full-length peptide sequences (NB-ARC) with 1000 replicates. Classification is shown based on a phylogenetic tree using differences into groups: b Exon–intron structure analyses of (NB-ARC) genes. The gray line represents introns, while the yellow boxes represent exons. The blue boxes represent the untranslated region (UTR). C: Conserved domains of soybean (NB-ARC) proteins. According to the scale, the conserved domains of (NB-ARC) proteins identified by MEME are shown with colored boxes. Gray lines represent the non-conserved sequences, and each domain is shown by a colored box numbered at the bottom.
3.2 Soybean NB-ARC description, gene structure, and conserved domains analysis
The soybean NB-ARC gene was classified using an un-rooted phylogenetic tree in nine major groups (Figure 2). This grouping followed the same trend as in other crop species (Zhang and Wang, 2005). The gene structure of all selected NB-ARC genes, i.e., the intron/exon distribution pattern, was also calculated to provide further insight into the soybean development NB_LRR family. An ordinary location and intron–exon distribution pattern in the genome area helped determine the gene family’s expansion pattern and evolutionary relationship with their ancestors. Soybean NB-ARC genes displayed a plurality of introns between one to seven. The phylogeny of the 103 NB-ARC genes was constructed using MEGA 7.0 software. The NB-ARC genes deduced full-length protein sequences were aligned with Clustal Omega, and a phylogenetic tree was constructed using an un-rooted maximum-likelihood process with 1000 bootstraps. The 103 genes were divided into (IX) groups. The tree was divided based on the specific groups that contain CC regions such as RX_N RX_CC + LRR (18), RX_CC_like (22), RX-N PLN 03,210 (1), NB-ARC + LRR (3), RX_N (8), RX_CC PLN00113 (1), RX_N NB-ARC LRR (1); RX_N NB-ARC RX_CC (21); RX + RX_CC (2); RX_N NB-ARC RX_CC (5); RX + RX_CC (18) NB-ARC genes, and they showed diversity among the same family members (Fig. 3). Although their genomic regions have different sizes, they showed the relatively conserved genetic structure within phylogenetic tree groups. The gene structures of glyma01g065800, glyma03g034800, glyma08g317700, glyma13g190300, glyma13g195600, glyma15g230700, glyma15g232800, and glyma20g195400 had only one intron site in the genomic region. However, all other chromosomes did not have any intron site.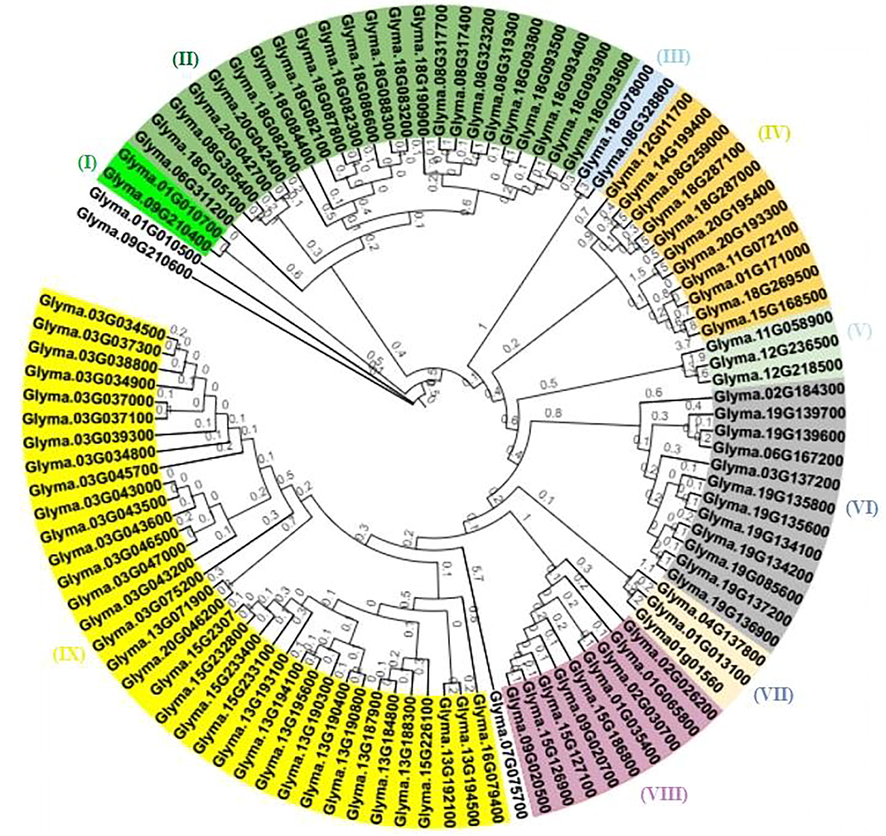
Phylogenetic tree of 103 NB-ARC genes based on maximum likelihood methods with 1000 bootstraps constructed in MEGA 7.0. The numbers on the nodes represent the percentage of bootstrap values from the 1000 replicates. Different colors are used to differentiate the significant cluster of orthologous genes (I-IX).
