Translate this page into:
Dermoscopic image classification using CNN with Handcrafted features
⁎Corresponding authors. rajasekharks@anu.ac.in (Kotra Sankar Raja Sekhar), long.ming@ubd.edu.bn (Long Chiau Ming)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Globally skin cancer is one of the main cause of death in humans. Early diagnosis plays a major role in increasing the prevention of death rate caused due to any kind of cancer. Conventional diagnosis of skin cancer is a tedious and time-consuming process. To overcome this an automated skin lesion classification must develop. Automated skin lesion classification is a challenging task due to the fine-grained variability in the visibility of skin lesions. In this work dermoscopic images are obtained from the International Skin Image Collaboration Archive 2016 (ISIC 2016). In the proposed method the analysis and classification of skin lesions is done with the help of a Convolution Neural Network (CNN) along with the hand crafted features of dermoscopic image using Scattered Wavelet Transform as additional input to the fully connected layer of CNN, which leads to an improvement in the accuracy for identifying Melanoma and different skin lesion classification when compared to the other state of the art methods. When raw dermoscopic image is given as an input to the CNN and feature values of segmented dermoscopic image as input to the fully connected layer as an additional information, the proposed method gives a classification accuracy of 98.13% for identification of Melanoma and the accuracy achieved for classification of skin lesions is 93.14% for Melanoma Vs Nevus, 95.4% for Seborrheic Keratosis (SK) vs Squamous Cell Carcinoma (SCC), 96.87% for Melanoma vs Seborrheic Keratosis (SK), 95.65% for Melanoma vs Basal Cell Carcinoma (BCC), 98.5% for Nevus vs Basal Cell Carcinoma(BCC).
Keywords
Scattered wavelet
Feature extraction
Classification
CNN
Fully connected layer
Concatenation
1 Introduction
Abnormal growth of cells in a human body causes cancer. Skin cancer is the abnormal growth of skin cells. Skin cancer may be caused due to excessive exposure to sun or it may occur due to some genetical disorders. Over the past two decades the occurrence of skin cancer is increasing rapidly and every year five million cases are occurring across all the countries (Albahar, 2019). There are various types of skin cancers like BCC, Melanoma, Nevus, SK, SCC, etc., Out of all these types Melanoma is the most dangerous and it originates from the pigments which produces melanocytes that arise infrequently but it is more harmful and globally around 10 thousand persons are dying with Melanoma every year (Al-Masni et al., 2020). It is a very significant and challenging task to classify skin lesions of dermoscopic images with the help of Computer Aided Diagnosis (CAD) system, hence it requires a deep study to introduce an automated CAD system to classify skin lesions using dermoscopic images. The dermoscopic images are acquired by using various sources like Microscopy, Electrical Impedance Spectroscopy (EIS) and Optical Coherence Tomography (OCT) etc. In this work Melanoma identification, along with classification of skin lesions are done because some skin lesions may flare up the malignancy in the other parts of human body.
2 Related works
Convolution Neural Networks is widely used in the diagnosis of various medical areas. (Su et al., 2020) have used CNN to improve the diagnosis of ultrasound images. Rabie (2021) have identified Ultra-Low Frequency signals in the geomagnetic records by extracting DWT features which are given as input to the CNN achieving accuracy of 91.11%.
(Rashmi and Bellary, 2020) have proposed classification of Melanoma into different stages using CNN with various loss functions. An accuracy of 96% is achieved for Similarity Measure for Text Processing (SMTP). Celebi M Emre et al., (Celebi et al., 2007) performed dermoscopic images classification utilizing both features of colour and texture. By using relief algorithm these features are optimized which are then classified using linear SVM resulting in a specificity of 92.34% and sensitivity of 93.33%.
(Lau and Al-Jumaily, 2009) proposed early detection of skin cancer using DWT features which are classified using three layers back propagation and auto associative neural network acheiving a recognition accuracy of 89.9%.
Elgamal (2013) proposed automatic skin cancer classification with features of DWT, that are reduced in dimension by applying PCA resulting an accuracy of 95% using feed-forward back propagation artificial neural network. ((Albahar, 2019) introduced a CNN with novel regulizer for the classification of skin lesions achieving an AUC of 0.77, 0.93, 0.85, and 0.86 for Melanoma vs Nevus, BCC vs SK, Melanoma vs SK, Melanoma vs Solar Lentigo respectively.
(Mahbod et al., 2019), have done an automated skin lesion classification system using CNN with ensemble technique resulting in an AUC of 87.3 %. (Yu et al., 2017c), have done dermoscopic images hybrid classification using ensemble of DCNN, SVM and Fisher vector results an accuracy of 83.09 %.
(Codella et al., 2017), introduced Melanoma identification in Dermoscopic images by fusing hand-crafted features like colour histogram, MSLBP, edge histogram. Dictionary based features acquired using sparse coding and CNN resulting in an AUC of 83.8 %.
