Translate this page into:
Genome-wide identification and analysis of B-box zinc finger gene family in sugarcane (Saccharum officinarum)
⁎Corresponding authors. sengar2022@gmail.com (R.S. Sengar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective and methods
B-box-containing proteins are a kind of zinc finger (ZF) transcript protein that contains 1 otherwise 2 B-box domains even, in some cases, an additional CCT motif that is crucial for plant growth development and controlling biotic and abiotic stress. There has yet to be a comprehensive identification of the BBX genes in sugarcane by using bioinformatics tools/softwares.
Results
Considered in the present research work, 21 Sugarcane genes were discovered by comprehensive bioinformatics analysis viz the phylogeny relationships, structures of genes, locations of chromosomes, gene duplication, subcellular localization, homology and codon analysis were analysed. Relying on phylogeny as well as domain constitution analysis, the identified genes must be divided into five clades. Eight out of ten Sugarcane chromosomes had an uneven distribution of Shbbxs. The significant proportion of sugarcane BBX proteins are expected to be found in nuclei and extracellular spaces.
Conclusion
Our findings will serve as a foundation for subsequent functional findings of the BBX gene family, highlighting its roles in sugarcane. The findings will aid our understanding in the B-box gene family's complexity towards the abiotic stress responses, as well as provide suggestions for the imminent functional annotation of specific genes in sugarcane.
Keywords
BBX Gene
Sugarcane
Phylogeny
Chromosomal localization
Gene Structures
- TF
-
Transcription factors
- ZF
-
Zinc finger
- A. thaliana
-
Arabidopsis thaliana
- GO
-
Gene ontology
- PDB
-
Protein data base
Abbreviations
1 Introduction
The primary regulatory proteins that contribute in higher plant life cycles by activating or inhibiting gene transcription are known as Transcription factors (TFs) (Zou et al., 2018). They usually have four distinct domains: “ site for DNA binding, activation domain for transcription, site for oligomerization and nuclear localization signal (NLS)”. These domains collaborate to govern some specialized actions like “physio-biochemical” activities at the same time (J. Ma et al., 2021). In recent years, the B-BOX zinc finger protein family has gotten a lot of attention. Because of plant genome sequencing, genome-wide studies of the Sugarcane “BBX gene” family are now feasible. zinc finger TFs are a class of B- box protein that comprise one or two B-box domains with distinct 3D structures which may be stabilized by the binding of Zn ion (Huang et al., 2012). Many researchers believe that the BBX protein plays a significant role in DNA binding as well as in protein–protein interactions. However many in plants BBX proteins have one or more than one B-box domains located near the amino terminus, and some have a CCT “(CONSTANS, CO-like, and TIMING OF CAB1)” domain located close to the carboxy terminus (Khanna et al., 2009). The researcher Khanna et al., 2009 re-identified and renamed approximately 32 BBX genes in A. thaliana. According to the literature, B-box proteins are classified into five subclades determined by the number of B-BOX domains and the existence of the CCT domains like (CO, CO-like, and TOC1) (Gangappa and Botto, 2014; Yang et al., 2014).
A family of B-box TFs engaged in circadian signalling which detect the light in A. thaliana was discovered (Khanna et al., 2009). The first A. thaliana BBX gene to be studied was CONSTANS (CO)/AtBBX1, which regulates time of flowering by activating the gene Flowering Locus T (FT) (Wei et al., 2020). Some studies have found that the BBXs gene plays a role in abiotic stress response, particularly in drought. Several stress-responsive Cis elements, including ABRE (ABA-responsive element), HSE (heat shock element), and MBS, were found in the majority of rice and tomato BBX promoters, implying that these genes may act in different abiotic stress responsive activities (Huang et al., 2012; J. Ma et al., 2021). In A. thaliana, low temperatures induce the transcription of several AtBBXs (Zou et al., 2018) Abiotic stress responses include “BlCOL13 in Betula luminifera, Ghd7 in rice (Oryza sativa), Ma-COL1 in banana (Musa sp.) and SsBBX24 in potato (Solanum tuberosum)”(Chen et al., 2012; Kie bowicz-Matuk et al., 2014; Weng et al., 2014).Overexpression of the CmBBX24 gene results in tolerance to freezing and drought, but in the absence of this gene, tolerance activities in chrysanthemums slow down (Yang et al., 2014). Similarly the expression of VvBBX32 gene in berry upsurges cold tolerance (Wei et al., 2020). Different species have been used to study the BBX gene family such as “A. thaliana, Oryza sativa, Solanum lycopersicum, pear (Pyrus bretschneideri Rehd) and Solanum tuberosum” (Gangappa and Botto, 2014; Huang et al., 2012; Khanna et al., 2009; J. Ma et al., 2021; Zou et al., 2018).
Sugarcane (Saccharum spp. hybrid) is a major crop grown around the world, primarily for its high content of sucrose and as a source of biomass for the production of biofuel (Li et al., 2020). Sugarcane accounts for 75% of global sugar output and 40% of global ethanol output, respectively. Environmental pressures that harm sugarcane can have a large economic impact. Drought stress reduces sugarcane production by 30 to 70% during growth, while sucrose formation and recovery are reduced by 5% (Manoj et al., 2019). This study identified 21 non-redundant Shbbx gene family members responsible for drought stress in sugarcane. Following that, the detailed gene structures and phylogenetic relationships, as well as chromosomal distribution and structural domains, were studied. This is the first study to identify Shbbxs to our knowledge. Our findings will lay the groundwork for identifying the BBX family in Saccharum spp. in gene-specific marker development and functional genomic studies that will benefit sugarcane breeding programmes for the tolerance of different kinds of abiotic stress.
2 Materials and methods
2.1 Identification and retrieval of BBX genes
The family of BBX gene in A. thaliana was previously conveyed by Khanna et al., in 2009 in his paper “The Arabidopsis B-Box Zinc Finger Family”. To identified homology in sugarcane we extracted all the protein sequences of A. thaliana BBX gene family from the phytozome database (https://phytozome.jgi.doe.gov/) and picked AtBBX22 (AT1G78600) as a queries sequence for protein BLAST (BLASTP) search with an e-value threshold of < 1e-5 in the Sugarcane genome hub database (https://Sugarcane-genome.cirad.fr/). Following that, the identified hits were screened by using the different online available tools to ensure authenticity and the occurrence of the target domains: SMART (https://smart.embl-heidelberg.de/), Inter Pro Scan programme (https://www.ebi.ac.uk/interpro/), and Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/). The ExPASyProtParam tool (https://web.expasy.org/protparam/) was used to calculate the isoelectric point (PI), molecular weight (Da), and grand average of hydropathicity (GRAVY) of each Shbbx.
