Translate this page into:
Copper and chromium binding by Pseudomonas aeruginosa strain PA01 for implications of heavy metal detoxification and soil remediation: A computational approach
⁎Corresponding author at: Department of Microbiology, PSG College of Arts & Science, Civil Aerodrome Post, Coimbatore - 641014, Tamil Nadu, India. priyajasper@gmail.com (Shanmuga Priya Ramasamy),
⁎⁎Corresponding author at: Department of Biomedical Science, Faculty of Science, University Tunku Abdul Rahman, Jalan University, Bandar Barat, Kampar 31900, Malaysia. sinouvassane@utar.edu.my (Sinouvassane Djearamane)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Heavy metal pollution poses significant environmental and health risks due to the toxic effects of metals like copper and chromium at elevated concentrations. Despite their essential roles in trace amounts, these metals can be highly toxic. Bacteria such as Pseudomonas aeruginosa are promising candidates for bioremediation due to their robustness and adaptability. The objective of this study was to analyze and identify potential copper and chromium binding genes involved in metal detoxification in Pseudomonas aeruginosa PA01. The heavy metal binding protein identified as ferredoxin using MALDI-TOF/PMF-MS analysis was further characterized. The structure of the ferredoxin protein was elucidated using the SWISS-MODEL tool. Metal-binding domains were validated through a pattern search against UniProtKB/Swiss-Prot and UniProtKB/TrEMBL databases using the ScanProsite tool. Comparative sequence alignments were conducted between the copper-binding NosD gene of P. aeruginosa, the ferredoxin gene of P. aeruginosa PA01, and the chromium-binding iron hydrogenase 1 gene of Clostridium chromiireducens. The SWISS-MODEL analysis revealed alpha helices and beta sheets with key metal-coordinating amino acids (cysteine, glutamic acid, aspartic acid, histidine, and methionine). The ScanProsite tool confirmed the presence of a 4Fe-4S ferredoxin-type iron-sulphur binding domain essential for coordinating chromium and copper ions. Sequence alignments showed a 64.29 % similarity between the NosD gene and ferredoxin gene, and a 67 % identity between the iron hydrogenase 1 gene and ferredoxin gene, with correlations in amino acid residues involved in metal binding. These findings suggest that the ferredoxin gene could effectively bind heavy metal ions, offering potential applications in bioremediation of metal-polluted soils using Pseudomonas species. This study contributes to sustainable agricultural productivity by facilitating the targeted remediation of heavy metal-contaminated soils through biological means.
Keywords
Copper
Chromium
Pseudomonas aeruginosa PA01
Ferredoxin
NosD
Iron hydrogenase 1
Pollution remediation
1 Introduction
Heavy metals exist naturally with high atomic weight, have a specific density of more than 5 g/cm3, and detrimentally affect the environment and living organisms. Heavy metals like cobalt, copper, iron, chromium, nickel, magnesium, selenium, manganese, and zinc are the essential micronutrients that source various physiological and biochemical processes in plants and animals. However, they become toxic due to an excessive supply of these micronutrients beyond their threshold concentration, resulting in various diseases or disorders. Based on the high degree of toxicity, arsenic, cadmium, chromium, lead, and mercury are ranked among the prioritized toxic heavy metals that have a significant concern for public health (Tchounwou et al., 2012a,b; Jaishankar et al., 2014a,b; Yap et al., 2023). For instance, copper is essential but becomes toxic at concentrations above 20–30 μM in plants (Yruel, 2005) and around 100 μM in humans (Linder and Hazegh-Azam, 1996; Mitra et al., 2022). Chromium, another essential micronutrient, is toxic in aquatic organisms at concentrations above 100 μM (Katz and Salem, 1993; Naseri et al., 2021). Similarly, nickel becomes harmful in plants at concentrations exceeding 10–20 μM (Seregin and Kozhevnikova, 2006; Mitra et al., 2022).