(Papanastasiou, 2017) introduced a model based on deep learning that is designed from basic as well as the pertained Inception v3 and VGG-16 models resulting in an accuracy of 91.4 % and 86.3 % for identification of Melanoma.
(Yu et al., 2017b), have done FCRN integrated with a very deep residual network which enables the classification of Melanoma identification resulting an AUC of 0.826, 0.801, and 0.783 using VGG-16, Google Net, DRN-50 networks respectively. Lei et al., 2017) performed classification of skin lesions as Melanoma, SK and Nevus using deep residual networks yielding an average AUC of 91.50%.
(Matsunaga et al., 2017) have proposed lesion classification by fine tuning of CNN with the training samples, using generic object recognition in Keras with RMSProp and AdaGrad optimization achieving an AUC of 0.924 with Melanoma classifier and 0.993 with Seborrheic Keratosis classifier.
Harangi, 2018) have done skin lesion classification using ensemble of features extracted from different pretrained models such as GoogLeNet, AlexNet, ResNet, VGGNet achieving an accuracy of 89.1% for classifying Melanoma, Nevus, and SK.
(Mahbod et al., 2019) proposed segmentation and classification of multiple skin lesions using integrated deep convolutional networks results in a two class classification accuracy of 81.29%, 81.57%, 81.34%, and 73.44% for Inception-v3, ResNet-50, Inception-ResNet-v2, and DenseNet-201 respectively using ISIC 2016 dataset.
3 Materials
International Skin Imaging Collaboration (ISIC) archive ISBI challenge 2016 dataset is considered in this work, which consists of 900 training images, 379 test images, that are used for the identification of Melanoma. For classification of various skin lesions, 587 images of BCC, 2169 images of Melanoma, 18,566 images of Nevus, 419 images of SK and 225 images of SCC are considered for the present work. Meta data like clinical findings, image dimensions, diagnosis and localization are provided in the data set. The dimension of dermoscopic image ranges from 722 × 542 to 4288 × 2848 in the form of jpeg. Fig. 1 shows the dermoscopic images obtained from ISIC2016.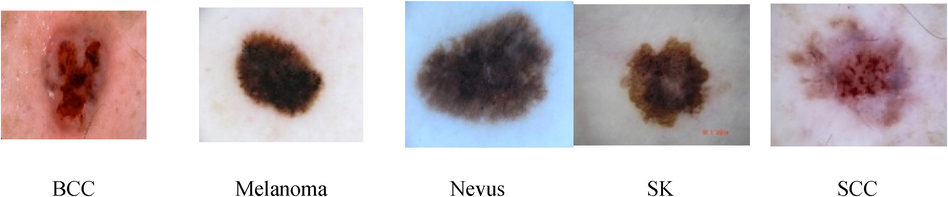
Different Skin Lesions in Dataset.
4 Method
The workflow of proposed methodology is depicted in Fig. 2. In the proposed work a segmented dermoscopic image is subjected to a CNN and the hand-crafted features which are extracted by decomposing the segmented dermoscopic image using scattered wavelet transform are given as additional input to the Fully connected layer of a CNN. The fully connected layer in the proposed method concatenates both the hand crafted features and CNN features which results in a better accuracy compared to the other state of the art methods.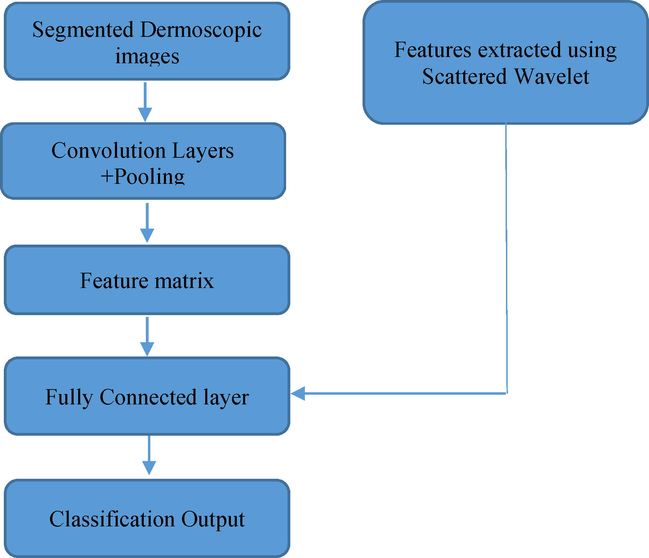
Block Diagram of Proposed Method.
