Biodegradation of atrazine using selected marine bacteria: Possibilities for treating pesticide - contaminated wastewater
⁎Corresponding author. ebtesamelbestawy@alexu.edu.eg (Ebtesam El Bestawy) ebtesamelbestawy@yahoo.com (Ebtesam El Bestawy) ebtesamelbestawy@gmail.com (Ebtesam El Bestawy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
The use of pesticides including atrazine can cause detrimental problems to the environment. Atrazine, 2-chloro-4-(ethylamine)-6- (isopropyl amine)-s-triazine, is one of the widely used herbicides. In this work, thirteen pure bacterial strains were isolated from water and sediments collected from different sites along Alexandria Mediterranean Coast and were evaluated for their efficacy to biodegrade atrazine at three elevated concentrations (I: 109, II: 299 & III: 438 mg/L) for 7 days. Atrazine residues were determined using gas chromatography (GC). Marine isolates exhibited very high atrazine biodegradation with removal efficacy ranging between 15.79 and 75.49, 77.97–97.13 and 27.4–87.6% at three elevated concentrations, respectively. The results indicated that 5 of the isolates (E7, 8, 9, 11 and 13) were the most efficient, and active as atrazine bio-degraders. They were affiliated as Bacillus pacificus strain MCCC 1A06182 (E7 and E8), Bacillus cereus strain ATCC 14579 (E9) and Bacillus paramycoides strain MCCC 1A04098 (E11 and E13). Results obtained provided evidence that marine environment is a natural, rich and renewable source of bacteria with marvelous metabolic capabilities for efficient bioremediation of atrazine-contaminated aquatic environments or wastewater.
Keywords
Atrazine biodegradation
Aquatic media
Bacillus
Marine ecosystem
Pesticide pollution
1 Introduction
Around 30 to 60% of pesticides will reach the soil eventually end up in the soil based on their use (Zhu et al., 2019; Ofaim et al., 2020). Pesticides and their metabolites have extremely drastic effects on human health, wildlife and the environment (food webs, soil, and aquatic organisms) and increase with increasing the concentration applied. They cause a major disruption to the biological inhabitants of the soil leading to disruption of all soil functions (Wirsching et al., 2020). This is attributed to the fact that the soil microbes and invertebrates have combined and critical effects to maintain soil functions, enhance food production and human health (Brussaard, 2021).
The biotic and abiotic processes result in the transformation and changes in the chemical composition of pesticides. In addition to pesticide solubility and microbial population, biodegradation of pesticides is highly influenced by various soil conditions including pH, temperature, moisture, and organic matter content (Houjayfa et al., 2020).
Atrazine, 2-chloro-4-(ethylamine)-6-(isopropyl amine)-s-triazine, is one of the widely used herbicides, although prohibited in the European Union in 2004 (Billet et al., 2019). It is a non-polar toxic compound and considered as a serious environmental contaminant that pollutes water resources and soil worldwide due to its long-term use in crop production (Jakinala et al., 2019; Li et al., 2019). Being both inexpensive and effective, atrazine is applied as pre- and post-emergence herbicide for the protection of major crops such as conifers, macadamia nuts, pineapples, chemical fallows as well as for industrial weed control especially during sorghum, corn production and sugarcane (El-Bestawy et al., 2013). Environmental fate of Atrazine (half-life: 13–261 day) depends on many effectives including the attachment to polar soil colloids, uptake, transport through the runoff and leaching and biodegradation. The original applied compound and biodegradation metabolites were frequently often reach raw drinking water and/or washed out from the root zone to ground water resources, particularly when applied prior to irrigation or heavy rainfall (Espín et al., 2020; Carpio et al., 2021). It is detected decades after application, sometimes at concentrations exceeding the maximum permissible limit (3 mg/L) according to the United States Environmental Protection Agency (USEPA) (Qu et al., 2020). Therefore, deleterious environmental consequences have emerged (El-Bestawy et al., 2013; El-Bestawy et al., 2014).
Although it is less toxic to humans compared to other chlorinated herbicides, problems arise such as reduced biodiversity, damaged future crops and food contamination (Fernandes et al., 2020, Carpio et al., 2021). It has long-term reproductive and endocrine-disrupting effects, interrupts regular hormonal functions and causes defects in human birth, reproductive tumors, and weight loss in both humans and amphibians (Ma et al., 2017). Atrazine leads to low birth weights, low sperm counts in men, menstrual problems and known as a probable human carcinogen (Houjayfa et al., 2020; Zhang et al., 2019).
As such, the atrazine removal from the environment is considered imperative and is of growing public health concerns. Atrazine removal from contaminated soils, sediments, and water involves either microorganisms mediated biotic transformation processes (He et al., 2019; Lihl et al., 2020), or abiotic processes via photochemical and chemical reactions (Rózsa et al., 2019; Shawky et al., 2020). However, bioremediation involving microbial communities is more effective and remains the most promising approach used for pesticide degradation (Houjayfa et al., 2020; Wirsching et al., 2020). Due to the large-scale agricultural utilization in Saudi Arabia, especially in corn cultivation, and the dangerous atrazine toxic effects to both humans and the environment, new naturally occurring microorganisms have been searched and sought to completely degrade atrazine. Marine environment is well known as a very attractive and rich natural resource for macro- and microorganisms with potent capabilities ranging from production of many bioactive compounds (antibiotics, anticancer and cardiovascular agents) to biodegradation of toxic environmental pollutants (Fenical, 2020; Lyu et al., 2021; Zhu et al., 2021). Therefore, the present study aimed to explore the ability and efficiency of selected marine bacterial isolates to degrade atrazine. The outcome of this research is to effectively remediate and control the widespread atrazine pollution in the environment by using potent indigenous marine bacteria with or without exogenous variants.
2 Materials and methods
2.1 Sampling
Water and sediment samples were collected from 4 chemically and biologically different sites along Alexandria Coast, extending from Abu-Qir in the Far East through Sidi Gaber, El-Selsela until El-Anfoushi in the central part of the coast (Fig. 1). Abu-Qir, the most industrialized area in Alexandria, has a total area of about 38,000 ha. It is a semi-circular, shallow water area with a depth ranging from less than one meter along the shore, increasing gradually to a maximum of 15 m around the middle (Maged and Mikhail, 2008). Sidi Gaber and El-Shatby are used for swimming and recreational purposes as well as residential area (El-Bestawy et al., 2011, 2017). In the central part of Alexandria Coast, the Eastern and Western harbours as well as El-Anfoushi lie. They are shallow, protected embayments, semi enclosed circular basins. The four sites were selected based on type and extent of pollution prevailed from industrial (Abu Qir) and domestic (Sidi Gaber, El-Selsela and El-Anfoushi) discharges.