Some genes contained untranslated regions on the gene structure i.e., glyma01g065800, glyma02184300, glyma03g034500, glyma03g046500, glyma03g075200, glyma06g167200, glyma13g187900, glyma13g190300, glyma13g193100, glyma13g195600, glyma15g186800, glyma16g079400, glyma18g078000, glyma18g093900, glyma18g105100, glyma18g190900, and glyma19g134200 (Fig. 3, Supplementary Fig. 1). Identifying conserved domains within a gene family also provides a way of checking and dissecting gene replication events during evolution. MEME was subjected to the peptide sequences of all NB-ARCs to classify the conserved domain (Table 2). For the 103 NB-ARC genes, twenty conserved domains with residue lengths of 12–42 were identified. Domains 1 and 2 reflect the NB-ARCs DNA-binding domain, which is completely conserved among the 103 NB-ARC genes (Fig. 3). Also, the results of conserved domain analysis directly corresponded with phylogenetic grouping and confirmed the conserved domain analysis results.
Domains
E value
Sites
width
Multilevel consensus sequence
1
3.6e-2764
99
50
NSIIPALRLSYHDLPSHLKRCFAYCSJYPKDYEFEKERLIRLWMAEGFLK
2
1.8e-2097
165
39
LPSSJGKLKHLRYLDLSNTGIEKLPESJGKLYNLQTLDL
3
8.3e-1788
515
20
ALPSLKTLSISDCPKLESLP
4
2.5e-1595
100
28
LSVISIVGMGGLGKTTLAKLVFNDPRVK
5
7.6e-1335
99
28
DIGKEIVKKCKGLPLAIVTJGGLLRRKS
6
5.3e-1118
117
28
MDLESLQDELRNKLKGKRYLLVLDDVWN
7
4.9e-1426
64
50
KKLKTTLRSVKAVLDDAEQKQFTBSRVKEWLRELKDAVYDAEDLLDEIET
8
2.6e-933
104
24
FDLKAWVCVSQDFDIEKLTRTIJE
9
2.3e-1048
98
28
SEEGKTLEEVGZQYLBELLSRSFFQVSS
10
1.3e-1151
96
36
GANGSKILVTTRSEKVASIMGTSSVYHLHLLSPEDC
11
1.1e-792
84
24
FVMHDLVHDLALYVAGDFCFRLEE
12
2.4e-626
53
28
DVLENLQPSQHLEKLSIRGYGGTQFPDW
13
2.3e-715
56
28
TTSLVDESDIYGREEDKEKIIKLLTSDN
14
1.0e-553
18
50
PFLKELSISGLDGIVSINADFFGSSSSSFTSLESLKFSDMKEWEEWECKG
15
3.5e-445
45
39
NFFKSSKHLVFRYKIASRMKDISERLEKLASERDKFGLK
16
5.5e-506
17
50
FIVGKHKENGIKELGGLSNLHGSLSIRNLENVTQSBEALEARMMDKKHIN
17
3.2e-421
70
24
VGGAFLSAFLQVLFDKLASPEVVD
18
5.0e-445
67
20
RPKGGEDWPKIAHIPHVRID
19
4.4e-413
31
39
APVLQKLRLVGRLKKFPNWISKLQNLVTLSLSGSRLTND
20
3.6e-414
93
40
QLPDDPGCAALLCKAIDFIKTTASRLQSAYKNQDVKSEFR
Overall, conserved domains and the intron–exon distribution pattern among soybean NB-ARC genes were group specific and confirmed NB-ARC domains' groupings within the phylogenetic tree.