4.1 Segmentation
Segmentation is the process of obtaining required image object which gives the location of object and boundaries like lines and curves. Before extracting the feature values, the dermoscopic images are subjected to a segmentation process. In this work, a region growing technique is used to extract the region of interest. In region growing technique initially a seed point will be taken, and the neighbouring pixels of the initial seed point are examined to determine whether the neighbouring pixels falls into that region or not. The neighbouring pixels which are having the properties similar to the seed will grow on and the segmentation of the required tumour part can be acquired. The segmented dermoscopic images are shown in Fig. 3.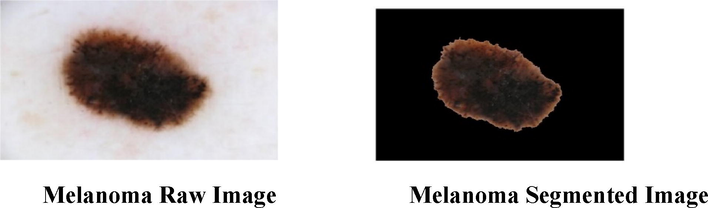
Segmentation of Dermoscopic Image.
4.2 Feature extraction
Feature extraction is the process of making a raw data into some numerical value features which can be processed by preserving the actual data. Simply an image feature is a characteristic which helps in the discrimination of one image from another image.
Various methods exist for feature extraction such as GLCM, Bag of Features, Local Binary Patters etc., (Paci et al., 2013) proposed multi local quinary patterns and multi local phase quantization with ternary coding for extracting texture features which are classified using Support Vector Machine and Gaussian Process Classifier achieving good classification accuracy with different texture datasets such as Broadatz, KTH-TIPS, OUTEX TC 00,000 dataset etc., (Tahir, 2018) proposed analysis of patterns in the images of proteins using GLCM and concluded that it is efficient for analysis of patterns in the biomedical area. (Xu and Li, 2020) used GLCM features for extraction of features for efficient detection of breast tumours.
In the present work the handcrafted features that are to be considered are Texture, Shape and colour features. From GLCM the texture features attained are autocorrelation, correlation, contrast, energy, dissimilarity, entropy, variance, homogeneity and normalized inverse difference moment. In addition to these texture features, the coefficients of Scattered Wavelet Transform obtained from the decomposed dermoscopic images are also considered. The shape features such as area, perimeter, form factor, major axis length and minor axis length is obtained from segmented dermoscopic images. Colour features such as mean, standard deviation, kurtosis, skewness, and smoothness index are calculated from each channel (RGB) of the colour histogram for color features.
4.3 Scattered wavelet transform
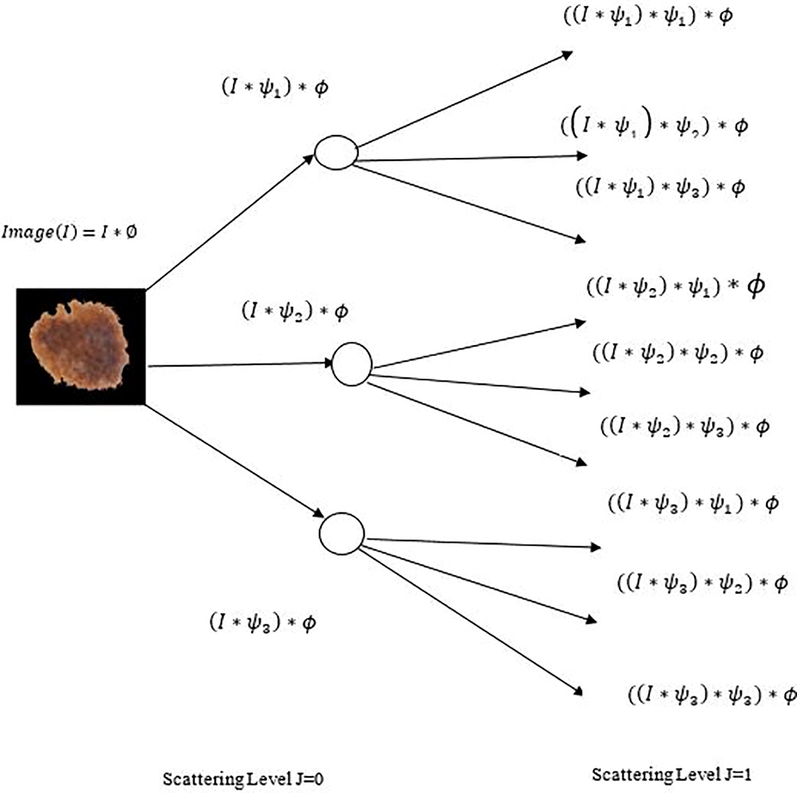
Wavelets techniques are an effective tool for feature extraction and an adequate data. Scattered wavelet transform proposed by (Bruna and Mallat, 2013), is capable to generate definitive features that are locally steady to small deformations. This transform builds a stable invariant representation by computing the cascade of wavelet modulus operator with filter coefficients. To construct a wavelet scattering transform of an input image (I) three successive important operations such as Convolution, Nonlinearity, and Averaging need to be performed as depicted in Fig. 4.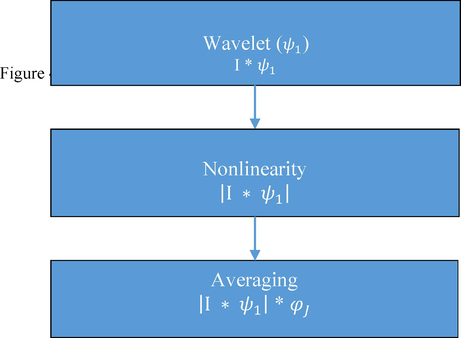
Construction of Scattered Wavelet Transform.