- Alexandria Coast and the four sampling sites (marked yellow).
Samples were collected according to specifications set by the International Organization for Standardization (ISO 5667/6, 2016) and ISO 5667/10 (2020). Samples collection was carried out at a depth of 25–35 cm below the seawater surface, 50 m off-shore in 250 mL-glass screw autoclaved capped bottles with wide mouthed openings. Special stainless steel sampling rod was used for this purpose. Bottles were opened at the time of collection. Sediment samples were collected using a piston corer at a depth of 20 cm and transferred in sterilized plastic bags that were tightly closed. All samples were analysed in triplicates. Samples were maintained on ice in an ice box at 4 °C, while being transported to the laboratory and were processed within 2 to 3 h post collection. These samples were used to isolate bacteria acquired high resistance, enzymatic, degradative capabilities and possess astonishing metabolic activities from their polluted environments.
2.2 Isolation of marine bacteria
Marine bacteria were isolated from water and sediment samples on nutrient agar plates (prepared with filtered marine water) using pour plate technique of the standard plate count method (Baird et al., 2017). Purification of heterotrophic bacterial isolates was performed using streaking method on nutrient agar plates (NA, Oxoid, England), transferred onto NA slants and incubated at 37 °C. Thirteen pure isolates (designated E 1–13) were obtained and kept refrigerated for later use.
2.3 Atrazine stock solution
Atrazine (2-chloro-4-(ethylamine)-6-(isopropyl amine)-s-triazine) stock solution was prepared by dissolving technical atrazine (80% active ingredient) in deionized water reaching a final concentration of 1,000 mg/L. Atrazine stock solution was sterilized by filtration using 0.22 µm polycarbonate membrane (Millipore, Waatman Limited, Maidstone, England) to avoid precipitation or chemical changes during autoclaving (Baird et al., 2017).
2.4 Synthetic wastewater
Concentrated synthetic wastewater seeded with atrazine was used in the bioremediation assays. It was prepared by dissolving the following chemical ingredients (g) in in one L distilled water: NaCl (40.7), CaCl2·2H2O (0.37), H2MoO4 (0.31), Tripton (122.1), Na2SO4 (4.46), MnSO4 (0.57), K2HPO4 (4.46), MgCl2·6H2O (0.37), and NaOH (0.08) (Özbelge et al., 2005; El-Bestawy et al., 2013). After that, a one liter of the working wastewater was prepared by adding 10 mL concentrated synthetic wastewater to 990 mL distilled water and was autoclaved at 121 °C for 20 min.
2.5 Bioremediation bioassays
Pure marine bacterial isolates were investigated for the removal of atrazine in liquid culture from synthetic wastewater to be able to identify the most promising candidates. They were individually activated by transferring a loop full into 250 mL flask contained 200 mL nutrient broth (NB) and incubated at 37 °C and 150 rpm shaking for 24 h. Activated inoculates (200 mL each) were individually transferred into one-liter conical flasks containing 800 mL synthetic wastewater reaching a final volume of one liter (3 replicas for each isolate), with definite aliquots of atrazine stock solution reaching elevated atrazine levels (I: 100, II: 250 & III: 500 mg/L) at pH 7. Immediately after bacterial inoculation of wastewater, all cultures were aseptically sampled and analyzed for total bacterial counts at the starting point (data not shown). In addition to the inoculated wastewater (39 cultures), three 1-L un-inoculated synthetic wastewater flasks amended with the 3 atrazine levels were prepared and used as control. Atrazine was determined in all inoculated (treated) and un-inoculated (control) cultures immediately after inoculation to determine the start-up concentration, after which they were incubated at room temperature ≈ 25–30 °C (late spring). The experiment was performed for 7 days and samples were aseptically collected at 24 h interval. Fifty mL from each culture was drawn and residual levels of atrazine were assayed at each exposure time. Removal efficiencies of atrazine by the tested bacteria were calculated to determine the efficacy of the remediation process and determine and identify the best degrading bacterial isolates (El-Bestawy et al., 2020, 2021):
RC = Residual Concentration after Treatment at each Exposure Time.
2.6 Atrazine residues analysis
2.6.1 Extraction, clean up and determination of atrazine residues
Extraction of atrazine from treated and control wastewater was done as previously described (EL-Saeid and Alghamdi, 2020). Cleanup of the extracted residue was done using the previously published method of Wang et al. (2020b). Determination of atrazine residues in wastewater samples was performed using gas liquid chromatography (GLC) according to previously described method (El-Bestawy et al., 2013, 2014 & 2017; EL-Saeid and Alghamdi, 2020).
2.7 Atrazine recovery efficiency study
2.7.1 Atrazine recovery
To define the efficiency of the determination method for the recovery of atrazine, untreated samples of water were spiked with known quantities of atrazine active ingredient solutions. Spiked samples were then undergone atrazine extraction, cleaning-up and determination.
2.7.2 Preparation of blank solution
The solvent and the anhydrous sodium sulphate used in the fractioning and clean up were checked for purity of atrazine and for the presence of any traces of the atrazine before their use.
2.8 Molecular characterization of marine bacteria
Five bacterial isolates out of the thirteen tested isolates showed the highest atrazine biodegradation capability during bioremediation processes, therefore, they were molecularly characterized. Total genomic DNA was extracted from 5 mL overnight NB culture of the purified isolates according to the method described by Sambrook et al. (1989). Then fragments of the 16S rDNA gene were amplified using the primers B341F (5′-CCTACGGGA GGCAGC), and 1392R (5′-ACGGGCGGTG TGTRC-3′) as described by Ausubel et al. (1999). Each of these purified products was sequenced and the resulting DNA sequences were phylogenetically analyzed using the BLAST search program (Hall, 1999).
2.9 Statistical analysis
Mean (3 replicates) and standard error values were determined for all parameters and the results were expressed as mean ± standard error. The data were analyzed using one-way analysis of variance (ANOVA) followed by Duncan multiple comparison (SPSS for windows, Version 20) in order to compare treated groups with control. The differences were considered significant at P < 0.05 (95% of the confidence level).
3 Results and discussion
Atrazine was tested at elevated concentrations as enrichment and acclimatization approach to select the resistant isolates from one side as well as testing bacterial abilities for handling high levels of atrazine to simulate situations during pollution accidents or disaster from the other.