2.2 Phylogenetic analysis and sequence alignment
For Phylogenetic analysis between sugarcane and A. thaliana full length protein sequences of BBX genes was selected for better understanding between their evolutionary relationship. Protein sequences from both species were imported into MEGA X (Version 10.2.4) to achieve multiple sequence alignment with the default parameters in order to construct the phylogenetic tree. The obtained alignment data file was used as input for the construction of a phylogenetic tree in MEGA X (Version 10.2.4) using the neighbor-joining algorithm with a boot-strap value of 1000, but before that we compute pairwise distance of alignment file. The phylogenetic tree was then visualised with the ITOL (version 6.4.1) software. The MEME suite (https://meme.nbcr.net/meme/cgi-bin/meme.cgi) tool was used to create the sugarcane B-box domain motif logos.
2.3 Chromosomal location, gene structure, and duplication analysis
The sugarcane Genome hub (https://sugarcane-genome.cirad.fr/) platform was used to determine the chromosomal location of all the identified Shbbx genes in this study. Genes were mapped to individual chromosomes using map chart software (version 2.32) (Huang et al., 2012; Voorrips, 2002). Furthermore, the genomic sequences were aligned with their corresponding coding sequences in GSDS software (gene structure display server, https://gsds.cbi.pku.edu.cn) to determine the exon–intron organisation of Shbbx genes. Tandem duplications of adjacent genes were also identified if their similarities exceeded 70%. The ratio of Ka/Ks (evolutionary selection pressure) was used to express positive selection (with a ratio greater than 1), negative or stabilising selection (Ka/Ks less than 1), and neutral selection (Ka/Ks equal to 1), respectively, to interpret the evolutionary relationship of Shbbx genes. TBtools software (v1.09854) calculated the Ka (nonsynonymous substitutions) and Ks (synonymous substitutions) values.
2.4 Subcellular localization analysis
WoLF PSORT (https://www.genscript.com/psort/wolf_ psort.html) tool was used to identified the sugarcane BBX proteins location in different organelles. Furthermore, the results of the analysis were represented by a heatmap created with the TBtools software (v1.09854).
2.5 Codon analysis
The cDNA sequences of identified Shbbx genes were retrieved from the Sugarcane genome hub database (https://Sugarcane-genome.cirad.fr/) and stop codons were removed from each gene sequence. The multiple sequence alignment of the Shbbx genes was carried out by ClustalW in MEGA X (Version 10.2.4). After that the multiple sequence alignment result file served as input for the codon model selection analysis in classical datamonkey (https://classic.datamonkey.org/) online tool.
2.6 Gene ontology (GO) annotation
The Blast2GO server was used to perform GO annotation of Shbbx genes (Zou et al., 2018). The GO terms were divided into three categories: biological process, molecular function and cellular component. WEGO (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) was used to plot GO classifications.
2.7 Protein structure analysis by homology modelling
The 3D structure of the Shbbx protein was predicted to probe the possible functions of the protein. 3D structures of Shbbx proteins were created by using homology modelling technique and the online available template-based program SWISS-MODEL (http://swissmodel.expasy.org) (R. Ma et al., 2021).
2.8 Structure validation
For the validation of protein structures, SWISS MODEL generated PDB file of the best models was used to create the Ramachandran plots using RAMPAGE server (https://zlab.umassmed.edu/bu/rama/) and assessing them under three factors as “favoured, allowed and outlier regions of amino acid residues”(das et al., 2019).
2.9 Energy minimization of the predicted structures
Energy minimization was applied to predicted and evaluated structures in order to achieve the lowermost energy conformation by S P B D V (SWISS PDBVersion 4.1.0).
2.10 Molecular dynamics simulation
The model with the lowest energy was considered for molecular dynamics simulations. iMODS (https://imods.iqfr.csic.es/)was used to understand the best model stability in terms of the torsion angles between molecules.
3 Results
3.1 Sugarcane BBX gene identification
BLAST searches were used to characterise the BBX gene family in sugarcane genome-wide analysis. The BBX genes discovered through similarity searches were confirmed the presence of number of B-box domain by the use of two different software’s are SMART (Simple Modular Architecture Research Tool) and Pfam databases. A local BLAST was performed in the sugarcane genome hub database to determine how many genes in the sugarcane genome encode BBX members with and without the CCT domain out of 21 Shbbxs genes. Nomenclature and consistency were maintained by naming these putative BBX genes as Shbbx1 to Shbbx21. Table 1 contains detailed information on 21 Shbbx genes, including “name of gene, length of protein, location of gene on chromosome, molecular weight, theoretical isoelectric point (pI) and GRAVY”. The 21 Shbbx proteins had varying molecular lengths and weights, with amino acid lengths ranging from 206 (Shbbx11) to 585 (Shbbx10). Shbbx had the smallest molecular weight (22.65 kDa), while Shbbx had the largest molecular weight (62.08 kDa). These Shbbx proteins' theoretical isoelectric points (pI) was ranged from 4.53 (Shbbx5) to 6.16. (Shbbx3). The hydrophilic nature of all Shbbx genes revealed that the gene's GRAVY value was less than zero (Table 1).WoLF PSORT predicted that the large number of Shbbx proteins would be present in the nucleus and extracellular space, but it also revealed that some Shbbx proteins show their presence in other subcellular compartments such as the chloroplast and cytoplasm (Table 1).