Dilated mother wavelets with different scaling levels are the wavelet functions used in the scattering transform (Pandey et al., 2016). Morlet wavelet denoted by
is considered as a mother wavelet. A two dimensional Image I considered to be a real number which can be represented as
and a low pass averaging filter in the frequency domain controlled by scaling factor J can be represented by
Convolving dermoscopic image with an averaging filter results in locally affine transformation given by
Majority of the high frequency components of the image are rejected and Eq. (1) produces exactly same representation up to
pixels. To acquire high frequency components of the image a set of constant high frequency bandpass filters can be applied to an image. Morlet filters or wavelets
with altering scales
and rotations
are constituted with a set
, and is given as
The constant set of wavelet bank
can be given a
Where with and and
Applying average filter, high frequency bandpass filters or wavelets over an image is done with the wavelet modulus
of an image
given by
For the higher frequency, locally translational invariant descriptors,
can be calculated by attaining the space average over the wavelet modulus coefficients,
Which are the coefficients of 1st order scattering calculated by using modulus of 2nd wavelet transform
that is applied to
for each
. It also gives complimentary wavelet coefficient of high frequency
Where and .
Here
and
represents two different filter banks and by using a low pass filter
the coefficients are averaged, which defines second order scattering coefficients and can be represented as
By applying a third wavelet modulus transform, the averages of all these are computed by
to for each and . By using third wavelet modulus the wavelet modulus coefficients are computed by convolving with a new set of wavelets which consists of scaling and orientation parameters defined by such that where can be the new wavelet bank. By repeatedly applying this process further layers can be computed for defining a scattering wavelet transform network, which is able to extend for any order m, with the help of at each step. A robust feature representation can be achieved with the help of higher order scattering coefficients.
At the mth layer the scattering wavelet coefficients can be defined as
for I = 1,2,…,m, and
is an ordered set of wavelets, where
and
Here X represents Cartesian product. During the scattering process the loss of information in one stage will be recovered in the next stage. The energy of the scattering coefficients decreases in proportion with the increasing level of scattering; hence an infinite level of decomposition is not required due to the decrease in the energy. Visualization of first order scattering disc on a dermoscopic image is shown in Fig. 5. Decomposing a dermoscopic image using scattered wavelet transform is shown in Fig. 6.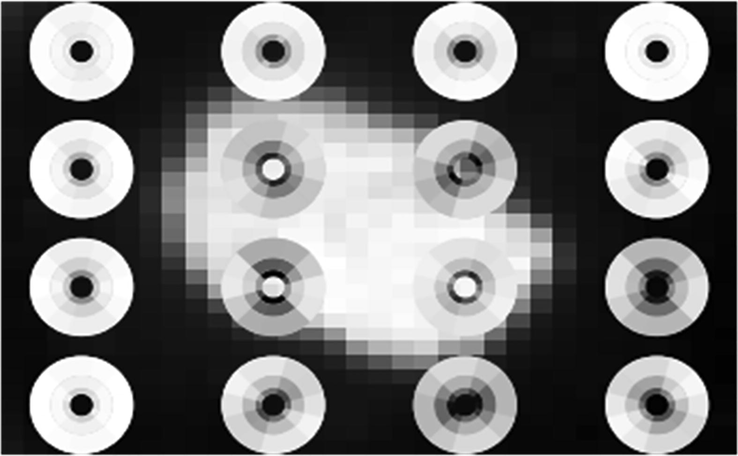
Visualization of first order Scattering Disk on Dermoscopic Image.
Decomposition of Dermoscopic Image At Different Scales Using Scattered Wavelet Transform.
4.4 Feature extraction using Scattered Wavelet Transform
The Statistical Texture features are extracted by applying the Scattered Wavelet Transform to the dermoscopic image along with the GLCM, colour and shape features for Melanoma as well as various Skin Lesions. The GLCM features are calculated by the mathematical relations shown in Supplementary File Table SI and the Statistical Texture features are calculated by the mathematical representations from the coefficients of different multiresolution transforms shown in Supplementary File Table SII. Supplementary File Table SIII–SVIII shows the features obtained for different skin Lesions.