The conventional remediation methods, including flocculation, solvent extraction, precipitation, coagulation, and ozonation, are widely adopted to recover and restore metal-contaminated effluent. Nevertheless, these methods are expensive and unreliable in removing heavy metals to attain expected effluent quality standards (Dawodu et al., 2020). Microorganisms play a pivotal role in detoxifying and removing heavy metals from the polluted ecosystem (Quintelas et al., 2008; Jobby et al., 2018). Heavy metal resistance genes of microbes are diverse and beneficial for heavy metal remediation from metal-polluted environments. Many biological and chemical processes require metal ion-binding proteins called metalloproteins. These genes play a significant role in the structural and functional stability of protein molecules. Understanding the metal binding motifs using an in-silico approach could help us to better understand the gene expression of microorganisms in heavy metal remediation (Akcapinar and Sezerman, 2017).
Bioremediation research is still hampered in the current scenario due to an incomplete understanding of the genome characterization of the microbes used in metal adsorption. Hence in this work, an attempt was made to identify the copper and chromium binding motifs that are responsible for the metal uptake in Pseudomonas aeruginosa PA01.
2 Experimental details
2.1 Identification of metal binding protein in P. aeruginosa PA01 using MALDI-TOF/PMF-MS
The protein spots were excised and washed twice with 100 mM ammonium bicarbonate and 100 % acetonitrile (ACN) and reduced. Then it was alkylated using 25 mM dithiothreitol (DTT) and 55 mM iodoacetamide and incubated with 200 ng of trypsin gold (Promega) in 25 mM ammonium bicarbonate for 3 h at 37 °C. After digestion, the samples were aspirated and eluted once with 50 % acetonitrile and 2.5 % trifluoroacetic acid (TFA) to stop the digestion process. The samples were spotted and overlaid on a MALDI matrix containing l5 mg/mL of α-cyano-4-hydroxycinnamic acid and 10 mM ammonium monobasic phosphate. The peptide mass spectrometric data were obtained using ABI 4800 MALDI-TOF/TOF tandem mass spectrometry (MS) (Applied Biosystems Inc., Foster City, CA). The data was acquired in reflector mode with a mass range of 600–4000 Daltons. The obtained protein spectra were submitted for database searching using the online MASCOT program (Matrix Science, Boston, MA) against databases like SwissProt and NCBI (National Centre for Biotechnology Information) (Zhang et al., 2016). Further, the structure of ferredoxin was elucidated using SWISS-MODEL software (https://swissmodel.expasy.org/interactive) and visualized in RasMol software (https://www.umass.edu/microbio/rasmol/index2.htm).
2.2 Validation of copper and chromium binding domain in ferredoxin of P. aeruginosa PA01
The metal ion binding domain in the ferredoxin of P. aeruginosa PA01 was validated using the ScanProsite tool (https://prosite.expasy.org//) at the PROSITE database against the UniProtKB/SwissProt (Release 50.0) and UniProtKB/TrEMBL (Release 33.0) databases (Gattiker et al., 2002; Thilakaraj et al., 2007; Tian et al., 2019).
2.3 In-silico analysis of copper binding motif in P. aeruginosa PA01
Based on a gene name search, the NosD (copper-binding gene sequence) of P. aeruginosa was retrieved from the UniProtKB 2022_03 database. The gene sequence alignment was performed between the NosD gene of P. aeruginosa and ferredoxin of P. aeruginosa PA01 using BLAST (Basic Local Alignment Search Tool) to identify the copper-binding motif. The correlation of amino acid residues of copper-binding motifs was determined for NosD and ferredoxin genes (Gattiker et al., 2002; Richard et al., 2007; Tian et al., 2019).
2.4 In-silico analysis of chromium binding motif in P. aeruginosa PA01
The chromium-binding gene sequence, iron hydrogenase 1 of Clostridium chromiireducens was retrieved from the UniProtKB 2022_03 database based on a gene name search. The gene sequence alignment was carried out between the iron hydrogenase 1 of C. chromiireducens and the ferredoxin of P. aeruginosa PA01 using BLAST (Basic Local Alignment Search Tool) to identify the chromium-binding motif. The correlation of amino acid residues of chromium binding motifs was determined for iron hydrogenase 1 and ferredoxin genes (Gattiker et al., 2002; Richard et al., 2007; Tian et al., 2019).