Transcript Id
Gene Name
Chromosome
Location Start- End
Strand
CDS (bp)
Protein Length
Protein Mol. Weight(kDa)
pI
GRAVY
No. of Intron/Exon
Subcellular Localization Seq
Sh_240N19
Shbbbx1
7
99983…100868
Forward
885
294
29.350
5.84
−0.049
0:01
Chloroplast, Extracellular
Sh_241P15_contig-1
Shbbx2
10
80078…81571
Reverse
1344
447
47.831
5.32
−0.52
1:02
Nuclear, Chloroplast
Sh_239A02_contig-2
Shbbx3
1
41070…42436
Forward
1257
418
45.917
6.16
−0.61
1:02
Nuclear
Sh_232B06
Shbbx4
10
90928…93703
Forward
1215
404
43.503
5.3
−0.454
3:04
Nuclear
Sh_225C07
Shbbx5
1
98249…105104
Forward
801
266
28.032
4.53
−0.456
1:02
Chloroplast
Sh_220L15
Shbbx6
1
84767…87301
Forward
1239
412
44.117
5.2
−0.604
3:04
Extracellular
Sh_246C13
Shbbx7
4
104138…106285
Reverse
867
288
29.655
5.24
−0.164
2:03
Nuclear
Sh_243P13
Shbbx8
10
55216…56736
Reverse
1371
456
48.922
5.26
−0.501
1:02
Nuclear, Chloroplast
Sh_220H07
Shbbx9
6
67395…68310
Reverse
729
242
25.618
5.15
−0.239
1:02
Cytoplasmic, Extracellular
Sh_211D19
Shbbx10
4
90955…94612
Reverse
1758
585
62.084
5.84
−0.566
3:04
Nuclear
Sh_240C06
Shbbx11
2
77033…80051
Forward
621
206
22.657
5.51
−0.714
4:05
Extracellular
Sh_212L22
Shbbx12
6
20440…21882
Reverse
768
255
27.401
5.63
−0.406
2:03
Extracellular, Nuclear
Sh_230M22
Shbbx13
4
4426…5222
Reverse
699
232
24.025
5.96
0.022
1:02
Nuclear
Sh_201E17
Shbbx14
3
90860…92325
Forward
1044
347
35.962
5.44
−0.199
2:03
Extracellular, Nuclear
Sh_237D16
Shbbx15
4
23980…24950
Forward
762
253
27.240
4.95
−0.261
2:03
Cytoplasmic, Extracellular
Sh_055F12
Shbbx16
10
16353…20853
Reverse
1107
368
39.598
5.23
−0.356
2:03
Extracellular, Nuclear
Sh_220I18
Shbbx17
7
33232…34126
Forward
894
297
29.619
5.71
−0.063
0:01
Nuclear
Sh_206L19
Shbbx18
8
70977…72667
Reverse
669
222
22.739
5.78
−0.205
0:01
Extracellular
Sh_239M04
Shbbx19
8
52840…54530
Forward
534
222
22.739
5.78
−0.205
5:06
Extracellular
Sh_250D18
Shbbx20
6
24053…25134
Forward
987
328
34.462
5.34
−0.304
1:02
Chloroplast, Cytoplasmic
Sh_224D12_contig-2
Shbbx21
10
161…4660
Forward
1107
368
39.672
5.32
−0.384
2:03
Extracellular, Nuclear
3.2 Protein sequence and phylogenetic analysis of the Shbbx gene family
Phylogenetic analysis was performed to understand the evolutionary relationship between the BBX genes of Sugarcane and A. thaliana. The phylogenetic analysis revealed 5 clades. The first, second, third, fourth, and fifth clade consists of 4, 2, 8, 9, and 23 genes respectively. Whereas 4 genes were not found in any clade. In the first clade, Shbbx20 showed association with 3 genes of A. thaliana. In contrast to the first clade, second clade genes belong only to Sugarcane. On the other hand, in the third clade, 4 genes of Sugarcane showed an evolutionary relationship with 4 genes of A. thaliana. Whereas, in clade 4, only 2 genes of Sugarcane i.e.., Shbbx6 and Shbbx4 were found. Maximum genes of Sugarcane i.e., 11genes were found in clade 5 (see Figs. 1,2).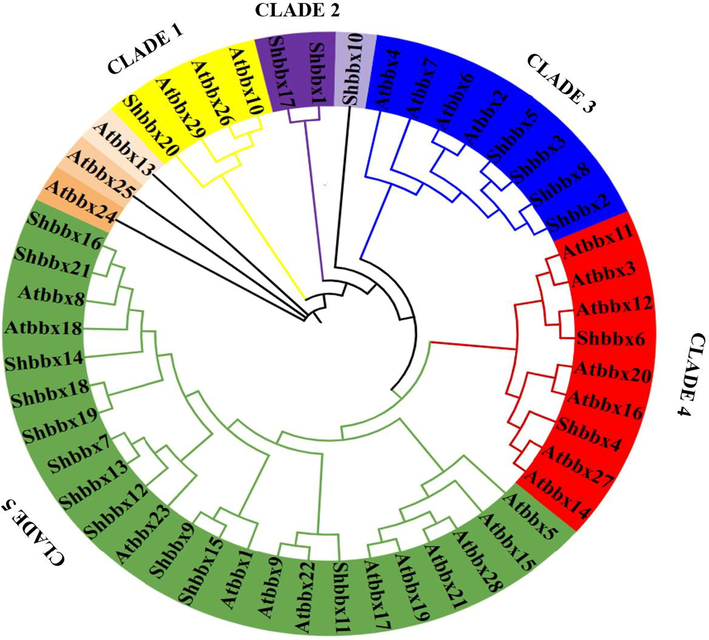
Sugarcane and A. thaliana BBXs phylogenetic relationships. A phylogenetic tree was constructed using BBXs among sugarcane and A. thaliana. The brown colour signifies AtBBXs in A. thaliana and the colour blue Shbbxs in Sugarcane. The sequence alignments and phylogenetic tree were used to classify the 21 Shbbxs.
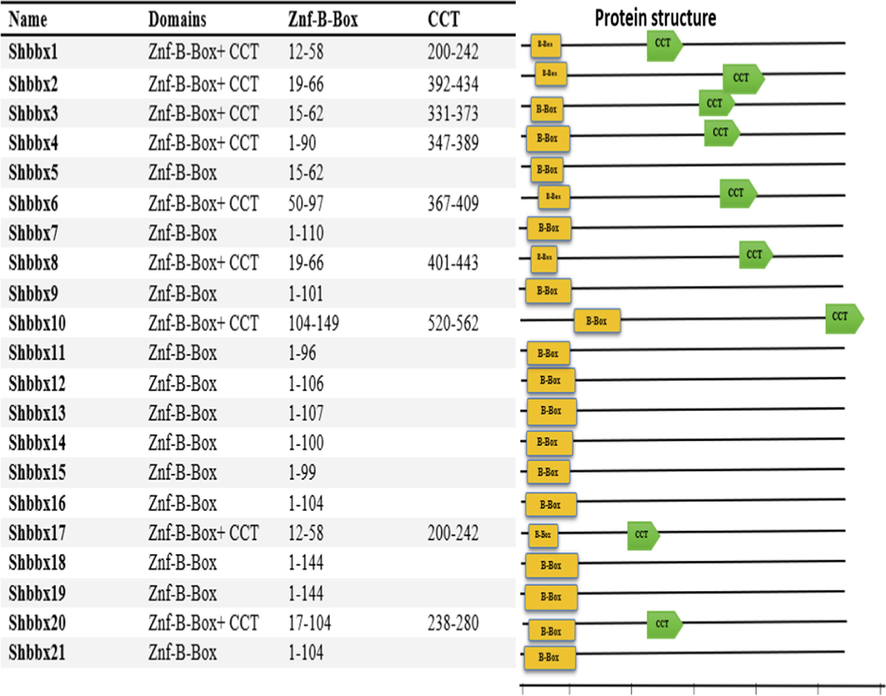
BBX proteins structure of sugarcane. Numbers In this figure show position of amino acid residues present in domains of sugarcane. The yellow rectangles signify B-BOX1domain and light green pentagons signify CCT domains in sugarcane. The bar scale represents 100 amino acids.