3.3 Synteny analysis of NB-ARC genes family of soybean with Arabidopsis thaliana
Synteny provides a framework in which conservation of homologous genes and gene order is identified between genomes of different crop species. This work revealed that several soybean NB-ARC genes are syntenic to those of A. thaliana, demonstrating an evolutionary relationship between both species. Twenty-four syntenic regions were identified in the genome of A. thaliana (Fig. 4) using a strict E-value of 1x10-50 for BLAST run and BLOSSUM scoring matrix. In the ideogram, many NB-ARC genes of G. max found best hit orthologous in the A. thaliana representing sequence conservation up to 80% (red in Figure). However, few genes in soybean did not show any orthologous relationship in the genome of both species, for instance, Glyma.18G093900. Furthermore, it has been observed that duplications, including segmental duplication, tandem duplication, and genomic duplication, played an essential role in the expansion of the NB-ARC gene family in both crop species.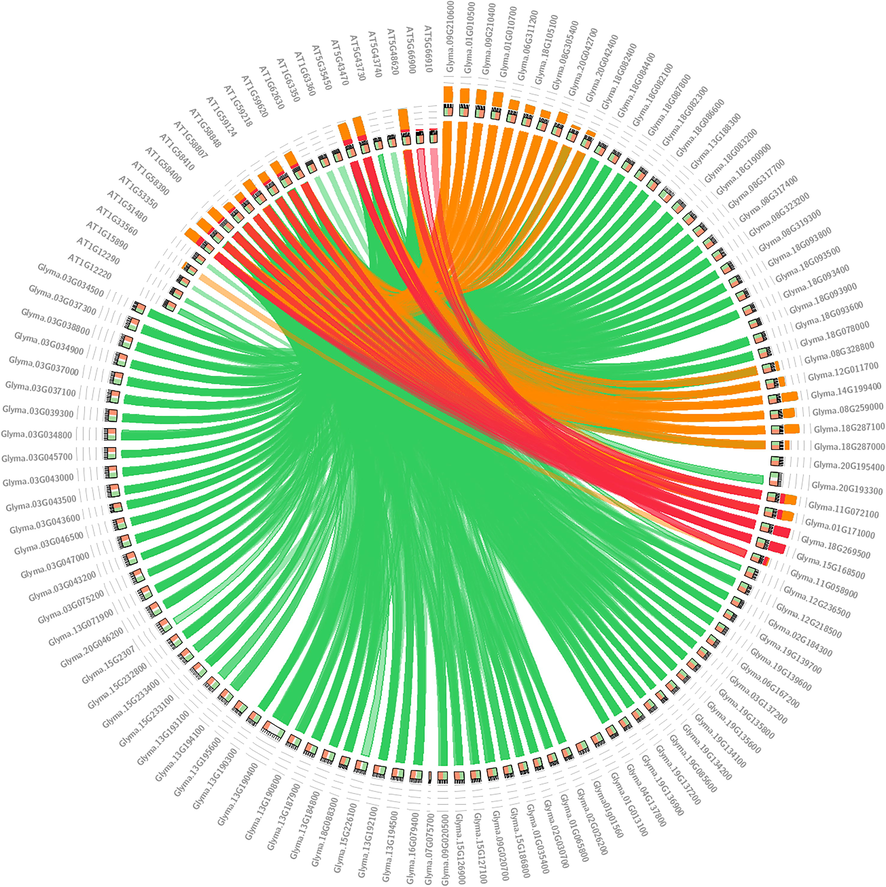
Homologous identification of the NB-ARC genes family of soybean and Arabidopsis thaliana synteny analysis was performed using the Circoletto online tool, which was practiced using the strict E-value of 1x10-50 and BLOSSUM matrix.
