Translate this page into:
Bifunctional neuraminidase inhibitory and simultaneously anti-Mycoplasma pneumoniae of flavan-3-ols and flavanones: Combined molecular docking, virtual screening, ADMET, and synthesis prediction
⁎Corresponding author. htkquy@ctu.edu.vn (Thi-Kim-Quy Ha)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
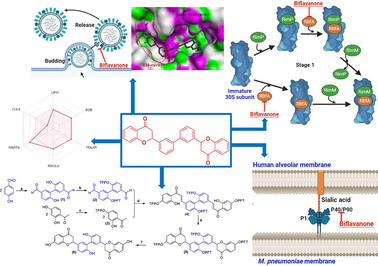
After lifting COVID-19 restrictions, pneumonia caused by influenza A virus coinfection with Mycoplasma pneumoniae bacteria emerges as a matter of concern. Only a few reports have recently proposed natural compounds for treating the coinfection of influenza A virus and M. pneumoniae, especially resistant strains. Hence, this study demonstrated in silico investigation of two natural classes (flavan-3-ol and flavanone) that could become future inhibitors for bifunctional neuraminidase and anti-M. pneumoniae. Combining the molecular docking with based-virtual screening strategies for over 900 ligands with neuraminidase protein of oseltamivir-resistant virus strain at H274Y mutation (PDB ID: 3CL0), 03 flavan-3-ols and 46 flavanones interacted with both the active site and the adjacent 430-cavity, and showed lower binding energies than laninamivir (−8.1 kcal/mol). The four most suitable candidates (FL22, FN166, FN316, and ChEMBL1779463) also exhibited potential characteristics for inhibiting M. pneumoniae through some vital targets (30S Ribosome-binding factor A (PDB ID: 1PA4) and immunodominant protein P40/P90 complex (PDB ID: 6TLZ)). Furthermore, the total-synthesis strategy of ChEMBL1779463 was successfully proposed using commercial reagents and modern techniques. Our results will offer potential insights for future methods to identify biflavonoids, especially biflavanone, to simultaneously suppress the resistant strains of influenza A virus and M. pneumoniae.
Keywords
In silico
Bifunctional neuraminidase inhibitors
Anti-Mycoplasma pneumoniae
ADMET
Total synthesis design
- 3D
-
Three-dimensional
- COCONUT
-
Collection of open natural products
- DUD-E
-
Directory of useful decoys-enhanced
- HBs
-
hydrogen bonds
- MeCN
-
Acetonitrile
- MM2
-
Molecular mechanics-2
- NA
-
Neuraminidase
- NAMD
-
Nanoscale molecular dynamics
- NP
-
Natural product
- ADMET
-
Absorption, distribution, metabolism, excretion, and toxicity
- PDB ID
-
Protein data bank identifier
- PFP
-
Pentafluoropyridine
- RBfA
-
Ribosome-binding factor A
- RMSD
-
Root mean square deviation
- RSV
-
Respiratory syncytial virus
- SARS
-
Severe acute respiratory syndrome
- TFP
-
Tetrafluoropyridyl
- THF
-
tetrahydrofuran
- VMD
-
Visual molecular dynamics
- WHO
-
World health organization
Abbreviations
1 Introduction
Influenza is a dangerous and widespread disease in human history (Peiris et al., 2012). The neuraminidase (NA) protein/antigen on the influenza A virus surface, which hydrolyzes sialic acid to release newly generated virions from the cell membrane, has received special research attention as an antivirus target (Gubareva and Mohan, 2022). However, frequent NA protein mutations emerge in drug-resistant strains and reduce the effectiveness of anti-neuraminidase drugs (Hussain et al., 2017; Shobugawa et al., 2012). The oseltamivir-resistant mutant with the replacement of histidine to tyrosine (H274Y mutation) at the hydrophobic binding site significantly reduced the clinical effectiveness (Woods et al., 2013). Many studies investigated the NA inhibitors targeting the active site of oseltamivir-resistant NA, but few reports revealed the role of new candidates could bind to both the active site and the adjacent 430-cavity through the unique arginine triad Arg118-Arg292-Arg371, which significantly enhances the binding affinity of complexes (Evteev et al., 2021; Ha et al., 2024). Therefore, there is still a gap need to find new bifunctional NA inhibitors to replace oseltamivir to improve the effectiveness of anti-influenza clinical therapy.
Mycoplasma pneumoniae is one of the most clinically significant (Meyer Sauteur and Berger, 2021). Common antibiotics, including macrolide, quinolone, and tetracycline, are used to treat this bacteria (Ma et al., 2015; Saraya et al., 2014). Mutations in the 23S rRNA gene led to the emergence of macrolide-resistance strains, limiting treatment options to the other two antibiotics (Cao et al., 2017; Zheng et al., 2015). Another study suggested that immunodominant protein P40/P90 is bound to sialylated oligosaccharides, and these complexes have an essential role in the function of “Nap” during gliding and adherence to host cells (Vizarraga et al., 2020). These findings have consequences for advancing prophylaxis and remedy.
Recently, M. pneumoniae has been reported as a potential coinfection with the influenza virus (Giannattasio et al., 2016; Olatunji et al., 2024; World Health Organization, 2023). This phenomenon leads to rough clinical conditions and requires an appropriate approach for the coinfection of bacteria and viruses (Sauteur et al., 2022). Until now, few studies have proposed natural compounds for treating pneumonia caused by M. pneumoniae coinfected with influenza A virus, requiring scientists to find a specific treatment that may result in better disease management and outcomes.