3.3 Conserved domain analysis
Motifs serve an important function in transcriptional regulation. To understand the diversity and similarity of the identified Shbbx genes, the MEME suite tools were used. Motifs are generally important because of their relationship with protein structure and function, as specific motifs regulate protein networks. We discovered 15 conserved protein motifs in this study (supplement file 1). According to our findings, motifs 1 and 6 were found in all 21 genes of the BBX gene family. In contrast, motifs 3 and 4 were found in 11 and 10 genes, respectively. Similarly, motifs 2 and 9 were found in 9 different genes. As a result, motif 1 and 6 may be the primary domains representing the BBX gene family (Fig. 3).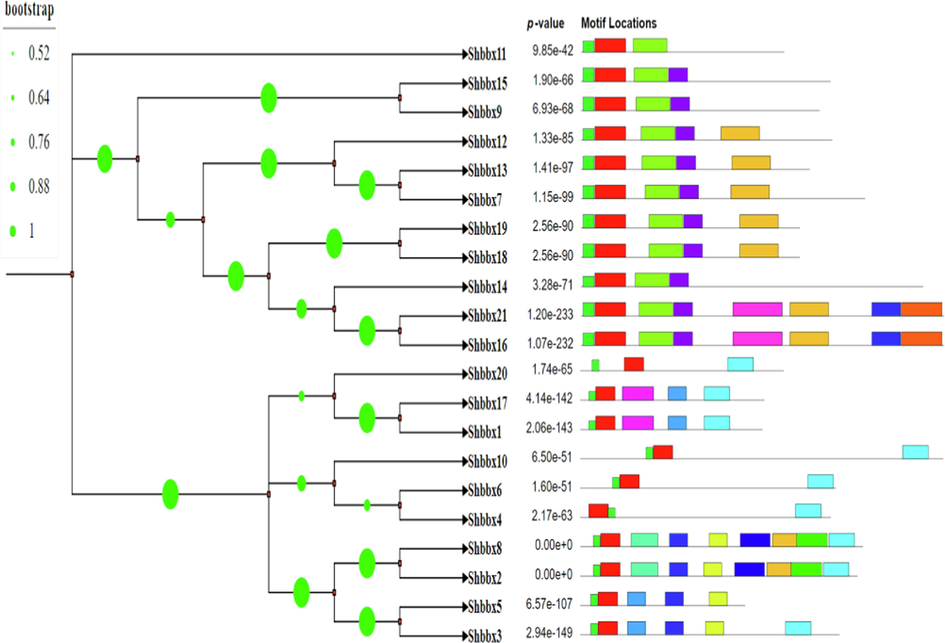
MEME online tools were used to create a schematic representation of the amino acid motifs of BBX proteins. On the left, you'll find a list of BBX gene family. The length of the corresponding BBX protein is signified by the black solid lines. The various coloured boxes represent various motifs and their positions in the BBX protein sequences.
3.4 Chromosomal Localization, gene duplication and gene structure analysis of Shbbxs
All of the Shbbx genes were mapped onto different sugarcane chromosomes to determine their genomic distribution. Gene distribution was found to be uneven. Excluding the chromosomes 5 and 9, the 21 Shbbx genes were found on all sugarcane chromosomes. The maximum number of genes is found on chromosome number 10, where five genes are found, followed by chromosome number 4, where four genes are found. On Chromosomes 1 and 6 three genes have found while on chromosomes 7 and 8 two genes. The minimum number of genes were mapped on chromosomes 2 and 3, which each had one gene. Fig. 4 depicts the location of the Shbbx genes on the sugarcane chromosomes.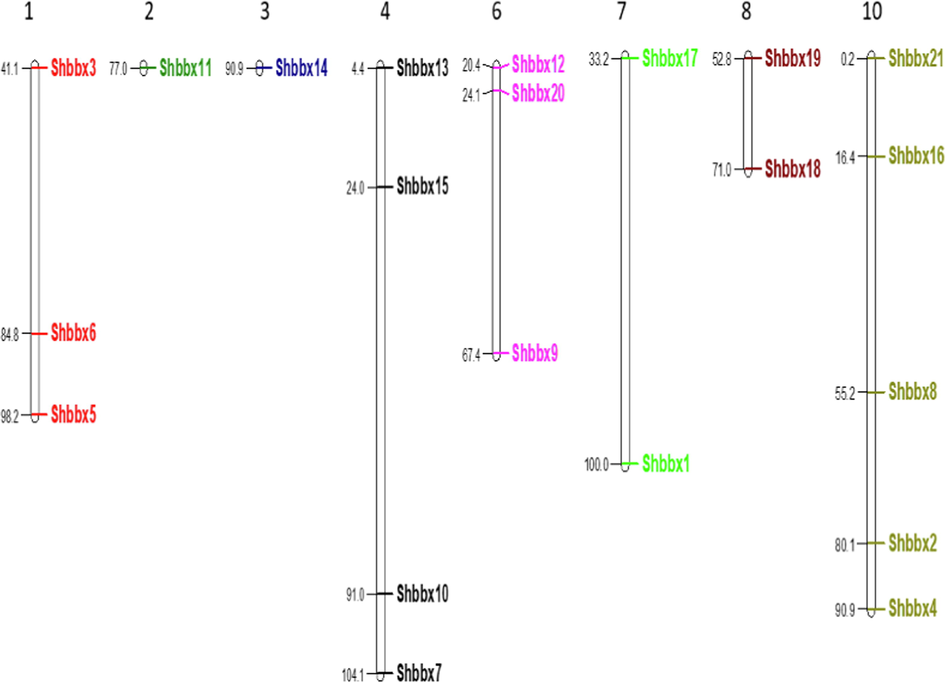
Sugarcane chromosomes contain BBX genes. The number of chromosomes (Chr 1-Chr 10) is designated at the top of each chromosome. The values following the BBX genes indicate their chromosomal location.