3 Results and discussion
3.1 Identification of metal binding protein in P. aeruginosa PA01 using MALDI-TOF/PMF-MS
The peak values for individual peptides were obtained through MALDI-TOF/TOF studies (Fig. 1). The manual interpretation of MS/MS data on charges ions at m/z 1534.7808 (MALDI), m/z 1858.8228 (MALDI), m/z 1874.8125 (MALDI), m/z 2236.2629 (MALDI), m/z 2298.0432 (MALDI) defined the partial peptide sequences. Based on the MASCOT search, the metal-binding protein of P. aeruginosa PA01 was identified as ferredoxin (Fig. 2), which was found to have an accession number of gi/15599966, the mass of 103928, a score of 26, matched queries of 5 with the sequence coverage of 23 %. Similarly, Yi-Min et al. (2003) identified copper-binding proteins, namely histone H2B, S100 calcium-binding protein, peroxiredoxin, and histone with sequence coverage of 28, 12, 30, and 22 %, respectively.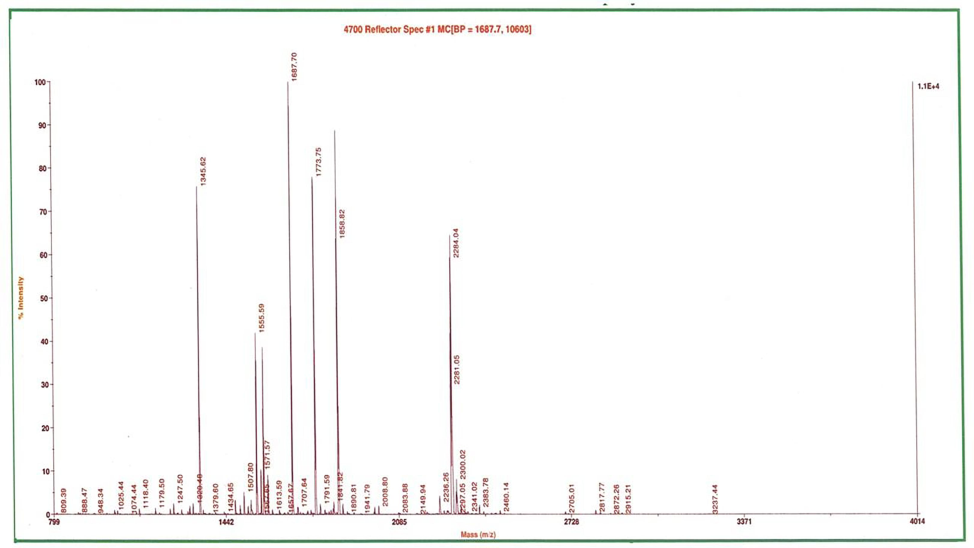
Identification of heavy metal binding protein of P. aeruginosa PA01 (Key: MALDI-TOF results showing the peak value for individual peptides present in heavy metal binding protein of P. aeruginosa PA01).

MASCOT search results indicating heavy metal binding protein as ferredoxin.