3.1 Molecular characterization of the most active bacterial isolates
The present study amid to evaluate the efficacy of thirteen marine bacterial isolates for atrazine degradation. Five of those isolates (E 7, 8, 9, 11 and 13) had the highest degradation capabilities even at the highest tested concentration. Table 1 compiles Gen Bank accession numbers of the highest sequence similarity of the most active atrazine degraders as well as the closest neighbor(s) to their 16S rDNA gene partial sequences. Sequences of the five isolates were affiliated to members of the genus Bacillus. Isolates E 7 and 8 were identified as Bacillus pacificus strain MCCC 1A06182 (100 and 99.85% sequence similarity), respectively, isolate E 9 was identified as Bacillus cereus strain ATCC 14579 (similarity 99.9 %) while both isolates E 11 and 13 were identified as Bacillus paramycoides strain MCCC 1A04098 (98.44 and 99.16 % similarity), respectively (Table 1). The phylogenetic relationships of the experimental isolates and closely related species were analyzed using the multi-sequence alignment program (MEGA 5) to evaluate the phylogenic relationship of the experimental isolates and their closely related species and the data are shown in the phylogenic tree (S1), while their 16S ribosomal RNA partial sequences and alignments are presented in the supplementary material (S2).
| Similarity% | Gen Bank accession of the Nearest Neighbor | Nearest Neighbor(s) | Isolate No. |
|---|---|---|---|
| 100 | NR157733.1 | Bacillus pacificus strain MCCC 1A06182 | 7 |
| 99.85 | NR157733.1 | Bacillus pacificus strain MCCC1A06182 | 8 |
| 99.9 | NR074540.1 | Bacillus cereus strain ATCC 14,579 | 9 |
| 98.44 | NR157734.1 | Bacillus paramycoides strain MCCC 1A04098 | 11 |
| 99.16 | NR157734.1 | Bacillus paramycoides strain MCCC 1A04098 | 13 |
It was reported that microorganisms such as Pseudomonas, Bacillus and Arthrobacter are well known natural degraders to aromatic compounds like aromatic amino acids, phenols, or quinones (Rotta et al., 2018; Kapoor and Saini, 2019; Kundu et al., 2019; Wang et al., 2020a) prevailed in pharmaceuticals wastewater (El-Bestawy et al., 2019) where they have evolved catabolic pathways. Results of the present study confirmed the astonishing efficiency of the tested Bacillus spp. for atrazine degradation and removal from contaminated wastewater, which is consistent with and supported by many workers (El-Bestawy et al., 2013, 2014; Swapna et al., 2016; Khatoon and Rai, 2018; Jakinala, et al., 2019), especially those previously exposed to the herbicide or its analogues (Houjayfa et al., 2020; Li et al., 2019; Ma et al., 2017; Yang et al., 2018; Ye et al., 2016). Atrazine biodegradation was remarkably enhanced through bioaugmentation of exogenous potent atrazine degraders into contaminated media and biostimulation of the indigenous microorganisms (Zhu et al., 2019; El-Bestawy et al., 2014; El-Bestawy and Zabermawi, 2017).
3.2 Atrazine biodegradation at the lowest tested concentration (I)
Biodegradation of atrazine at the lowest tested concentration I (initial concentration (IC): 109 mg/L) showed regular trend of decreasing the residual concentration (RC) with time reaching the lowest after 7 exposure days by all the tested bacteria except isolate 4 (after 3 exposure days) with no clear variations in its metabolic activity till the end of the experiment. As shown in Table S3 and Fig. 2, the highest REs % (Removal Efficiency) values of atrazine at concentration I (ranged between 15.79 and 75.49 %) were achieved by the tested bacteria at the last exposure day with relative variations among them. REs of 75.49, 75.41, 70.61 and 67.90 % were reached by isolates E 9, 11, 6 and 8, respectively. Isolates E 5, 3, 12, 4 and 13 recorded intermediate atrazine removals (62.77, 57.0, 51.18, 51.49 and 50.18 %, respectively) compared to the other tested bacteria during the treatment duration. However, isolates E 10, 1, 7 and 2 were the least active in atrazine biodegradation recording 47.83, 36.93, 33.09 and 21.23 % RE, respectively. On the other hand, the control recorded very low atrazine removal (15.79 %) after 6 days confirming the active role of the tested bacteria towards atrazine biodegradation.
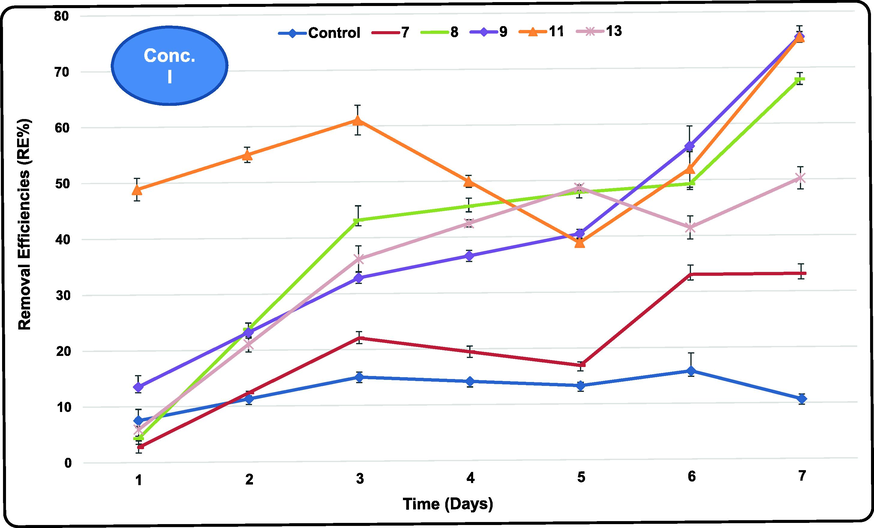
- Removal efficiencies (%) of Atrazine at the lowest tested concentration using the most efficient marine bacterial isolates for 7 exposure days.
3.3 Atrazine biodegradation at the intermediate tested concentration (II)
Atrazine II recorded 299.0 ± 2.02 mg/L as the initial IC (Table S2). Biodegradation showed irregular removal trend by most of the tested cultures except for isolates E 1, 2, 4, 8 and 13 that showed their highest RCs after one exposure day followed by regular decrease reaching their lowest RCs after 6 and 7 exposure days. At concentration II, the tested cultures including the control showed generally higher removal ranges of atrazine (Fig. 3) compared to those obtained by the same cultures at the lowest concentration I. The highest achieved atrazine REs ranged between 77.97 and 97.13 %. REs of 97.13, 89.86, 87.80, 86.52 and 86.10 were achieved by isolates E 8, 9, 11, 13 and 7, respectively. Other tested cultures reached considerable atrazine removal ranged between 77.97 % by E 5 and 83.36 % by E 1. Surprisingly, at this concentration, the control culture recorded considerably high removal range (65.52 % after 24 h to 88.44 % after 6 days).
- Removal efficiencies (%) of Atrazine at the intermediate tested concentration using the most efficient marine bacterial isolates for 7 exposure days.