Tandem and segmental duplications gone up significantly during the evolution and expansion of the gene family. Tandem duplication typically results in gene clusters, whereas segmental duplication may result in dispersed family members. Within the sugarcane genome, hardly two duplicated segments (Shbbx4/Shbbx6 and Shbbx9/Shbbx15) in the Shbbx gene family were found. Then, on chromosomes 4, 7, 10, 1, and 8, six pairs of tandem duplications (Shbbx7 / Shbbx13, Shbbx1 / Shbbx17, Shbbx2 / Shbbx8, Shbbx16 / Shbbx21, Shbbx3/ Shbbx5 and Shbbx18 / Shbbx19) in the Shbbx gene family were discovered (Shbbx (Supplementary file 2). The Ka/Ks ratios of the 21 Sugarcane BBX gene pairs were computed depending on the BBX family's comparative sequence homology, among which 10 Sugarcane homologous pairs gene were discovered, and the Ks values have been used to predict the time of Sugarcane BBX gene duplication activities. We accomplished a Ka/Ks (evolutionary selection pressure) analysis on Sugarcane Shbbx homologous gene pairs and observed that Shbbx17/Shbbx1 had the lowest Ka/Ks ratio (0.1798), along with Shbbx5/Shbbx3 (0.2786), and Shbbx21/Shbbx16 had the highest ratio (9567550). Sugarcane BBX homologues showed a Ka/Ks ratio of less than one, indicating strong purifying selection during the development of the sugarcane BBX gene family (Supplementary file 3). The ks values derived from this analysis were used to calculate the time sugarcane took to mimic the developing BBX gene. According to the findings of this study (Maya), the replication time of the sugarcane B-box gene was mainly between 0 and 44 million years.
The intron/exon structure of the Shbbx gene were studied by matching cDNA sequences with corresponding genomic sequences for well understand their evolutionary relationships. As shown, the number of exons ranged from 1 to 6 (Fig. 5). Shbbx19 had the most exons (6), but Shbbx1, Shbbx17 and Shbbx18 only had one Shbbx exon. These data indicate that exon acquisition or loss occurred during the course of evolution.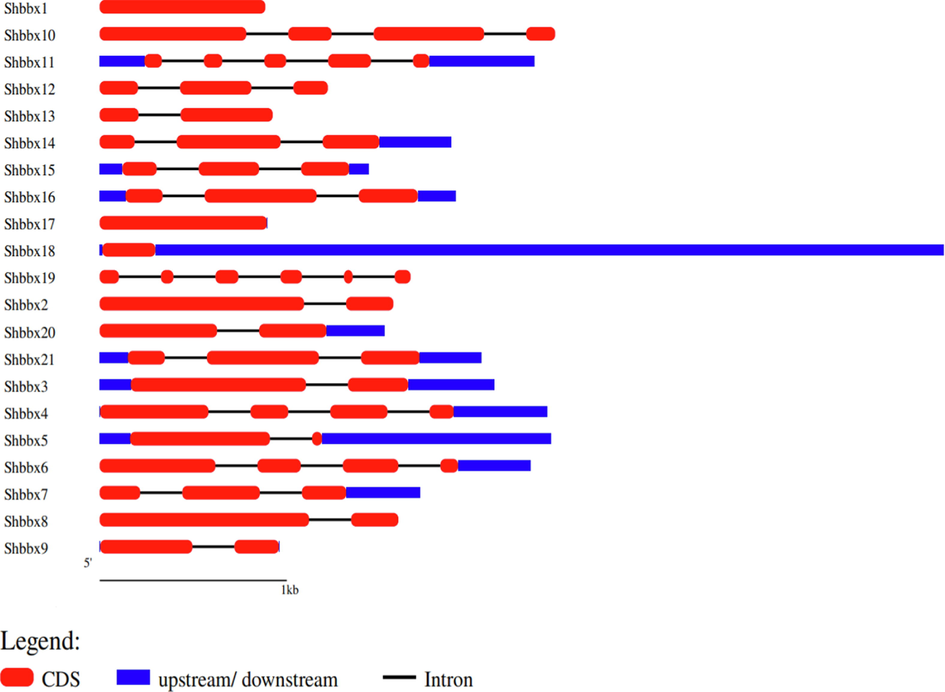
GSDS-generated gene structure of the sugarcane BBX family. The red colour block represents the coding sequence (CDS), the blue colour block represents the genes' upstream or downstream, and the black line represents the intron. The length of the DNA sequences is indicated by the scale bar.