3.4 Insilico expression analysis
The expressions of 86 (NB-ARC) genes were investigated in root tissues under biotic stress conditions by Lanubile et al. (2015). Differential expression of these (NB-ARC) genes in root tissues under biotic stress was revealed by heat map-based expression profiles (Fig. 5). Heat map was divided into non-pathogenic Oxysporum isolate F036, and pathogenic F. oxysporum isolates FO40 at 72- and 92-hours post-inoculation (hpi). In the analysis, a significant expression variation was observed in the spectrum of highly pathogenic to non-pathogenic isolates collected from F. oxysporum that was collected from roots. The expression-based hierarchical clustering of genes was presented to show various gene clusters. The normalized gene expression in each group to the expression levels from dark green (downregulated) to dark red (upregulated) (Fig. 5).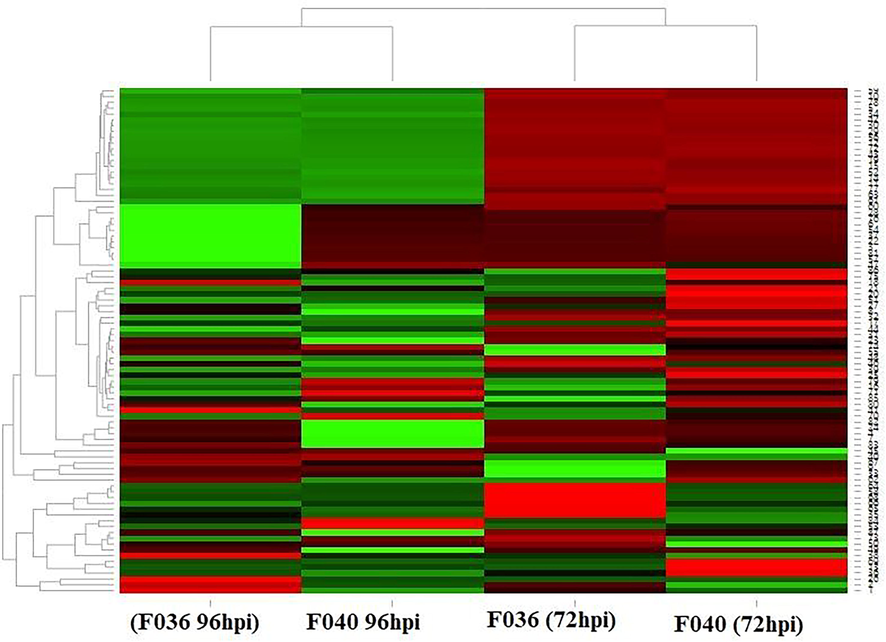
Heat map of 86 genes (NB-ARC) inoculated with non-pathogenic Oxysporum isolate F036 and pathogenic F. oxysporum isolates FO40 hours post-inoculation (hpi). The expression-based hierarchical clustering of genes was presented to show various gene clusters (Supplementary Table 2). Down-regulated genes are shown in green, and upregulated are shown in red, with the color intensity corresponding to the degree of change.
Overall, 68 orthologous gene pairs were identified between soybean and A. thaliana. A detailed analysis helped us classify expression data for the gene’s NB-ARC (86 soybean) gene expression (Supplementary Table 2). The expression of non-pathogenic Oxysporum F036 isolates showed (NB-ARC) gene downregulation relative to pathogenic F040 Oxysporum isolates at 72 h post-inoculation (hpi). Conversely, the pathogenic oxysporum isolates (F040) were recorded more upregulated (NB-ARC) gene expression (Fig. 5). Similarly, the expression pattern was differently recorded at 90 hpi as compared to the 72 hpi level. The more significant number of downregulated genes was recorded at 90 hpi under both pathogenic and non-pathogenic fungal isolates (Fig. 5).
4 Discussion
R genes are a crucial element of the gene interaction between biotrophic bacteria, fungi, and other plants, and they are also used to control resistance to bacterial invasion (Flor, 1955). The fungal genome sequences speed up the process for identifying more avirulence (AVR) genes in plant pathogenic fungi and infecting essential agriculture crops. As single AVR genes are characterized by their R allele, AVR and R gene interaction have become more complex (Petit-Houdenot and Fudal, 2017). Pathogens can become virulent by developing their AVR gene repertoire under the selection pressure of R genes (Guttman et al., 2014). The main identified R proteins are intercellular nucleotide-binding and leucine-rich repeat receptors (NLR). In the sense of a reciprocal transition between invader and host, other studies have suggested the 'zigzag model' to explain plants' resistance mechanism (Jones et al., 2006). Pathogenic molecular pattern-triggered immunity (PTI) is the first step of plant defense, whereby the immune system of the plant identifies a wide range of pathogenic agents with keeping molecular patterns that provide non-host resistance. In the second step, effector-caused immunity (ETI) is observed by the type III secretion system (TTSS), injecting into plant cells. ETI typically contributes to an intensified PTI reaction, which is also called the (HR). Among the known types of R-protein, those containing an NBS-LRR are the most common (Dangl and Jones, 2001). In several monocot and dicot species, including Arabidopsis (Meyers et al., 2003). Genome wide association-based identification provides a closer look into gene structure and conserved motifs lends credibility classification system. Furthermore, evolutionary pattern can be seen in gene expression analyses and subcellular localization such type of study (Ayaz et al., 2021). The similar identification, characterization, and functional validation of the expression study of using genome-wide association were also confirmed in legume crop (Waqas et al., 2019). In comparison with other crop species, soybean recorded 103 CC-NBS-LRR genes, which differs from that of other crop species, such as 149 in Arabidopsis, 315 in cotton (Shi et al., 2018), 148 in common bean (Wu et al., 2017), 29 in orchards (Xue et al., 2020), and 104 in chickpea (Sharma et al., 2014). From this comparison, we can infer that the number of NB-ARC encoding genes does not appear to be proportional to the genome size of the individual plant species.