3.4 Atrazine biodegradation at the highest tested concentration (III)
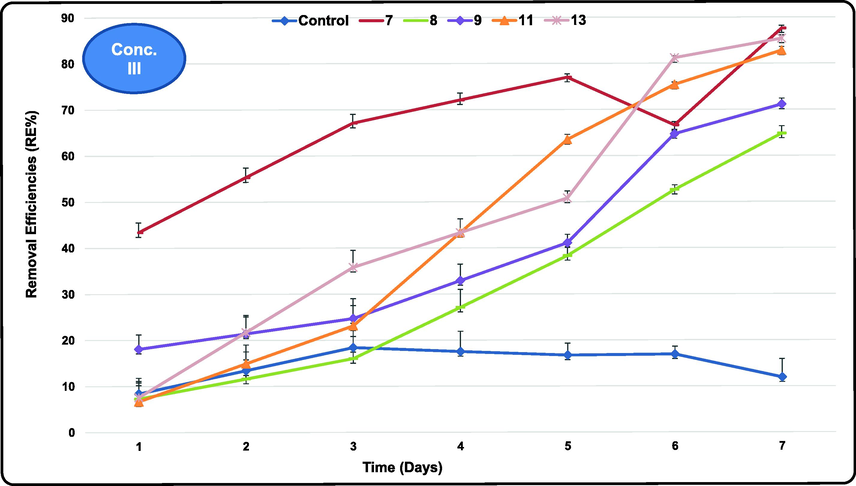
Atrazine III recorded 438.8 mg/L as the IC (Table S5). Atrazine biodegradation showed very clear and regular trend at this concentration with decreasing concentration with time reaching the lowest RCs after 7 exposure days. Unexpectedly, very high removals exceeding 85% were obtained by some of the tested cultures at this very high atrazine level. The highest atrazine REs ranged between 27.4 and 87.6 % were achieved by E 7, 13 and 11 (87.6, 85.4 and 82.8 %, respectively). Seven isolates (E 12, 5, 10, 9, 8, 6 and 2) had intermediate REs (75.8, 73.8, 73.0, 71.1, 64.8, 57.3 and 56.5 %, respectively) compared to the other tested bacteria. While isolates E 1, 4 and 3 reached 45.8, 31.8 and 27.4%, respectively that considered relatively the minimum achievements of the highest atrazine RE range (Fig. 4). Active atrazine degrading cultures at this concentration considered highly resistant against its toxicity and possess all the required enzymes for its biodegradation. However, as expected the control (unseeded wastewater) showed the least removal range (8.4–18.4 %) after 1 and 3 exposure days. The lowest atrazine RCs in the treated wastewater recorded 26.7, 8.6 and 63.9 mg/L achieved by isolates E 9, 8 and 13 at I, II and III atrazine concentrations tested, respectively. Such RCs are much higher than the maximum permissible limit (MPL) of atrazine (≤0.1 mg/L) because atrazine initial concentrations tested in the present study were also very high. This limit is set by environmental laws in Egypt and Saudi Arabia to protect the aquatic life from any ecological disturbances and from hazardous discharges from the soil environments. However, the five promising atrazine degraders could be immobilized using any supporting media and used as a continuous treatment system as individual or serial units.
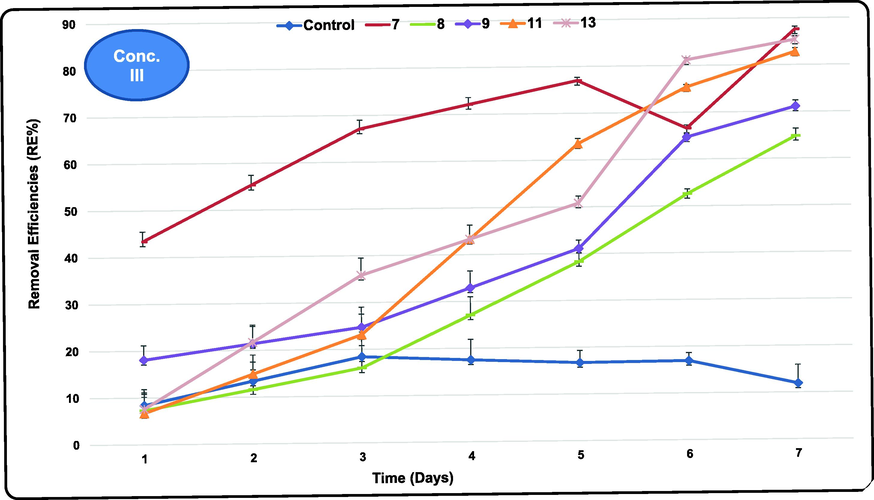
- Removal efficiencies (%) of Atrazine at the highest tested concentrations using the most efficient marine bacterial isolates for 7 exposure days.
Statistical analysis (Table 2) revealed that at the first atrazine concentration isolate E 9 is the most effective for atrazine degradation and significantly (P < 0.05) different compared to the control and other tested cultures. Values denoted by different letters within same column represent significant differences (P < 0.05). There was a significant decrease in atrazine concentration after treatment by all the tested strains when compared to the control. Wastewater treated with isolates E 6, 8, 9 and 11 showed significant decrease in atrazine concentration compared to the other isolates confirming that such isolates have high atrazine removal efficiency rather than other isolates. At the intermediate atrazine concentration, there was a significant (P < 0.05) decrease in atrazine concentration after treatment using isolates E4, 8, 9 and 13 compared to the control and all other tested groups but treated groups of isolates E1, 2, 3, 5, 7, 10 and 12 showed significant increase in atrazine concentration as compared to the control groups. On the other hand, the treated groups of isolates E 6 and 11 showed insignificant changes in comparison to the control. Finally, at the highest tested atrazine concentration, groups treated with isolates E 7, 11 and 13 showed significant (P < 0.05) decrease in atrazine concentration as compared to all other treated groups. Moreover, there was a significant decrease in atrazine concentration in all the treated groups (by different isolates) compared to the control group.
| Concentration | Isolate | ||
|---|---|---|---|
| III | II | I | |
| 387.71 ± 4.0a | 54.05 ± 0.57e | 97.4 ± 0.57a | Control |
| 239.28 ± 2.4d | 65.07 ± 0.63c | 68.8 ± 0.63c | 1 |
| 191.25 ± 1.9e | 58.90 ± 0.66d | 85.9 ± 0.66b | 2 |
| 319.71 ± 3.3b | 60.25 ± 0.54d | 46.9 ± 0.54ef | 3 |
| 300.06 ± 3.1c | 50.13 ± 0.52f | 68.3 ± 0.52c | 4 |
| 115.33 ± 1.2gh | 82.21 ± 0.98a | 40.6 ± 0.98 fg | 5 |
| 187.94 ± 1.9e | 53.03 ± 0.63ef | 32 ± 0.63gh | 6 |
| 54.56 ± 0.6j | 71.04 ± 0.84b | 72.9 ± 0.84bc | 7 |
| 154.74 ± 1.6f | 8.60 ± 0.12i | 35 ± 0.12gh | 8 |
| 127.16 ± 1.3 g | 30.42 ± 0.35 h | 26.7 ± 0.35 h | 9 |
| 118.74 ± 1.2gh | 58.74 ± 0.69d | 56.9 ± 0.69d | 10 |
| 75.82 ± 0.8i | 54.65 ± 0.64e | 26.8 ± 0.64 h | 11 |
| 106.60 ± 1.1 h | 61.61 ± 0.73d | 53.2 ± 0.73de | 12 |
| 64.08 ± 0.7ij | 40.43 ± 0.46 g | 54.3 ± 0.46de | 13 |
Results are Expressed as Mean of 3 Replicates ± SE.