3.5 Subcellular localization of sugarcane BBX protein
WoLF PSORT was used to predict sugarcane BBX protein subcellular localization (Table 1). The large number of Shbbx proteins were identified in the nucleus, with only a few found in another subcellular compartments like chloroplasts and cytoplasm. We used a heatmap to represent the subcellular localization of each gene, with the abundance of organelles in each gene represented by a graph scale ranging from 0 to 14 (Fig. 6). The colours red, green, and orange indicate high abundance, medium abundance, and low abundance, respectively. According to our findings, Shbbx4, Shbbx12, and Shbbx14 were abundant in the nucleus, whereas Shbbx2 was abundant in the chloroplast. Sugarcane has a medium to low abundance of the BBX gene in the remaining organelles.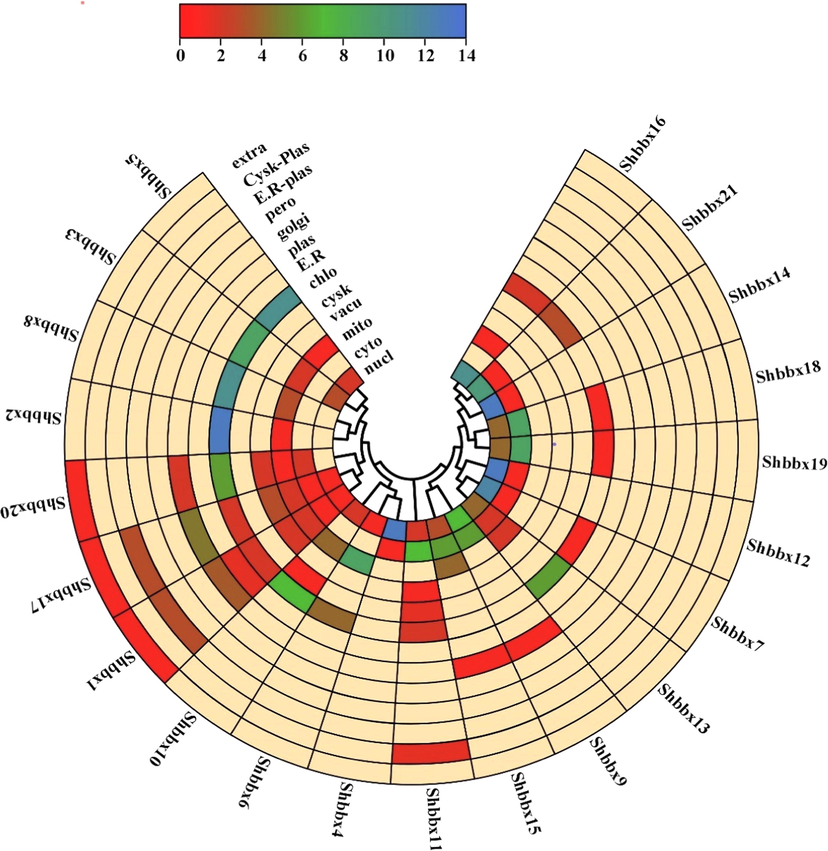
The BBX genes' subcellular localization. The heatmap was created using WoLF PSORT data, and the colour scale depicts the abundance of organelles. Higher levels of expression were shown in blue, while lower levels were shown in dark red.
3.6 Codon analysis of sugarcane BBX protein
To observe whether the Shbbx genes have been subjected to adaptive evolution, an investigation of variation in selective pressure was carried out. A codon-based MSA was evaluated for multiple models by using Datamonkey web server (https://classic.datamonkey.org/). In total, codon model selection procedure had examined 15052 models. The best model (log(L) = -19293.9, mBIC = 39638.65), has 4 rates (Supplementary file 4). This model provides an improvement of 377.44 log(L) and 697.55 mBIC points over a single rate model, where all non-synonymous substitutions occur at the same rate. According to the table (2) dN/dS ratio of 0.67 has highest rates in class (32), Stanfel class changes (15), polarity changes (14) and charge changes (11) (Supplementary file 5). show correlation between inferred rates and biochemical properties Fig. 7.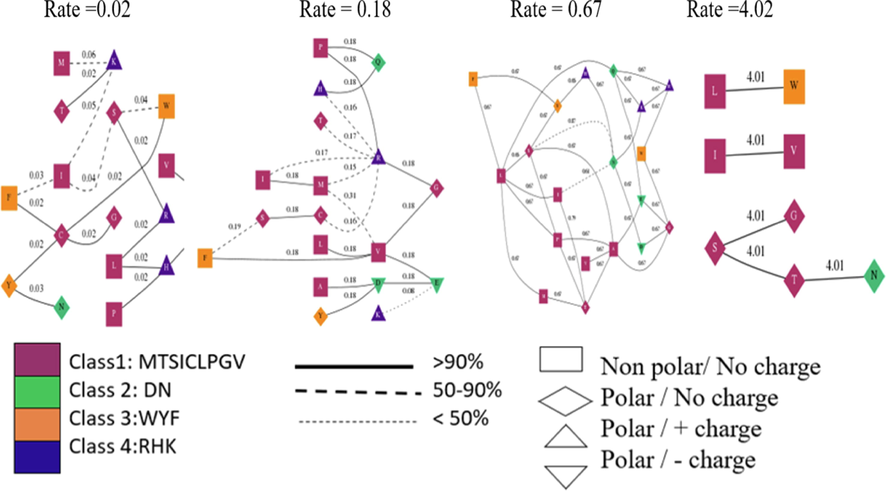
codon analysis of BBX gene family members in Sugarcane.
3.7 GO annotation analysis
To understand the function of BBX gene found in sugarcane, GO annotation was used. Based on the GO footnote analysis, 21 Shbbx gene family were distributed into 13 functional categories from three major ontologies are: cellular components, molecular function, and biological processes (Supplementary file 6). Cell parts and organelles were the most common in these categories. Examination of molecular function annotations revealed that the majority of Shbbx is involved in binding of DNA, along with chromatin binding. Examination of the cellular component annotations show that BBX proteins were found primarily in the nucleus.
3.8 Protein structure analysis by homology modelling
The tertiary structure of the protein was predicted using the Swiss model chosen to analyze the function of the protein. We selected Shbbx20 for subclass I, Shbbx17 for subclass II, tertiary structural homology using pair and multiple alignments. The 3D structures of the five subclasses showed different tertiary structure (Fig. 8). The Shbbx20 sequence comprises of 3 helices, 6 sheets and 6 amino acid residues particularly bind to zinc ions (C.65, C.62, C.85, C.82, C.34, C.74). The Shbbx17 sequence contained two helices and triple sheets and five amino acid residues that particularly bound to Zn ions (C.39, C.20, C.36, C.17, C.28). The Shbbx5 sequence comprised of two coils, two folded shells, and five amino acid residues particularly bound to Zn ions (C.23, C.40, C.43, C.20, C.32). The Shbbx4 sequence contained only four coils, six folded strands, and five zinc-binding amino acid residues (C.17, C.51, C.48, C.68, C.71). The Shbbx14 sequence comprised mainly of four helices, five folded sheets, and five amino acid residues linked by Zn ions (C.81, C.58, C.78, C.70, C.17).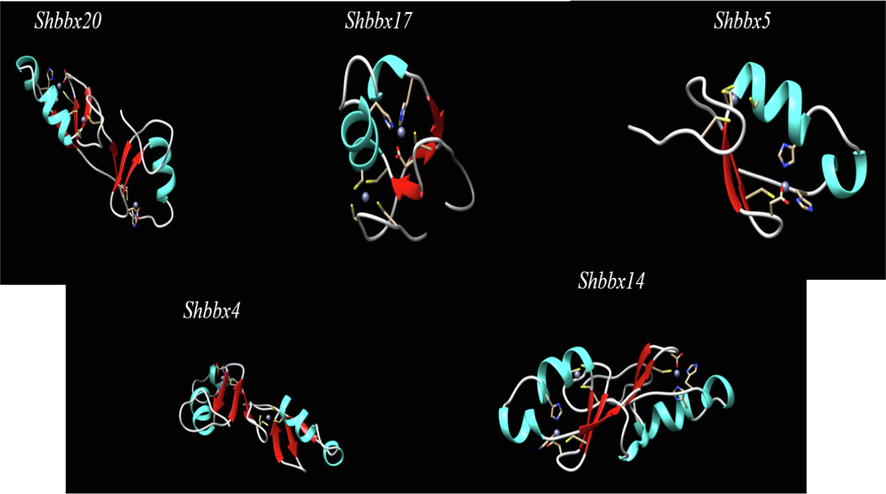
Five sugarcane BBX protein sequences in three dimensions. SWISS-MODEL software was used to create the models. The red areas represent -folds, while the bright blue areas represent -helices. Cysteines bound to a zinc ion are indicated by small yellow spikes surrounding a blue ball.