The structure of ferredoxin elucidated through SWISS-MODEL was found to have alpha helix and beta sheets (Fig. 3), which were responsible for the structural stability of the protein. Proteins with mainly local interactions (such as α-sheets) have rapid folding transitions, whereas proteins with more complex topologies (such as β-helices) usually fold more slowly. Thus, the protein folding helped to maintain the native topology and offered stability to the protein, as indicated in the earlier work done by Fersht, (2000). Ferredoxin also has cysteine, glutamic acid, aspartic acid, histidine, and methionine as predominant metal-coordinating amino acid residues (Fig. 4). These metal-coordinating amino acids would play a paramount role in copper and chromium binding in P. aeruginosa PA01. Similarly, Sano et al. (2006) reported that the isolated heavy metal binding protein of bacteria was known to contain several metal-coordinating amino acids like aspartic acid, glutamic acid, serine, and methionine that project from the water phase plays a significant role in the binding of metal ions.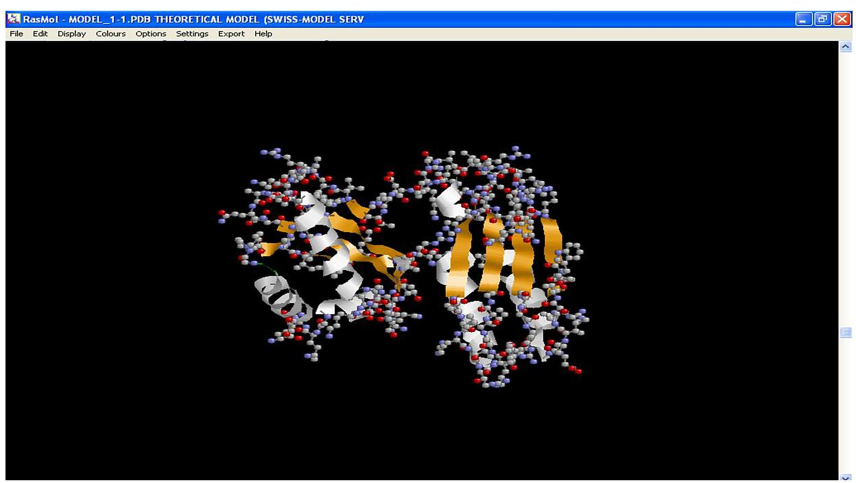
Structure of Ferredoxin present in P. aeruginosa PA01 obtained through SWISS-MODEL (Key: White – alpha helices; Orange – beta sheets; No coils found).
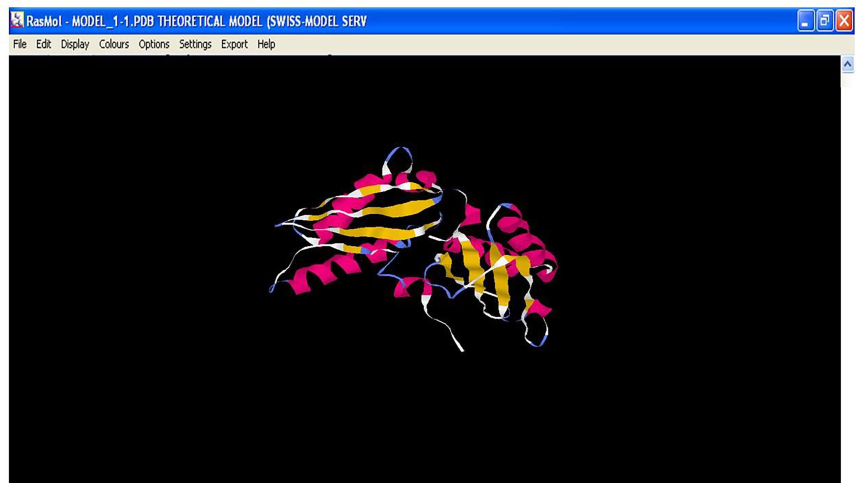
Structure of ferredoxin containing metal coordinating amino acids (Key: Predominant metal coordinating amino acids – Pink-Cysteine, White-Glutamic acid; Aspartic acid, Histidine, Methionine).