RC mean values denoted by different letters (a-j) within same column.
represent significant differences (at P < 0.05).
Means with the same letters are not statistically significant.
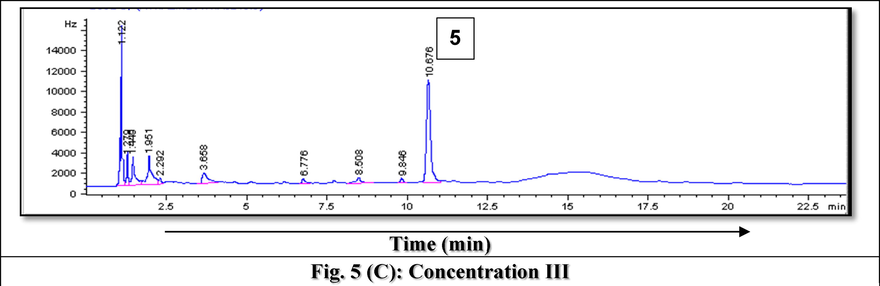
Fig. 5(A-C) represents example GC chromatograms of atrazine biodegradation at the three tested concentrations where average recovery from spiked samples recorded 89%. Biodegradation results of atrazine by the 13 tested isolates at the tested elevated concentrations concluded that 5 isolates (7, 8, 9, 11 and 13) considered the most resistant, efficient and active in the removal of the target herbicide, atrazine, therefore, they were molecularly identified.
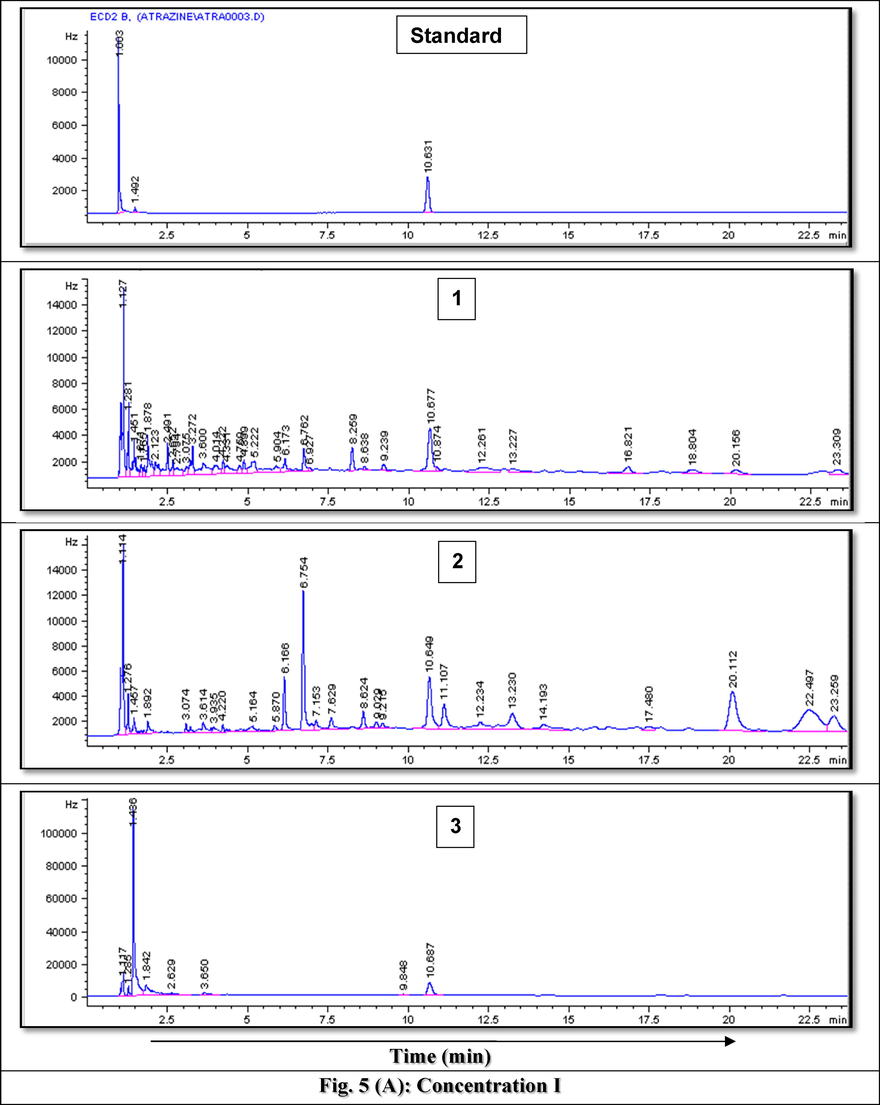
- GC Chromatograms of Atrazine residues after rreatment using A) Bacillus paramycoides strain MCCC 1A04098 at the lowest tested concentration after 1) 3 days, 2) 5 days and 3) 7 days, B) Bacillus pacificus strain MCCC 1A06182 at the intermediate tested concentration after 1) 2 days, 2) 3 days, 3) 5 days and 4) 6 days and C) Bacillus cereus strain ATCC 14,579 at the highest tested concentration after 1) 2 days, 2) 3 days, 3) 5 days, 4) 6 days and 5) 7 days.