4.5 Convolution neural network
Convolution Neural Networks are a kind of deep feed-forward artificial neural networks which can be able to analyse visual imagery (LeCun and Bengio, 1998). A CNN is a combination of local receptive fields, shared weights and spatial or temporal subsampling. A CNN is constructed by combining convolutional, pooling and fully connected architectures. The neural networks which are having one or more convolutional layers are said to be convolutional neural networks. The convolution layers in the CNN consist of filters with a capability of learning.
A convolution layer processes a 2-dimensional convolution on the inputs. CNN perform feature extraction and classification. Feature extraction involves three phases, Convolution, Activation and Pooling. The features from the raw images are extracted automatically which yields better accuracy. The convolution and the pooling layers in the CNN makes the model computational efficient. CNN has the flexibility to handle large data sets which makes it superior to other methods. According to the size of the input, each and every filter is having a small equal height, width and depth. Convolution can be done by applying appropriate filters. Convolution is a mathematical operation which produces a third function (z) from two functions x and y.
CNN uses a hierarchical model to get a fully connected layer, in this all the neurons are connected to each other to process the output. A simple CNN model consists of two hidden layers (convolution layers). To create feature maps CNN uses various kinds of kernels to convolve the input image which results in the output matrix of a convolution layer.
Where the input image I is convolved with kernel
. The bias
is added to each element obtained to the sum of all convoluted matrices. The size of the input image I should be increased by padding with zeros to maintain the output image same as that of the input image. The output of the convolution consists of both negative and positive values. The negative values are removed using Activation or Nonlinear functions such as Hyperbolic, Softmax, Rectified Linear Unit (ReLU), Exponential Linear Unit (ELU), Scaled Exponential Linear Unit (SELU) which are given in the Eqs. (12)–(16) respectively.
The convolution layer is followed by pooling layer which is used to reduce the size of the feature maps and parameters of the network that increases computational speed. Max pooling is used widely to select the maximum of the value. The hyper parameters of pooling layer are filter size, stride and max pooling. Regularization methods need to be used to introduce dropouts not only on the training data but also on the new entries to reduce the problem of over fitting. Loss function gives the difference between the output and target variable helps in the connection of various inputs to the next layer independently. The basic architecture of a CNN is shown in Fig. 7.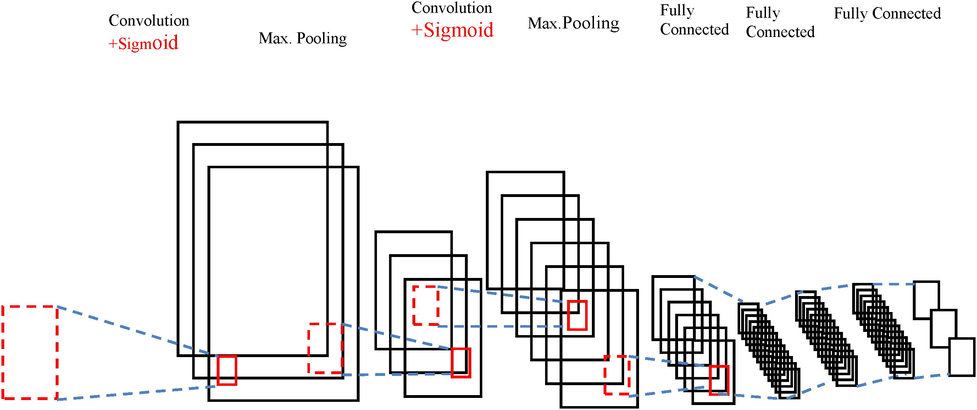
Basic Architecture of CNN.