The classification based on the phylogenetic tree followed the same pattern as in other crop species. The CC-NBS-LRR characterization in terms of the intron/exon distribution and conserved domains analysis results revealed that the conserved domain and genetic structure were present among the same group members. The TNL genes were at the predicted boundaries of the encoded protein domain TIR, NBS, and LRR, which are indicatives of the production of a modular protein comprising separate structural units with different functions. It was also suggested that a specific structure was achieved with NB-ARC-LRR would help with the trans CC domain. However, the Cis site's CC domain would help for cross-domain interaction for autoactivation (Rairdan et al., 2008).
The number of CC-NBS-LRR exon/introns ranged from one to seven, which corresponds to the gene structure of most NB-ARC-LRR genes in other plant species, such as chickpea (Sharma et al., 2014). Similarly, different intron positions related to the CC terminal were also reported in crop plants, such as Arabidopsis (Meyers et al., 2003) and the N gene in tobacco (Whitham et al., 1994). Like a shred of supporting evidence, structural diversity between exons and introns is considered a valuable tool for the phylogenetic grouping of these genes. Moreover, diversity is a significant part of gene families' evolution, development, diversification, and neo-functionalization (Han et al., 2016). Additional introns were also reported in Arabidopsis for both encoded LRR, and non-LRR-CC-terminus domains were present at the 3′ ends of the TNL genes. In some species, such intron-less genes have been recorded (Ross et al., 2007), which may be caused by intron losses during growth. The ancient fusion of independent genes that encoded proteins may represent the R gene configuration. CNL genes are more ancient and have lost the modular gene structure but may have been stable at the modular protein activity. The demonstration that the domains of the potato CNL protein Rx will work in trans to generate the hypersensitive response phenotype is confirmed by the distinct functions of the different domains when either the CC or the LRR is expressed from distinct genes (Moffett et al., 2002). The phylogenetic tree made the grouping based on gene clusters, i.e., TIR and CC motif, was also recorded (Zhou et al., 2004). Both groups are involved with pathogen identification but vary in their signaling pathway and amino acid sequences (Meyers et al., 2003). In the ideogram, many NB-ARC genes of soybean found best hit orthologous in the A. thaliana representing sequence conservation up to 80% (red in Figure). Zhang et al. (2019) also used synteny analysis to determine the synteny relationship. They suggested that R genes are essential to figuring out novel resistance traits among the two-genome data that have been functionally mapped are often found in tandem duplication (TDs), and their syntenic orthologous are strongly conflicting. The comparative synteny analysis results among soybean and Arabidopsis thaliana may deduce the NB-ARC gene role, as they have been presented with AtNB-ARC in an orthologous relation. The orthologous gene pairs usually depend on the species diversity (Blanc et al., 2004). In contrast, the TDs function played a significant role in expanding the NB-ARC family in other crop plants (Zhang et al., 2019). Furthermore, the purifying selection removed the harmful effect of alleles during the selection process (Biswas and Akey, 2006). Thus, it suggests that the critical nucleotide sequences in NB-ARC should be preserved to play an essential role in the survival of plants.
As a model plant, significant efforts have been made to characterize A. thaliana genes functionally. The AtNB-ARC was thus defined and functionally characterized. The resistance genes such as Glyma.09G020500.1 are significant for disease resistance in soybean and reported for systemic gained resistance (SAR). Similarly, another gene (Glyma.09G020700.1) that is important for defense response against disease resistance was also recorded significant in soybean, respectively (Smallwood et al., 2018). The transcriptome and expression data analysis results predicted that the similar gene Glyma.09G020500.1 was upregulated when FO36 (72hpi) was applied, while Glyma.09G020700.1 was upregulated when FO36 (96hpi) and F040 (96hpi) were applied respectively in soybean and functionally validate (Yang et al., 2008). The discovery of AtNB-ARC orthologs in soybean will aid in the functional validation of their roles in the plant. The NB-ARC can then be used for functional genomics in soybean and biotic stress breeding programs.