Ramachandran plot structure validation: To analyse the anticipated models, the PDB files for the Shbbx genes were obtained from the SWISS MODEL and put forward to the online database SAVES 6.0, which created the Ramachandran plot (Fig. 9). In this graph, the residues of amino acids were dispersed in 3 zones. Favored, allowed regions and generously allowed regions were the three distinct regions in (supplementary file 7).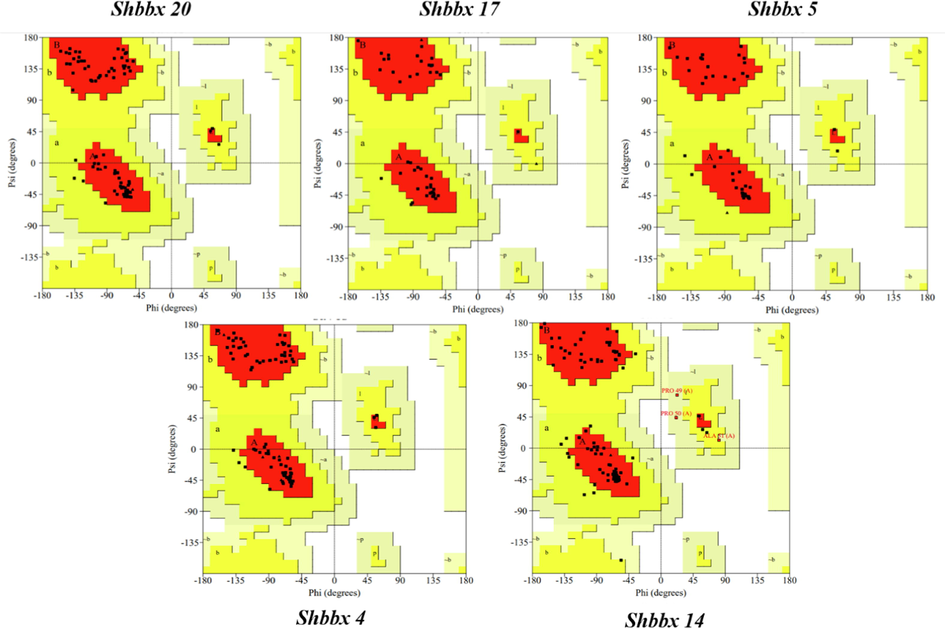
Ramachandran plots for different Shbbx genes.
Energy minimization: The energy minimizations of the predicted models were accomplished using the SPBDV9 software. The energy minimization values were compared to find the best and foremost predicted models. Shbbx 17 (E = -809.338KJ/mol) was the least energetic model shown in (supplementary file 8).
MD simulation: After comparing the energy minimization outcomes, we can see that the best predicted protein model (Swiss-Model) has the lowest value for minimization. As a result, the SWISS-MODEL was further interpreted for molecular dynamics simulation in order to examine the stability and physical activities utilizing the iMODS webserver's normal mode analysis (NMA). Even with significantly large macromolecules, iMODS enabled the exploration of such modes and produced plausible transition paths between two homologous structures. The unique internal coordinate formulation increases NMA's efficiency and broadens its applicability while maintaining stereochemistry implicitly.
Each normal mode's variance is inversely related to its eigenvalue. Individual (red) and cumulative (green) variances are represented by coloured bars (supplementary file 9 A). The elastic network model specifies which atom pairs are linked by springs. Each dot in the graph represents a spring between the corresponding atom pair. The stiffness of the dots is indicated by their colour; darker greys indicate stiffer springs and vice versa (supplementary file 9B). All above mentioned points denote the higher stability of the complex.
4 Discussion
Throughout their growth and development, plants are frequently confronted with a variety of unfavourable environmental conditions. A series of physiological and biochemical changes occur in response to these environmental changes. These changes necessitate a wide range of different genes, with transcription factors (TFs) playing a key role (Singh et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2005). There are “32 Arabidopsis members, 30 rice members, 29 tomato members, 30 potato members, 25 grapevine members, 64 apple members, 36 maize members, 24 sorghum members, 22 stiff brome members, 19 millet members, and 37 pear members” (Chu et al., 2016; Huang et al., 2012; Khanna et al., 2009; Liu et al., 2018; Shalmani et al., 2019; Talar et al., 2017; Zhang et al., 2021) They are all involved in flowering, seedling photomorphogenesis, shade avoidance, and biotic or abiotic stress responses (Gangappa and Botto, 2014). However, there is currently no information on BBXs in sugarcane. We wanted to identify Shbbxs and study their phylogeny relationships, so we looked at gene structures, chromosomal locations, gene duplication, subcellular localization, homology, and codon analysis.