4.6 Convolution neural network with Handcrafted features
The architecture of CNN considered in this work is shown in Fig. 8. The proposed architecture consists of two convolution layers followed by two max pooling layers and dropouts to randomly discard the nodes during the training phase which eventually reduces the overfitting and improves generalization error. Next to dropout, the 2-Dimensional features are flattened to 1-Dimensional array and given to fully connected layer consisting of 128 neurons. The output layer consists of one neuron per class.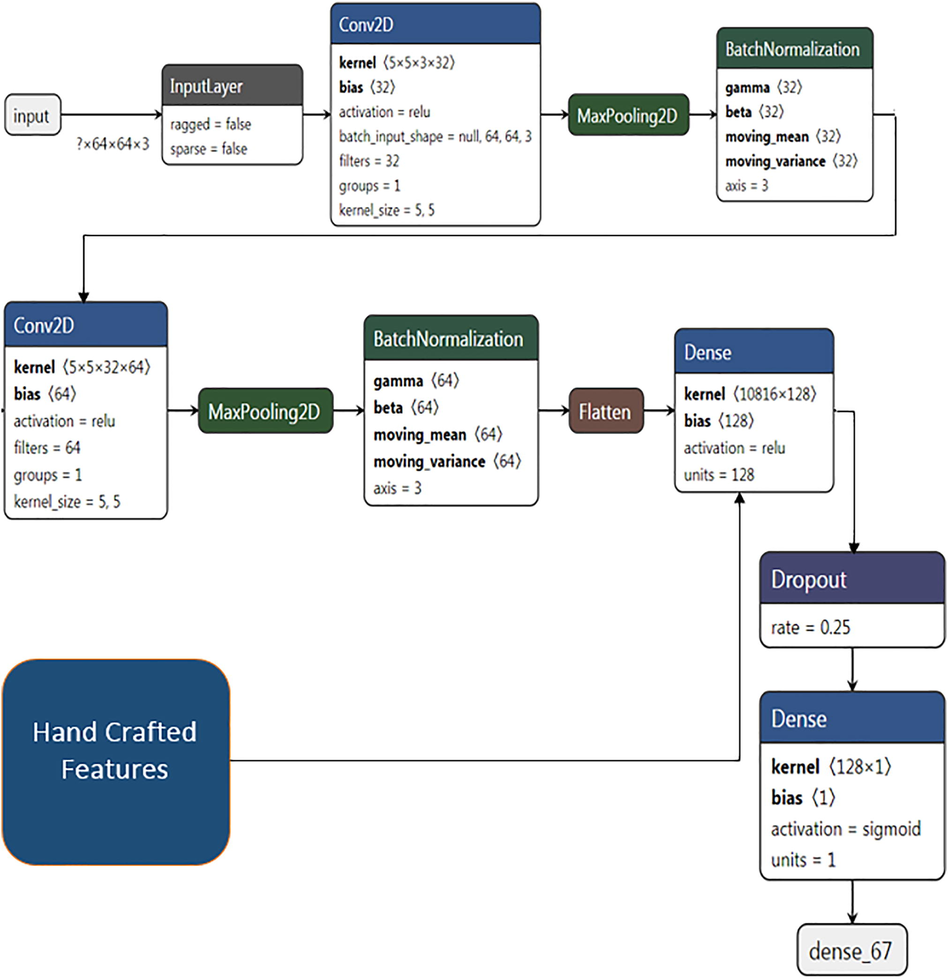
Architecture of CNN with Handcrafted features.
In the training process of the proposed architecture Skin Lesions are resized to 64X64 and given as input to the CNN with RGB channels. The filter should be 3-dimensional if the input image is a color image. Filters can occur in various sizes. The job of a filter is to identify an image specific feature. When a trained filter gets multiplied with a portion of the image, the area of the image containing that feature will produce very high values in the feature maps. All the values in the filter start with random initialization, just like the weights in the neural networks. First few convolution layers consist of filters that will identify common features such as lines, edges and curves which are part of Skin Lesion. Subsequent layers contain filters that can identify complex and image specific features.
The first convolution layer is assigned with 32 filters with a 5X5 kernel, producing 32 channels with a size of 60X60. Pooling can be performed after activation to reduce the dimensionality of the dataset. It reduces the height and width but keeps the depth. After applying the first convolution on the input images a maxpooling layer with a size of 2X2 is applied on the convolved output to select the maximum elements with prominent features using a stride of 1. First max pool layer consists of a size of 2X2 which is applied on first convolution layer output gives 32 channels with a size of 30X30. The second convolution layer is assigned with 64 filters with a 5X5 kernel, size which produces 64 channels with a size of 26X26. On applying the maxpool to the second convolved output, it gives 64 channels with a size of 13X13. Batch normalization is performed at the output of each maxpool layer to standardize the inputs. An activation function Rectified Linear Unit (ReLu) is used in this architecture to introduce nonlinearities to the network.
A dropout of 0.25 is applied before the flattening of 2-Dimensional to 1-Dimensional to reduce the complexity and increase the computation time by discarding some nodes. The discarded nodes weights and bias values are not considered during the training phase. The 1-Dimensional features are given to the fully connected layer with 128 neurons. In addition to the 1-Dimensional features which are obtained by the convolution process, hand-crafted features which are acquired by using Scattered Wavelet are also given as additional features to the fully connected layer. These additional features will be concatenated in the fully connected layer with the features obtained by the convolution layer, leads to a better accuracy when compared with the single input convolution neural network model.