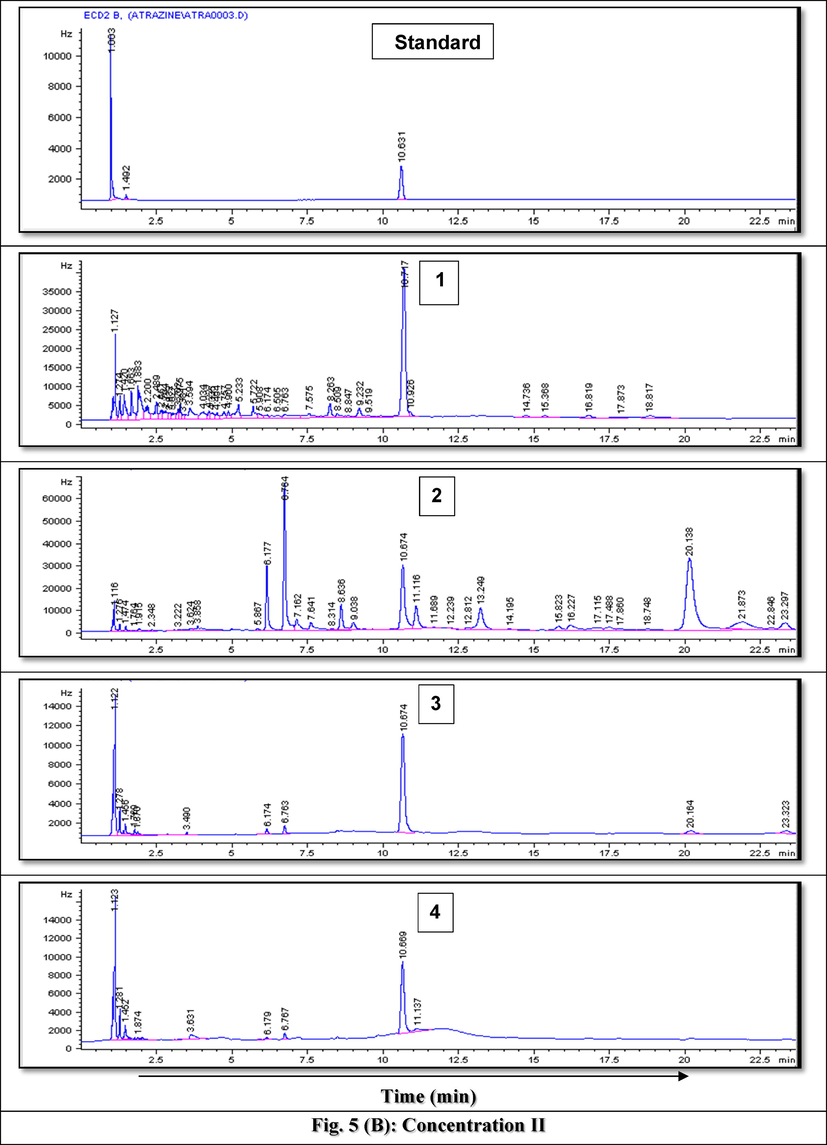
- GC Chromatograms of Atrazine residues after rreatment using A) Bacillus paramycoides strain MCCC 1A04098 at the lowest tested concentration after 1) 3 days, 2) 5 days and 3) 7 days, B) Bacillus pacificus strain MCCC 1A06182 at the intermediate tested concentration after 1) 2 days, 2) 3 days, 3) 5 days and 4) 6 days and C) Bacillus cereus strain ATCC 14,579 at the highest tested concentration after 1) 2 days, 2) 3 days, 3) 5 days, 4) 6 days and 5) 7 days.
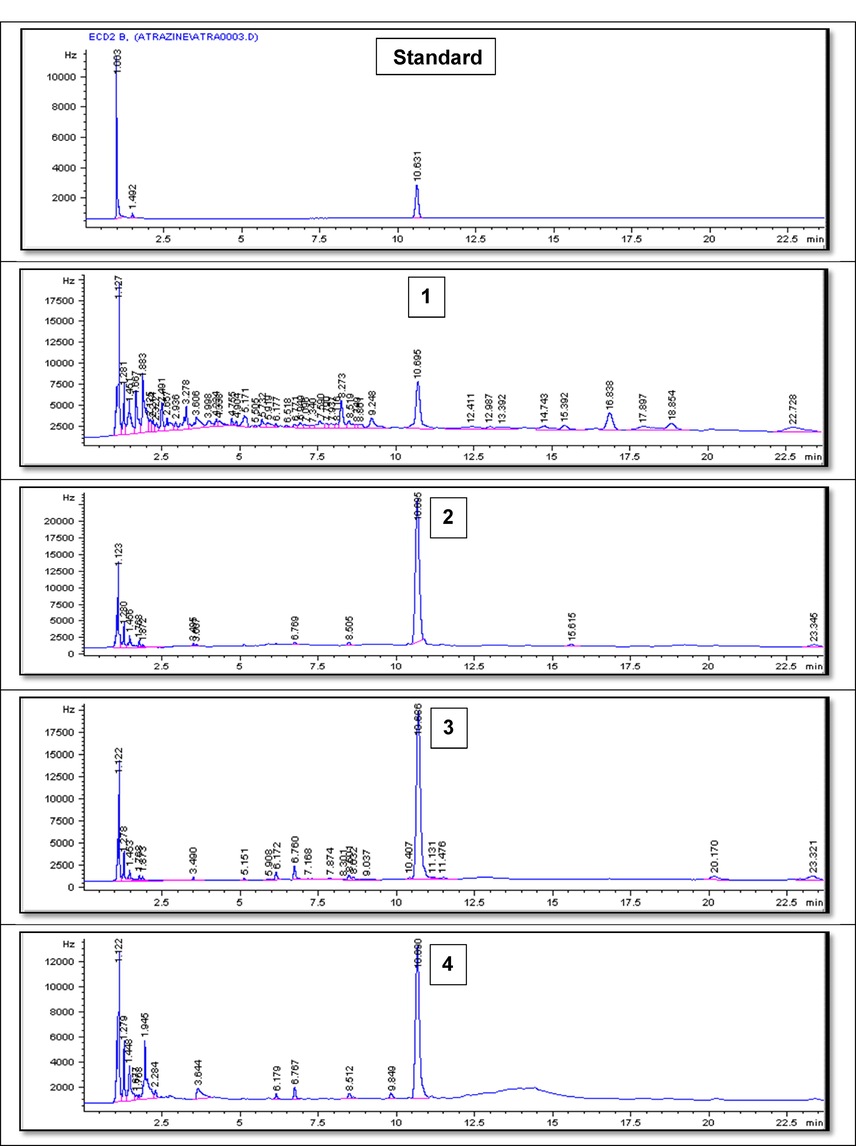
- GC Chromatograms of Atrazine residues after rreatment using A) Bacillus paramycoides strain MCCC 1A04098 at the lowest tested concentration after 1) 3 days, 2) 5 days and 3) 7 days, B) Bacillus pacificus strain MCCC 1A06182 at the intermediate tested concentration after 1) 2 days, 2) 3 days, 3) 5 days and 4) 6 days and C) Bacillus cereus strain ATCC 14,579 at the highest tested concentration after 1) 2 days, 2) 3 days, 3) 5 days, 4) 6 days and 5) 7 days.