The sugarcane genome contained 21 Shbbx genes, which was significantly fewer than the 64 Shbbx genes found in other plants, such as Malus domestica. Phylogenetic analysis revealed that sugarcane and A. thaliana BBX genes were divided into five mixed subgroups/clades. Previous research found BBX genes to be clustered in five subgroups, including A. thaliana, rice, tomato, Moso bamboo, and 12 other plant species (Crocco and Botto, 2013; Huang et al., 2012; Khanna et al., 2009; R. Ma et al., 2021, 2021). The survival of these groups across species indicates that BBXs played important physiological roles in growth as well as development of plants. According to the analysis of conserved sequences, 13 Shbbxs had one B-box domain and nine Shbbxs had one B-box domain and one CCT domain. Arabidopsis has 11 AtBBXs with one B-box domain and 17 AtBBXs with one B-box domain and one CCT domain (Khanna et al., 2009). 13 OsBBXs in rice have one B-box domain, while 9 BBXs have B-box 1 domain and one CCT domain (Huang et al., 2012). 11 SlBBXs in tomato have B-box1 domain, while 5 BBXs have B-box1 domain and one CCT domain (Chu et al., 2016). In Bamboo, one PeBBX contains B-box1 domain, while five BBXs contain B-box1 domain and one CCT domain (R. Ma et al., 2021). In pear, ten PbBBX each have B-box1 domain, while four BBXs each have B-box 1 domain and one CCT domain (Zou et al., 2018). On the basis of reported literature suggested that the B-box domains are highly conserved in different plants species.
As previously reported in tomato, berry, pear, and bamboo, the majority of Shbbxs were found at the tops or bottoms of chromosomes (Chu et al., 2016; R. Ma et al., 2021; Wei et al., 2020; Zou et al., 2018). These findings could be attributed to plant evolution's conservation.
During the time of plant evolution Tandem and segmental duplication actions are critical for gene family expansion (Wei et al., 2020). To better understand the Shbbx gene family's expansion mechanism in sugarcane, researchers looked into both tandem and segmental duplication events. According to the findings, two Shbbx genes (Shbbx4/Shbbx6 and Shbbx9/Shbbx15) are involved in segmental duplication. Furthermore, six gene pairs (Shbbx7 / Shbbx13, Shbbx1 / Shbbx17, Shbbx2 / Shbbx8, Shbbx16 / Shbbx21, Shbbx3/Shbbx5, and Shbbx18 / Shbbx19) had tandem duplications, indicating that tandem duplications were more common in Shbbx genes. These findings suggested that both segmental and tandem duplications contributed to the evolution of the sugarcane Shbbx gene family. The grapevine SBP-box, B-box, and WRKY gene families have all been linked to a certain possible mechanism of gene family evolution (Hou et al., 2013; Wang et al., 2013; Wei et al., 2020). The Ka/Ks value of segmental gene pairs was also calculated. Positive, neutral, and negative selection are represented by Ka/Ks ratios greater than one, equal to one, and less than one. Surprisingly, all sugarcane gene pairs had Ka/Ks ratios less than one, indicating that these gene pairs underwent significant purifying selection during evolution.
Acknowledgments
We are grateful to Professor R.S. Sengar of the Sardar Vallabhbhai Patel University of Agriculture and Technology Meerut for his encouragement and guidance. We also thank the editor and reviewers for their constructive criticism and suggestions. This work was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R62), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit. Gene. 2012;496:110-117.
- [CrossRef] [Google Scholar]
- Genomic Organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci.. 2016;7
- [CrossRef] [Google Scholar]
- BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene. 2013;531:44-52.
- [CrossRef] [Google Scholar]
- das, S., Johri, P., Sharma, R., Kashyap, M., Singh, Shivani, Singh, Sachidanand, 2019. Comparative modeling, characterization and energy minimization studies of aquaporin 9: an exclusive target protein for rheumatoid arthritis | Int. J. Pharmaceut. Invest. [WWW Document]. URL https://www.jpionline.org/index.php/ijpi/article/view/369 (accessed 5.20.22).
- The BBX family of plant transcription factors. Trends Plant Sci.. 2014;19:460-470.
- [CrossRef] [Google Scholar]
- Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS One. 2013;8:e59358.
- [Google Scholar]
- The rice B-Box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One. 2012;7:e48242.
- [Google Scholar]
- The Arabidopsis B-box zinc finger family. Plant Cell. 2009;21:3416-3420.
- [CrossRef] [Google Scholar]
- Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot.. 2014;113:831-842.
- [CrossRef] [Google Scholar]
- Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L.) BMC Genomics. 2020;21:685.
- [CrossRef] [Google Scholar]
- Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genomics. 2018;293:303-315.
- [CrossRef] [Google Scholar]
- The BBX gene family in Moso bamboo (Phyllostachys edulis): identification, characterization and expression profiles. BMC Genomics. 2021;22:533.
- [CrossRef] [Google Scholar]
- Genome-wide and expression analysis of B-box gene family in pepper. BMC Genomics. 2021;22:883.
- [CrossRef] [Google Scholar]
- Comparative analysis of glyoxalase pathway genes in Erianthus arundinaceus and commercial sugarcane hybrid under salinity and drought conditions. BMC Genomics. 2019;19:986.
- [CrossRef] [Google Scholar]
- Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genomics. 2019;20:27.
- [CrossRef] [Google Scholar]
- Singh, K.B., Foley, R.C., Oñate-Sánchez, L., 2002. Transcription factors in plant defense and stress responses - ScienceDirect [WWW Document]. URL https://www.sciencedirect.com/science/article/abs/pii/S1369526602002893 (accessed 5.20.22).
- Genome-wide survey of B-box proteins in potato (Solanum tuberosum)—Identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS One. 2017;12:e0177471.
- [Google Scholar]
- MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered.. 2002;93:77-78.
- [CrossRef] [Google Scholar]
- Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep.. 2013;40:2679-2688.
- [CrossRef] [Google Scholar]
- Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol.. 2020;20:72.
- [CrossRef] [Google Scholar]
- Weng, X., Wang, lie, Wang, J., Hu, Y., Du, H., Xu, C., Xing, Y., Li, X., Xiao, J., Zhang, Q., 2014. Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response - PubMed [WWW Document]. URL https://pubmed.ncbi.nlm.nih.gov/24390391/ (accessed 5.20.22).
- Yamaguchi-Shinozaki, kazuko, Shinozaki, K., 2005. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters - ScienceDirect [WWW Document]. URL https://www.sciencedirect.com/science/article/abs/pii/S1360138504002985 (accessed 5.20.22).
- A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell. 2014;26:2038-2054.
- [CrossRef] [Google Scholar]
- Genome-wide identification and expression analysis of the B-box transcription factor gene family in grapevine (Vitis vinifera L.) BMC Genomics. 2021;22:221.
- [CrossRef] [Google Scholar]
- Genome-wide identification, phylogenetic analysis, and expression profiling of the BBX family genes in pear. J. Hortic. Sci. Biotechnol.. 2018;93:37-50.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102720.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