5 Conclusions
In summary, 103 NB-ARC non-redundant genes were identified in soybean as legumes in the present study. Their classification, gene structure, and conserved domain characterization; and comparative phylogenetic analyses propose conservation among NB-ARC groups of the plant species. In this response, many well-known defense genes were triggered more strongly. Furthermore, most of the genes were upregulated in stress situations, implying that they play their role in the mediation of stress responses in soybeans. These studies help to speed up the functional analysis of NB-ARC under biotic stress. Overall, the candidate CC-ARC genes can be used in the laboratory studies in the future to elaborate gene function against stress breeding program.
Funding
The Deanship of Scientific Research funded This research at King Saud University, grant number research group NO. RG-1441-513.
CRediT authorship contribution statement
Muhammad Afzal: Conceptualization, Methodology, Software, Data curation, Writing – original draft. Salem S. Alghamdi: Writing – review & editing. Hira Nawaz: Conceptualization, Writing – review & editing. Hussein H. Migdadi: Writing – original draft, Writing – review & editing. Muhammad Altaf: Methodology, Software. Ehab El-Harty: Data curation. Suleiman A. Al-Fifi: Writing – review & editing. Muhammad Sohaib: Writing – review & editing.
Acknowledgements
This work was supported by Grants from Deanship of Scientific Research at King Saud University through research group NO. RG-1441-513.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol. Plant Microbe. 2003;16(9):817-826.
- [CrossRef] [Google Scholar]
- Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem.. 2021;161:1-11.
- [Google Scholar]
- MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res.. 2009;37:W202-W208.
- [Google Scholar]
- MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res.. 2006;34:W369-W373.
- [Google Scholar]
- Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667-1678.
- [Google Scholar]
- A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Ann. Review Plant Biol.. 2009;60:379-406.
- [Google Scholar]
- Chen, Z., Vu, J. L., Vu, B. L., Buitink, J., Leprince, O., & Verdier, J. 2021. Genome-wide association studies of seed performance traits in response to heat stress in Medicago truncatula uncover MIEL1 as a regulator of seed germination plasticity. Frontiers in plant science, 12.
- Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124(4):803-814.
- [Google Scholar]
- Plant pathogens and integrated defense responses to infection. Nat.. 2001;411:826-833.
- [Google Scholar]
- Host-parasite interactions in flax rust-its genetics and other implications. Phytopathol.. 1955;45:680-685.
- [Google Scholar]
- Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell Proteomics. 2013;12:3690-3703.
- [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A.2005. Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook, Springer:571-607.
- Dynamic evolution of NBS–LRR genes in bread wheat and its progenitors. Mol. Genet. Genom.. 2015;290:727-738.
- [Google Scholar]
- Microbial genome-enabled insights into plant–microorganism interactions. Nat. Rev. Genet.. 2014;15:797-813.
- [Google Scholar]
- Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci.. 2016;17:161.
- [Google Scholar]
- GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296-1297.
- [Google Scholar]
- The rapid generation of mutation data matrices from protein sequences. Bioinfor.. 1992;8:275-282.
- [Google Scholar]
- RPW8 and resistance to powdery mildew pathogens in natural populations of Arabidopsis lyrata. New Phytol.. 2009;182:984-993.
- [Google Scholar]
- Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol.. 2012;12(1):1-13.
- [Google Scholar]
- Identification of novel recessive gene xa44 (t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet.. 2018;131(12):2733-2743.
- [Google Scholar]
- Knudsen, B., Knudsen, T., Flensborg, M., Sandmann, H., Heltzen, M., Andersen, A., Dickenson, M., Bardram, J., Steffensen, P., Mønsted, S. 2011. Clc sequence viewer. A/S Cb, version 6.
- Künstler, A., Bacsó, R., Gullner, G., Hafez, Y.M., Király, L. 2016. Staying alive–is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant P. 93, 75-84.
- Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom.. 2015;16:1-14.
- [Google Scholar]
- Transcriptional and posttranscriptional regulation of the tomato leaf mould disease resistance gene Cf-9. Biochem. Bioph. Res. Co.. 2016;470:163-167.
- [Google Scholar]
- Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE. 2012;7:e34775
- [Google Scholar]
- Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Micro.. 2011;9(3):187-199.