- GC Chromatograms of Atrazine residues after rreatment using A) Bacillus paramycoides strain MCCC 1A04098 at the lowest tested concentration after 1) 3 days, 2) 5 days and 3) 7 days, B) Bacillus pacificus strain MCCC 1A06182 at the intermediate tested concentration after 1) 2 days, 2) 3 days, 3) 5 days and 4) 6 days and C) Bacillus cereus strain ATCC 14,579 at the highest tested concentration after 1) 2 days, 2) 3 days, 3) 5 days, 4) 6 days and 5) 7 days.
Batch treatment demonstrated a basic trend of increasing atrazine degradation in relation to increasing exposure time. Five strains belonged to 3 Bacillus spp. (B. pacificus, B. cereus and B. paramycoides) exhibited very high atrazine biodegradation ability with the highest achieved atrazine removal ranges recorded 15.79–75.49, 77.97–97.13, and 27.4–87.6% at 109, 299 and 438 mg/L atrazine IC, respectively at room temperature without shacking. These results indicated that these strains could utilize atrazine as a sole carbon, nitrogen and energy source with remarkably higher capability compared to other workers. For example, other studies reported atrazine degradation by other microorganisms at lower ICs of 50 mg/L by Citricoccus sp. strain TT3 after 66 h (Yang et al., 2018), 100 mg/L by Ensifer sp., Shewanella sp. YJY4 and Rhodococcus sp. BCH2 after 30, 36 h and 7 days, respectively (Ma et al., 2017; Ye et al., 2016; Khatoon and Rai, 2018). They were isolated from industrial soil, cornfield soil and atrazine long-term treated soil. These examples clearly confirmed the superiority of marine Bacillus spp. isolated during the present study for degradation of atrazine reaching as high as 75.49, 97.13 and 87.6% at atrazine initial concentrations of 109, 299 and 438 mg/L, respectively in 7 days. As shown here the peak REs of atrazine were achieved at concentration II and continued to III (97.13 and 87.6%, respectively). This may be attributed to the stimulation in their metabolic activity and induction of the required degradation enzymes with increasing atrazine concentration from I to II and III (almost 3 and 4 folds, respectively) by the resistant and atrazine degrading isolates. This is confirmed by atrazine removal range at concentration III (27.4–87.6 %) where 3 resistant isolates E 7, 13 and 11 could achieve 87.6, 85.4 and 82.8 %, respectively which considered remarkable removal at such high initial concentration tested.
Moreover, such degradation efficiencies were achieved at ambient temperature without any other requirements or adjustment (i.e. pH, agitation, temperature, carbon and nitrogen amendments…etc.) as with other workers (Yang et al., 2018; Ma et al., 2017; Khatoon and Rai, 2018), which make the present selection even better for decontamination of open environments as well as industrial wastewater. However, the lowest atrazine RCs in the treated wastewater (26.7, 8.6 and 63.9 mg/L at I, II and III tested atrazine concentrations, respectively) were not compatible with the MPL set by environmental regulation. But this problem can be solved reaching acceptable limits for safe discharge by immobilization of the promising atrazine Bacillus spp. degraders forming biofilm on or in suitable carriers to be used as a continuous treatment system in individual or serial units. This suggestion is supported by Khatoon and Rai (2018) who reported that α-Fe2O3 immobilized Bacilli cells degraded atrazine at a wide range of physicochemical factors (temperature: 20 to 45 °C, and pH: 4.0 to 9.0, atrazine concentration: 50 to 300 mg/L and stirring speed: 50 to 300 rpm). This illustrates that Bacilli cells modified by fixation as biofilm or decorated with specific nanoparticles such as α-Fe2O3 could tolerate a higher range of atrazine concentration compared to their free cells. The data obtained in this study will assist to effectively remediate and control the widespread of atrazine and possibly other pesticides in the environment.
4 Conclusion
Atrazine biodegradation using thirteen pure marine bacterial isolates concluded the following points:
Among the thirteen isolates, five {Bacillus pacificus (E7 and E8), Bacillus cereus (E9) and Bacillus paramycoides (E11 and E13)} were found to be the most effective and active in the degradation and removal of the target herbicide.
Atrazine removal ranged between 15.79 and 75.49, 77.97–97.13 and 27.4–87.6% equivalent to 26.7, 8.6 and 63.9 mg/L RCs were achieved by the tested bacteria at atrazine concentrations I, II and III (109, 299 & 438 mg/L), respectively. Atrazine residues are still much higher than the maximum permissible limit (MPL) of atrazine (≤0.1 mg/L).
To reach acceptable limits for safe discharge, atrazine degraders can be immobilized and used in a continuous treatment system as individual or serial units.
The achieved results provide an excellent potential for manipulating marine environment as a natural, rich and renewable source for bacteria with marvelous metabolic capabilities for efficient atrazine biodegradation.
Acknowledgement
This work was supported by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, Saudi Arabia under Grant No. G- 430-247-38. The authors, therefore, acknowledge with thanks DSR technical and financial support.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Short Protocols in Molecular Biology. New York: John Willey and Sons Inc.; 1999. p. :66-77.
- Standard methods for the Examination of Water and Wastewater. DC: American Public Health Association Washington; 2017.
- Labour sharing promotes coexistence in atrazine degrading bacterial communities. Sci. Rep.. 2019;9:1-8.
- [Google Scholar]
- Biodiversity and ecosystem functioning in soil: The dark side of nature and the bright side of life. Ambio. 2021;50(7):1286-1288.
- [Google Scholar]
- Effect of organic residues on pesticide behavior in soils: a review of laboratory research. Environments. 2021;8:32
- [CrossRef] [Google Scholar]
- Identification and distribution of antimicrobial-active marine bacteria along the heavily polluted coastal area of Alexandria. Egypt. Water Sci. Technol.. 2011;63:1081-1092.
- [Google Scholar]
- Antimicrobial phenolic compounds from Enterococcus faecium S29 (EU 158188): characterization and production optimization. Int. J. Environ. Sci. Technol.. 2017;14:497-508.
- [CrossRef] [Google Scholar]
- Isolation, identification and acclimatization of atrazine-resistant soil bacteria. Ann. Agric. Sci.. 2013;58:119-130.
- [CrossRef] [Google Scholar]
- Comparison among the efficiency of different bioremediation technologies of atrazine–contaminated soils. J. Biorem. Biodeg.. 2014;5(3)
- [CrossRef] [Google Scholar]
- Treatability of pharmaceutical effluents using free-living bacteria in a batch mode. Desalin. Water Treat.. 2019;152:316-327.
- [CrossRef] [Google Scholar]
- Integration between bacterial consortium and magnetite (Fe3O4) nanoparticles for the treatment of oily industrial wastewater. World J. Microbiol. Biotechnol.. 2020;36:141.