3.2 Validation of copper and chromium binding domain in ferredoxin of P. aeruginosa PA01
The metal binding motif of ferredoxin was validated, and it was observed to have a 4Fe-4S ferredoxin-type iron-sulphur binding domain (Suppl. Fig. 1) which was responsible for the coordination of both copper and chromium ions. It was found that Iron-sulphur (Fe-S) domains were responsible for protein folding and interaction of metallochaperones (deliver metal ions directly to the target protein and detoxify the metals) in the biological system, as stated by Ranawat and Rawat (2017). The earlier findings of Wittung-Stafshede (2002) also indicated that 4Fe-4S iron sulphur binding domain has a significant effect on protein folding, and further, the beta sheets of the 4Fe-4S cofactor offer stability for metal binding. Due to the stability of the 4Fe-4 s binding domain, the copper and chromium ions are so firmly bound to the binding sites of ferredoxin through intact protein folding. Zheng et al. (2021) reported that the 4Fe-4S ferredoxin-type iron-sulphur binding domain was associated with heavy metal resistance and removal by Pseudomonas cashew SRB007. Thus, it could be stated that 4Fe-4S clusters in ferredoxin of P. aeruginosa PA01 played a predominant role in protein folding and coordination with copper and chromium ions.
3.3 In-silico analysis of copper binding motif in P. aeruginosa PA01
There was a 67 % sequence similarity NosD gene sequence of P. aeruginosa (Suppl. Fig. 2) and the ferredoxin of P. aeruginosa PA01. This confirmed the existence of a copper-binding motif in P. aeruginosa PA01 (Suppl. Fig. 3). The correlation of copper binding motifs in the NosD gene sequence and ferredoxin gene showed the presence of homologous amino acid residues in them, which includes alanine, arginine, asparagine, aspartate, cysteine, glutamate, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine which are responsible for copper ion interaction (Table 1). Similarly, Acidothiobacillus caldus SM-1 and Acidothiobacillus caldus ATCC51757 showed a sequence similarity between 50 % and 90 %, respectively, for the putative copper resistance proteins like CusABC, CopB, CopZ, CueO of Acidothiobacillus ferroxidans (Navarro et al., 2013). The comparison with