- [Google Scholar]
- Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci.. 2013;14(4):7302-7326.
- [Google Scholar]
- Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. The Plant J.. 1999;20(3):317-332.
- [Google Scholar]
- Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell. 2003;15:809-834.
- [Google Scholar]
- Interaction between domains of a plant NBS–LRR protein in disease resistance-related cell death. The EMBO J.. 2002;21:4511-4519.
- [Google Scholar]
- Plant innate immunity: an updated insight into defense mechanism. J. Biosci.. 2013;38:433-449.
- [Google Scholar]
- Histopathological features of infections caused by Fusarium oxysporum and F. solani in purple passionfruit plants (Passiflora edulis Sims) Summa Phytopathologica.. 2014;40:134-140.
- [Google Scholar]
- Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol.. 2000;50:203-213.
- [Google Scholar]
- Functional markers based molecular characterization and cloning of resistance gene analogs encoding NBS-LRR disease resistance proteins in finger millet (Eleusine coracana) Mol. Biol. Rep.. 2011;38(5):3427-3436.
- [Google Scholar]
- Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci.. 2017;8:1072.
- [Google Scholar]
- The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20(3):739-751.
- [Google Scholar]
- The WRKY gene family in rice (Oryza sativa) J. Integr. Plant Biol.. 2007;2007(49):827-842.
- [Google Scholar]
- towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res.. 2012;41:D1167-D1171.
- [Google Scholar]
- Genome-wide identification and tissue-specific expression analysis of UDP-glycosyltransferases genes confirm their abundance in Cicer arietinum (Chickpea) genome. PLoS ONE. 2014;9(10):e109715.
- [CrossRef] [Google Scholar]
- Extraction and characterization of microcrystalline cellulose from waste cotton fabrics via hydrothermal method. Waste Manag.. 2018;82:139-146.
- [Google Scholar]
- Genome-wide distribution, organisation and functional characterization of disease resistance and defence response genes across rice species. PLoS ONE. 2015;10:e0125964
- [Google Scholar]
- Oxidative stress in autoimmune rheumatic diseases. Free Radical Bio. Med.. 2018;125:3-14.
- [Google Scholar]
- Evolution of the rice Xa21 disease resistance gene family. Plant Cell. 1997;9(8):1279-1287.
- [Google Scholar]
- Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.. 2013;30:2725-2729.
- [Google Scholar]
- Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp. Funct. Genomics. 2012;2012:1-12.
- [Google Scholar]
- MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered.. 2004;93:77-78.
- [Google Scholar]
- Analysis of TIR-and non-TIR-NBS-LRR disease resistance gene analogous in pepper: characterization, genetic variation, functional divergence, and expression patterns. BMC genom.. 2012;13(1)
- [CrossRef] [Google Scholar]
- The rpg4-mediated resistance to wheat stem rust (Puccinia graminis) in barley (Hordeum vulgare) requires Rpg5, a second NBS-LRR gene, and an actin depolymerization factor. Mol. Plant Microbe Interact.. 2013;26:407-418.
- [Google Scholar]
- Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum L.) reveal their role in abiotic stress-responses. Genes genom.. 2019;41(4):467-481.
- [Google Scholar]
- The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78(6):1101-1115.
- [Google Scholar]
- Genome-wide association study identifies NBS-LRR-encoding genes related with anthracnose and common bacterial blight in the common bean. Front. Plant Sci.. 2017;8:1398.
- [Google Scholar]
- Genome-wide analysis of the nucleotide binding site leucine-rich repeat genes of four orchids revealed extremely low numbers of disease resistance genes. Front. Genet.. 2020;10
- [Google Scholar]
- Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics. 2014;15:3.
- [Google Scholar]
- Comparative genomics analysis in grass species reveals two distinct evolutionary strategies adopted by R genes. Sci. Rep.. 2019;9:1-10.
- [Google Scholar]
- The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol.. 2005;5:1-12.
- [Google Scholar]
- Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genom.. 2004;271:402-415.
- [Google Scholar]
- R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U S A.. 2010;107(43):18735-18740.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary Materials: Table S1: The conserved domain and each gene's architecture presenting the location of the domain of interest in each gene. Table S2: Expression data for the gene’s NB-ARC (86 G. max). Figure S1: Conserved domain analysis grouping. Table S3: Hit-data-conserved domain analysis.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101758.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