- [CrossRef] [Google Scholar]
- Bacterial bioaugmentation as an efficient approach to enhance the quality of activated sludge-treated effluent. Desalination Water Treat.. 2021;239:68-83.
- [CrossRef] [Google Scholar]
- Residue analysis and biodegradation of Atrazine in open field and indoor cultures. Merit Res. J. Agri. Soil Sci.. 2017;5(7):128-141.
- [Google Scholar]
- Identification of pesticide residues and prediction of their fate in agricultural soil. Water, Air, Soil Pollut.. 2020;231:1-10.
- [Google Scholar]
- Microbial community and atrazine-degrading genetic potential in deep zones of a hypersaline lake-aquifer system. Appl. Sci.. 2020;10(20):7111.
- [Google Scholar]
- Marine microbial natural products: the evolution of a new field of science. J. Antibiot. (Tokyo). 2020;73(8):481-487.
- [Google Scholar]
- Impact of atrazine exposure on the microbial community structure in a Brazilian tropical latosol soil. Microbes Environ.. 2020;35(2):ME19143.
- [Google Scholar]
- Hall, B.K., 1999. Homology. Introduction, in: Novartis Foundation Symposium. pp. 1–4.
- A review on recent treatment technology for herbicide atrazine in contaminated environment. Int. J. Environ. Res. Public Health. 2019;16(24):5129.
- [CrossRef] [Google Scholar]
- Mobility studies of atrazine in the soil-plant system in two Cameroonian vegetables Amaranthus hybridus and Corchorus olitorius. Environ. Sustain. Indic.. 2020;6:100036.
- [Google Scholar]
- International Organization for Standardization, 2016. Water quality- Sampling - Part 6: Guidance on sampling of rivers and streams (ISO 5667-6).
- International Organization for Standardization, 2020. Water quality- Sampling - Part 10: Guidance on Sampling of Wastewater (ISO 5667-10).
- Enhancement of atrazine biodegradation by marine isolate Bacillus velezensis MHNK1 in presence of surfactin lipopeptide. Ecotoxicol. Environ. Saf.. 2019;182:109372
- [CrossRef] [Google Scholar]
- Is selection necessary for evolution? A dynamics of coastal population of India. Orient. Anthropol.. 2019;19:284-299.
- [CrossRef] [Google Scholar]
- Augmentation of Atrazine biodegradation by two Bacilli immobilized on α-Fe2O3 magnetic nanoparticles. Sci. Rep.. 2018;8:1-12.
- [CrossRef] [Google Scholar]
- Defining lower limits of biodegradation: atrazine degradation regulated by mass transfer and maintenance demand in Arthrobacter aurescens TC1. ISME J.. 2019;13(9):2236-2251.
- [Google Scholar]
- Isolating and identifying the atrazine-degrading strain Arthrobacter sp. LY-1 and applying it for the bioremediation of atrazine-contaminated soil. Polish. J. Environ. Stud.. 2019;28(3):1267-1275.
- [Google Scholar]
- Compound-specific chlorine isotope fractionation in biodegradation of atrazine. Environ. Sci. Process. Impacts. 2020;22:792-801.
- [CrossRef] [Google Scholar]
- CMNPD: a comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res.. 2021;49:D509-D515.
- [Google Scholar]
- Rapid biodegradation of atrazine by Ensifer sp. strain and its degradation genes. Int. Biodeterior. Biodegradation. 2017;116:133-140.
- [CrossRef] [Google Scholar]
- Notes on the “Ahl al-Dīwān”: the Arab-Egyptian Army of the Seventh through the Ninth Centuries C.E. J. American Orient. Soc.. 2008;128(2):273-284.
- [Google Scholar]
- Genome-scale reconstruction of Paenarthrobacter aurescens TC1 metabolic model towards the study of atrazine bioremediation. Sci. Rep.. 2020;10:1-11.
- [Google Scholar]
- Effects of hydraulic residence time on metal uptake by activated sludge. Chem. Eng. Proc.. 2005;44(1):23-32.
- [Google Scholar]
- Fate of atrazine and its relationship with environmental factors in distinctly different lake sediments associated with hydrophytes. Environ. Pollut.. 2020;256:113371
- [CrossRef] [Google Scholar]
- Isolation and characterization of phenol degrading Bacillus species from a southeast Brazilian Mangrove sediment. African J. Microbiol. Res.. 2018;12(46):1032-1038.
- [CrossRef] [Google Scholar]
- Transformation of atrazine by photolysis and radiolysis: kinetic parameters, intermediates and economic consideration. Environ. Sci. Pollut. Res.. 2019;26(23):23268-23278.
- [Google Scholar]
- Molecular Cloning. A laboratory Manual (2nd ed.). New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1989.
- Magnetically separable and visible light-active Ag/NiCo2O4 nanorods prepared by a simple route for superior photodegradation of atrazine in water. Prog. Nat. Sci. Mater. Int.. 2020;30:160-167.
- [CrossRef] [Google Scholar]
- Bioreduction of Cr (VI) by biosurfactant producing marine bacterium Bacillus subtilis SHB 13. J. Sci. Ind. Res.. 2016;75:432-438.
- [Google Scholar]
- Multi-residue screening of pesticides in aquaculture waters through ultra-high-performance liquid Chromatography-Q/qrbitrap mass spectrometry. Water. 2020;12(5):1238.
- [Google Scholar]
- Novel Variation and Evolution of AvrPiz-t of Magnaporthe oryzae in field isolates. Front. Genet.. 2020;11:746.
- [Google Scholar]
- Biodegradation of pesticides at the limit: kinetics and microbial substrate use at low concentrations. Front. Microbiol.. 2020;11:2107.
- [Google Scholar]
- Biodegradation of atrazine by the novel Citricoccus sp. strain TT3. Ecotoxicol. Environ. Saf.. 2018;147:144-150.
- [CrossRef] [Google Scholar]
- Isolation and characterization of atrazine-degrading strain Shewanella sp. YJY4 from cornfield soil. Lett. Appl. Microbiol.. 2016;63:45-52.
- [CrossRef] [Google Scholar]
- Biodegradation of atrazine by the novel Klebsiella variicola strain FH-1. Biomed Res. Int.. 2019;2019:1-12.
- [Google Scholar]
- Study on the isolation of two atrazine-degrading bacteria and the development of a microbial agent. Microorganisms. 2019;7(3):80.
- [Google Scholar]
- Characteristics of an atrazine degrading bacterium and the construction of a microbial agent for effective atrazine degradation. Water Environ. J.. 2021;35(1):7-17.
- [Google Scholar]
Further reading
- Bioremediation of heavy metal-contaminated effluent using optimized activated sludge bacteria. Appl. Water Sci.. 2013;3(1):181-192.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102721.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