these Acidithiobacillus caldus and Serratia sp. bacteria highlights the diversity of metal resistance mechanisms and underscores the importance of understanding these mechanisms at a molecular level. Further, by identifying conserved domains and motifs, such as the 4Fe-4S ferredoxin-type iron-sulphur binding domain, and it possible to develop more effective bioremediation strategies that control the natural abilities of various bacterial species to detoxify and remove heavy metals from contaminated environments.
Name of the residue
Number of residues
in NosDNumber of residues
in ferredoxin
Alanine
16
104
Arginine
4
77
Asparagine
2
23
Aspartate
4
58
Cysteine
5
24
Glutamate
4
59
Glutamine
8
33
Glycine
11
72
Histidine
2
21
Isoleucine
6
41
Leucine
13
111
Lysine
1
31
Methionine
4
11
Phenylalanine
1
30
Proline
6
54
Serine
5
49
Threonine
6
47
Tryptophan
2
9
Tyrosine
2
20
Valine
7
64
3.4 In-silico analysis of chromium binding motif in P. aeruginosa PA01
The sequence alignment between the iron hydrogenase 1 gene sequence of C. chromiireducens (Suppl. Fig. 4) and the ferredoxin gene of P. aeruginosa PA01 showed an alignment score of 64.29 %, which revealed the existence of a chromium-binding motif in P. aeruginosa PA01 (Fig. 5). The correlation between the chromium-binding motif of iron hydrogenase 1 gene sequence and ferredoxin gene showed the presence of homologous amino acid residues of alanine, arginine, asparagine, aspartate, cysteine, glutamate, glutamine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tyrosine, and valine which are responsible for chromium ion interaction (Table 2). Deng et al. (2015) reported that the chromium-binding flavoprotein (ChrT) of Serratia sp. CQMUS2 had a sequence similarity of 85.6 % to ChrR gene of E. coli with homologous amino acid residues like Tyr128, Glu146, Arg125, and Tyr85, which were responsible for chromium ion interaction. Similarly, Sreeshma and Sudandiradoss (2021) have also observed that metal-coordinating amino acid residues like histidine, aspartic acid, and glutamic acid play a prominent role in Chromium VI biosorption by the potent strains like E. coli and Saccharomyces cerevisiae.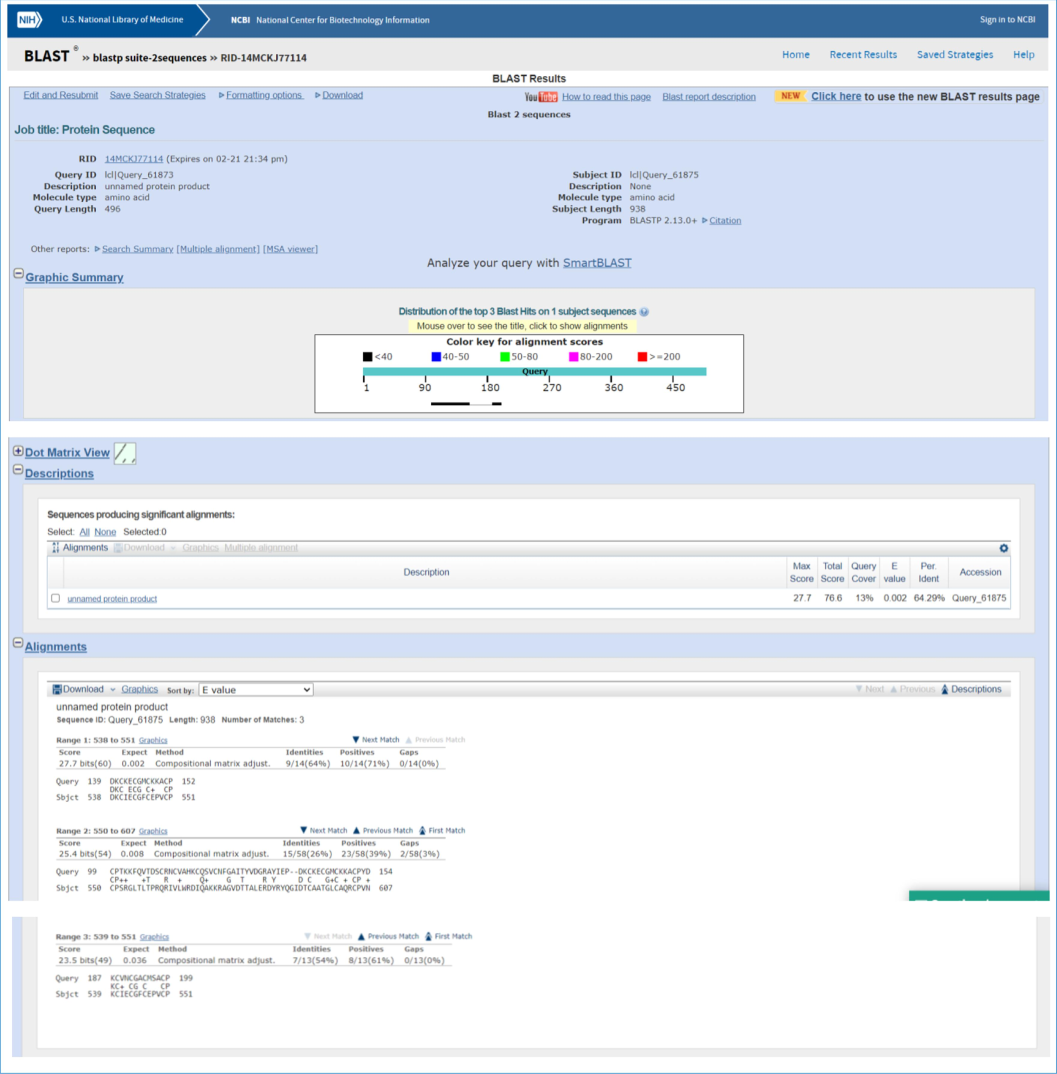
Sequence alignment of Iron hydrogenase 1 and ferredoxin.