The proposed architecture was trained with 80% data and tested with 20% data. The number of images used in the training and test data for Melanoma recognition and different skin lesion classification are listed in Supplementary File Table SIX. Sample Images given to CNN are shown in Fig. 9.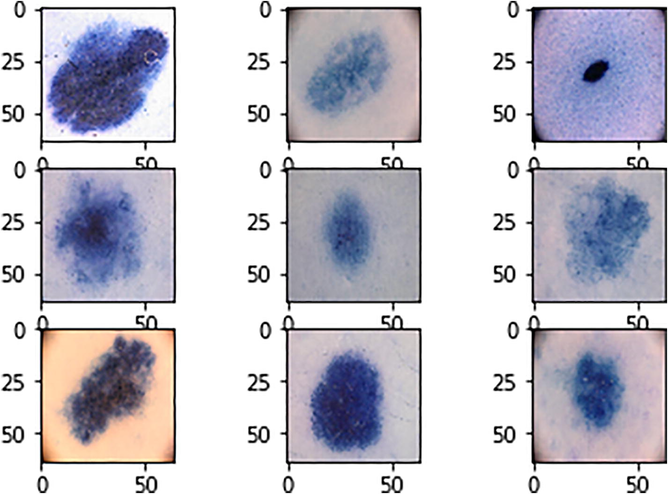
Sample input images to CNN.
5 Results
The proposed method achieves a classification accuracy for the recognition of Melanoma as 98.13% which is a better result when compared to the other state of the art methods. The validation accuracy and loss graph are shown in Supplementary File Fig. SI–SII. The classification accuracy for the classification of different Skin Lesions like Melanoma vs Nevus, Seborrheic Keratosis vs Squamous Cell Carcinoma, Melanoma vs Seborrheic Keratosis, Melanoma vs Basal Cell Carcinoma and Nevus vs Basal Cell Carcinoma is 93.14%, 95.4%, 96.87%, 95.65%, 98.5%, The accuracy graphs for the classification of various Skin Lesions are shown in Supplementary File Figs. SIII–SVII.
6 Discussions
The aim of this work is a deep study on the analysis and classification of dermoscopic images. It was found that the proposed method gives a better result when compared with the other state-of-the-art methods. The discussions based on Melanoma recognition and different skin lesion classifications are shown in Supplementary File Table SX as compared with different existing works described below.
(Nida, 2019) optimized the performance of a CAD system using CNN and fuzzy C-means for the classification of Melanoma through a proper segmentation is nothing but the Region Of Interest (ROI), achieves a performance measure in terms of accuracy is 94.08%. Majtner et al., 2016 have proposed a fully automated melanoma classification system by combining the Deep Learning features and hand crafted features which are derived using Rsurf and Local Binary Patterns together results in a classification accuracy of 82.6%. Yu et al., 2017a have proposed a novel framework to develop an automated Melanoma recognition using a pre-trained DRNN on ImageNet. The deep representation of a Skin Lesions are extracted using Local Descriptors and aggregated by Fisher Vector achieved a classification accuracy of 86.81%, Sensitivity of 42.67% and specificity of 97.70%.
Bakkouri and Afdel, 2018 have proposed a novel discriminative feature extraction for Melanoma recognition using the combination between the weights learning from a pretrained network and the weights trained on the CNN to classify as Malignant Melanoma or Benign Nevus achieved a classification accuracy of 97.40%, Specificity of 97.30% and Sensitivity of 97.50%. An automated skin lesion classification using transfer learning theory and pre-trained Alex Net deep neural network by fine tuning the weights and replacing the classification layer with softmax layer have been proposed by Hosny et al. (2019), achieving an accuracy of 95.91% for Melanoma, Seborrheic Keratosis and Nevus. The comparision of the Skin Lesion classification is shown in Supplementary File Table SXI.
7 Conclusion
In this work, the classification of different Skin Lesions and Melanoma Recognition is done using a Convolution Neural Network. Texture Features are extracted by applying Scattered Wavelet Transform from the skin lesions and the extracted Handcrafted features are given as additional information to the fully connected layer of a CNN. The Hand crafted features and the features extracted by CNN are concatenated at the fully connected layer resulting in good classification accuracy in the Recognition of Melanoma and various skin lesion classifications, compared to the other related works. Further the present work may be improved by considering multiple input data to the CNN with different optimization techniques.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Skin lesion classification using convolutional neural network with novel regularizer. IEEE Access. 2019;7:38306-38313. [Accessed: 25 January 2021]
- [CrossRef] [Google Scholar]
- Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed.. 2020;190:105351
- [CrossRef] [Google Scholar]
- Convolutional neural-adaptive networks for melanoma recognition. Springer, Cham: International conference on image and signal processing; 2018.
- Invariant scattering convolution networks. IEEE Trans. Pattern Anal. Mach. Intell.. 2013;35(8):1872-1886.
- [CrossRef] [Google Scholar]
- Celebi, M. Emre, et al. “A methodological approach to the classification of dermoscopic images” Computerized Medical Imaging and (2007):362-373. https://www.sciencedirect.com/science/article/abs/pii/S0895611107000146 [Accessed: 25 January].