Name of the residue
Number of residues in iron hydrogenase 1
Number of residues in ferredoxin
Alanine
48
104
Arginine
13
77
Asparagine
23
23
Aspartate
27
58
Cysteine
23
24
Glutamate
44
59
Glutamine
16
33
Glycine
33
72
Histidine
5
21
Isoleucine
29
41
Leucine
27
111
Lysine
48
31
Methionine
17
11
Phenylalanine
19
30
Proline
18
54
Serine
23
49
Threonine
24
47
Tryptophan
−
9
Tyrosine
16
20
Valine
43
64
An alignment score of 64.29 % between the copper-binding NosD gene and the ferredoxin gene, and a 67 % identity between the chromium-binding iron hydrogenase 1 gene and the ferredoxin gene, indicate a significant level of sequence similarity. In general, an alignment score above 50 % is considered to be indicative of functional or structural conservation. The presence of conserved domains, such as the 4Fe-4S ferredoxin-type iron-sulphur binding domain identified by ScanProsite, further supports the functional relevance of these alignment scores. This domain is critical for coordinating chromium and copper ions, suggesting that the ferredoxin gene in P. aeruginosa PA01 may have similar metal-binding capabilities. A high degree of sequence identity (e.g., 64.29 % or 67 %) often implies that the proteins share common ancestors and may perform similar functions. For metal-binding proteins, this similarity can indicate that they bind metals using similar mechanisms and structures. The alignment scores also suggest structural conservation between these proteins and also indicators of potential functional conservation.
The findings from the study on the copper and chromium binding capabilities of P. aeruginosa PA01, particularly through the ferredoxin protein, have significant implications for practical applications in metal-contaminated environments. Further, this strain used in bioaugmentation to enchase the natural bioremediation process. Finally, it contributes significantly to the development of effective, sustainable and cost-efficient bioremediation strategies for heavy metal contaminates environments.
4 Conclusions
The expression of metal-binding proteins in bacteria enhances heavy metal biosorption, and hence it plays a greater potential in metal binding. The heavy metal binding protein was isolated and identified as ferredoxin through MALDI-TOF/PMF-MS analysis. The protein sequence of ferredoxin validated in the PrositeScan tool revealed the presence of a 4Fe-4S cluster domain involved in the structural stability and coordination of copper and chromium with ferredoxin. Moreover, a comparative sequence alignment between the copper-binding NosD gene sequence and ferredoxin gene showed a sequence similarity of 67 %, and the sequence alignment of the chromium-binding iron-hydrogenase 1 gene sequence and ferredoxin gene showed a similarity of 64.29 %. Based on the sequence alignment, it was conferred that P. aeruginosa PA01 has both copper and chromium binding motifs, so it could be potentially exploited for enhanced coordination of copper and chromium ions from metal-polluted sites. Furthermore, it was observed that amino acids present in the ferredoxin of P. aeruginosa PA01 play a paramount role in copper and chromium binding. Thus, it could be concluded that acquiring heavy metal binding proteins like ferredoxin could be an ideal way to establish copper and chromium binding in a metal-polluted environment.
5 Consent to participate
All authors consented to participate.
CRediT authorship contribution statement
Shanmuga Priya Ramasamy: Validation, Methodology, Conceptualization. Priya Sundararajan: Writing – original draft, Data curation. Muthukrishnan Pallikondaperumal: Validation, Formal analysis. Ponmurugan Karuppiah: Visualization, Software. Saminathan Kayarohanam: Validation, Formal analysis. Natarajan Arumugam: Writing – review & editing. Ling Shing Wong: Writing – review & editing. Sinouvassane Djearamane: Validation, Methodology, Conceptualization.
Acknowledgment
The project was funded by Researchers Supporting Project number (RSP2025R143), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Computational approaches for de novo design and redesign of metal-binding sites on proteins. Biosci. Rep.. 2017;37(2)
- [CrossRef] [Google Scholar]
- Sequestered capture and desorption of hexavalent chromium from solution and textile wastewater onto low cost Heinsia crinita seed coat biomass. Appl. Water Sci.. 2020;10(1):1-15.