- Codella, N., Nguyen, Q. B., Pankanti, S., Gutman, D., Helba, B., Halpern, A., Smith, J. R. “Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images”, IBM Journal of Research and Development, 61(4–5), pp.1–28,2017.[online] https://arxiv.org/abs/1610.
- Elgamal, Mahmoud. “Automatic skin cancer images classification.” IJACSA) International Journal of Advanced Computer Science and Applications 4.3 (2013):287-294. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.299.3590&rep=rep1&type=pdf[Accessed: 25 January 2021].
- Skin Lesion Classification with ensembles of deep convolutional neural networks. J. Biomed. Informat.. 2018;86:25-32.
- [CrossRef] [Google Scholar]
- Classification of skin lesions using transfer learning and augmentation with Alex-net. PloS One. 2019;14(5)
- [Google Scholar]
- Lau, Ho Tak, and Adel Al-Jumaily. “Automatically early detection of skin cancer: Study based on neural network classification.” Soft Computing and Pattern Recognition, 2009. SOCPAR’09. International Conference of. IEEE, 2009.10.1109/SoCPaR.2009.80 [Accessed: 25 January 2021].
- LeCun, Yann, and Yoshua Bengio. “Convolutional networks for images, speech, and time series”, The handbook of brain theory and neural networks.”(1998). https://www.researchgate.net/profile/Yann_Lecun/publication/2453996_Convolutional_Networks_for_Images_Speech_and_Time-Series/links/0deec519dfa2325502000000.pdf
- B. Lei et al. “Automatic Skin Lesion Analysis Using Large-Scale Dermoscopy Images And Deep Residual Networks.”ARXIV: 1703.04197(2017). https://arxiv.org/abs/1703.04197
- Mahbod, A., Schaefer, G., Ellinger, I., Ecker, R., Pitiot, A., Wang, C. “Fusing fine-tuned deep features for skin lesion classification”, Computerized Medical Imaging and Graphics, 71, pp. 19–29, 2019. 10.1016/j.compmedimag.2018.10.007[Accessed: 22 Nov 2020].
- “Combining deep learning and hand-crafted features for skin lesion classification”. Sixth International Conference on Image Processing Theory. IEEE: Tools and Applications (IPTA); 2016. p. :2016.
- Matsunaga, Kazuhisa, et al. “Image Classification Of Melanoma, Nevus And Seborrheic Keratosis By Deep Neural Network Ensemble.” Arxiv Arxiv: 1703.03108 (2017). https://arxiv.org/abs/1703.03108.
- Melanoma lesion detection and segmentation using deep region based convolutional neural network and fuzzy C-means clustering. Int. J. Med. Informat.. 2019;124:37-48.
- [Google Scholar]
- An ensemble of classifiers based on different texture descriptors for texture classification. J. King Saud Univ.-Sci.. 2013;25(3):235-244.
- [Google Scholar]
- Pandey, Prateekshit, Richa Singh, and Mayank Vatsa. “Face recognition using scattering wavelet under Illicit Drug Abuse variations.” 2016 International Conference on Biometrics (ICB). IEEE, 2016. 10.1109/ICB.2016.7550091
- Papanastasiou, M. “Use of Deep Learning in Detection of Skin Cancer and Prevention of Melanoma”, MSc Thesis, KTH Royal Institute of Technology School of Technology and Health, 2017. [online] https://www.diva-portal.org/smash/get/diva2:1109613/FULLTEXT01.pdf
- Patil, Rashmi, and Sreepathi Bellary. “Machine learning approach in melanoma cancer stage detection.” Journal of King Saud University-Computer and Information Sciences (2020).
- Geomagnetic micro-pulsation automatic detection via deep leaning approach guided with discrete wavelet transform. J. King Saud Univ.-Sci.. 2021;33(1):101263
- [Google Scholar]
- Ultrasound image assisted diagnosis of hydronephrosis based on CNN neural network. J. King Saud Univ.-Sci.. 2020;32(6):2682-2687.
- [Google Scholar]
- Pattern analysis of protein images from fluorescence microscopy using Gray Level Co-occurrence Matrix. J. King Saud Univ.-Sci.. 2018;30(1):29-40.
- [Google Scholar]
- Image feature extraction in detection technology of breast tumor. J. King Saud Univ.-Sci.. 2020;32(3):2170-2175.
- [Google Scholar]
- Aggregating deep convolutional features for melanoma recognition in dermoscopy images. Cham: Springer; 2017.
- Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imag.. 2017;36(4):994-1004.
- [CrossRef] [Google Scholar]
- “Hybrid dermoscopy image classification framework based on deep convolutional neural network and Fisher vector”. IEEE 14th Int. Sympos. Biomed. Imag. (ISBI 2017) Melbourne, Australia. 2017;2017:301-304.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101550.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