- [CrossRef] [Google Scholar]
- Cloning and sequence analysis demonstrate the chromate reduction ability of a novel chromate reductase gene from Serratia sp. Exp. Ther. Med.. 2015;9(3):795-800.
- [CrossRef] [Google Scholar]
- Transition-state structure as a unifying basis in protein-folding mechanisms: contact order, chain topology, stability, and the extended nucleus mechanism. PNAS.. 2000;97:1525-1529.
- [CrossRef] [Google Scholar]
- ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics.. 2002;1(2):107-108.
- [Google Scholar]
- Toxicity, mechanism, and health effects of some heavy metals. Interdiscip. Toxicol.. 2014;7(2):60-72.
- [CrossRef] [Google Scholar]
- Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol.. 2014;7(2):60-72.
- [Google Scholar]
- Biosorption and biotransformation of hexavalent chromium [Cr(VI)]: A comprehensive review. Chemosphere.. 2018;207:255-266.
- [CrossRef] [Google Scholar]
- The Biological and Environmental Chemistry of Chromium. VCH Publishers; 1993.
- Copper biochemistry and molecular biology. Am. J. Clin. Nutr.. 1996;63(5):797S-811S.
- [Google Scholar]
- Impact of heavy metals on the environment and human health: Novel therapeutic insights to counter the toxicity. J. King Saud Univ. Sci.. 2022;34(3):101865
- [Google Scholar]
- Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol.. 2021;12:643972
- [CrossRef] [Google Scholar]
- Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol. Res.. 2013;46(4):363-371.
- [CrossRef] [Google Scholar]
- Biosorption of Cr (VI) by three different bacterial species supported on granular activated carbon—a comparative study. J. Hazard. Mater.. 2008;153(1–2):799-809.
- [CrossRef] [Google Scholar]
- Metal-tolerant thermophiles: metals as electron donors and acceptors, toxicity, tolerance, and industrial applications. Environ. Sci. Pollut. Res.. 2017;25(5):4105-4133.
- [CrossRef] [Google Scholar]
- In silico identification of putative metal binding motifs. Bioinformatics.. 2007;23:267-271.
- [CrossRef] [Google Scholar]
- Cloning of a heavy-metal-binding protein derived from activated-sludge microorganisms. Appl. Environ. Microbiol.. 2006;72(9):6377-6380.
- [CrossRef] [Google Scholar]
- Physiological role of nickel and its toxic effects on higher plants. Russ. J. Plant Physiol.. 2006;53(2):257-266.
- [Google Scholar]
- Identification of metal binding motifs in protein frameworks to develop novel remediation strategies for Hg 2+ and Cr (VI) Biometals.. 2021;34:621-638.
- [CrossRef] [Google Scholar]
- Heavy metal toxicity and the environment. Mol., Clin. Environ. Toxicol.. 2012;101:133-164.
- [Google Scholar]
- Prediction of protein–metal ion-binding sites using sequence homology and machine-learning methods. Adv. Bioinforma. Chem.. 2019;1(1):025-036.
- [CrossRef] [Google Scholar]
- Role of cofactors in protein folding. Acc. Chem. Res.. 2002;35(4):201-208.
- [CrossRef] [Google Scholar]
- Heavy metal exposures on freshwater snail Pomacea insularum: Understanding its biomonitoring potentials. Appl. Sci.. 2023;13(2):1042.
- [CrossRef] [Google Scholar]
- Identification of metal-binding proteins in human hepatoma lines by immobilized metal affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1306-1318.
- [CrossRef] [Google Scholar]
- Analysis of copper-binding proteins in rice radicles exposed to excess copper and hydrogen peroxide stress. Front. Plant Sci.. 2016;7:1216.
- [CrossRef] [Google Scholar]
- Pseudodesulfo vibrio cashew sp. Nov., a novel deep-sea sulfate-reducing bacterium, linking heavy metal resistance and sulfur cycle. Microorganisms. 2021;9:429.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103552.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