The ribosome of prokaryote consists of two subunits: a small 30S subunit containing 16S rRNA, and a large 50S subunit containing 5S and 23S rRNA. Azithromycin (belonging to the macrolides group) is the most effective and widely used antibiotic in treating M. pneumoniae (Saraya, 2017; Wen et al., 2018). The target of Azithromycin is 50S rRNA, thereby inhibiting protein synthesis in M. pneumoniae. Previous studies indicate that the surging mutation ratio in 23S rRNA and careless use of antibiotics led to the emergence of M. pneumoniae macrolide-resistance strains. However, other anti-M. pneumoniae agents are both less effective (more extended regimen) and also have the potential risk of drug resistance (Cao et al., 2017; Zheng et al., 2015). Therefore, this study aims for another critical target for bacteriostatic. The Ribosome-binding factor A (RBfA) is a molecule that plays a crucial role in the completion and maturation of the 30S subunit. In addition, adhesins and cytoadherence accessory proteins of M. pneumoniae play mediated roles in adherence into respiratory tract cells (Krause, 1996, Vizarraga et al., 2020). Among these factors, protein P1, in conjunction with the P40/P90 polypeptides, constitutes a transmembrane complex known as “Nap”, and it has critical roles in both gliding and attachment to host cells. The P40/P90 complex possesses the specific binding site for sialic acid cell receptors, which is absent in P1 (Vizarraga et al., 2020). Therefore, RBfA and P40/P90 complex should be a critical targets for treatments for M. pneumoniae.
Flavonoids are a group of pharmacological substances that have significantly contributed to basic biomedical and clinical research because of their proven effectiveness and safety (Alzaabi et al., 2021). This class could be a potential candidate for treating M. pneumoniae (Ding et al., 2023). However, based on our knowledge, no in silico studies have investigated the chemical structures of a subclass of flavonoid (flavan-3-ols and flavanones) that simultaneously inhibited NA of influenza virus and M. pneumoniae via RbfA and the P40/P90.
With the development of modern techniques, medicinal chemists are inspired by natural products (NPs) and their molecular frameworks. In the field of computational research using open database sources, numerous publications recently showed that NPs and NP-like molecules were collected from the COlleCtion of Open Natural ProdUcTs (COCONUT) database (Sorokina et al., 2021) and bioactive compounds from the ChEMBL database (Zdrazil et al., 2024). The COCONUT database, with more than 407,000 NP molecules, is beneficial for researchers in silico screening applications (Sorokina et al., 2021). Meanwhile, the ChEMBL database has about 2.4 million bioactive molecules with drug-like properties (collected in January 2024). Because of the massive number of bioactive molecules containing flavanol and flavanone skeletons in the ChEMBL database, this study first screened the molecular docking of over 900 flavanols and flavanones from the COCONUT database and the based-virtual screening for the most potential ligands were then carried out using the ChEMBL database. This strategy could reduce the time spent investigating bifunctional NA inhibitors against oseltamivir-resistant virus strain at H274Y mutation and inhibit M. pneumoniae simultaneously. Following this, the effective approach for the total synthesis of the potential candidate is based on absorption, distribution, metabolism, excretion, and toxicity (ADMET) predictions.
While studies looked at NA blockers and anti-Mycoplasma drugs separately, little work was done on drugs targeting both pathogens, mainly for coinfections. The main goal of this in silico study is to find possible drugs that can block both oseltamivir-resistant influenza A virus NA (H274Y mutation) and azithromycin-resistant M. pneumoniae using computer methods. The results of our research will offer potential insights for future strategies in identifying biflavonoids from natural sources for the suppression of oseltamivir-resistant influenza A virus and M. pneumoniae, simultaneously.
2 Experimental
2.1 Ligand preparation
The structures of 53 flavan-3-ols and 889 flavanones were obtained from the COCONUT database (https://coconut.naturalproducts.net/) (Listing in Supporting data files). The 3D structures were minimized energy using ChemOffice v.16.0 software based on the molecular mechanics-2 (MM2) method until the root mean square (RMS) gradient value lower than 0.001 kcal/mol Å (El Aissouq et al., 2020). These ligands with *.sdf file format were converted to *.pdb file format using OpenBabel v.2.4.1 software.
2.2 Protein preparation
3D structures of target proteins, including N1-H274Y-oseltamivir complex (PDB ID: 3CL0 – oseltamivir-resistant NA) (Ha et al., 2024); 30S ribosome-binding factor (PDB ID: 1PA4 − RBfA) (Rodrigues et al., 2019) and immunodominant protein P40/P90 complex (PDB ID: 6TLZ) (from M. pneumoniae) (Osigbemhe et al., 2022; Rubin et al., 2003), were obtained from the Protein Data Bank (https://www.pdb.org). These structures were removed the residues, undesirable chains, and water using Discovery Studio Visualizer v.2.5 software. They were then optimized based on the AMBER ff14SB force field with 500 steps of steepest descent and 100 steps of the conjugate gradient using Chimera v.1.16 software. The grid of the docking site of these proteins was provided in Table S1 of Supporting data.
2.3 Molecular docking screening for 3CL0 protein
The binding affinities between the ligands and 3CL0 protein were predicted using AutoDock Vina parameters based on the PyRx software (Trott and Olson, 2010). Firstly, the protein was input into the PyRx software and over 900 ligands were then docked into 3CL0 protein at the catalytic active site (Table S1 – S4) with the exhaustiveness and number of modes of 8, and scoring function based on the X-Score function. The candidates were selected when binding affinity values were equal to or lower than the binding energy at −8.4 (kcal/mol) of laninamivir, positive control of 3CL0 protein successfully identified from a previous study (Ha et al., 2024). The interaction effects between these structures and 3CL0 protein were visualized and analyzed to investigate the bifunctional NA inhibitors using Discovery Studio Visualizer v.2.5 and Chimera v.1.16 software.
2.4 Prediction of drug-likeness and ADMET values
Drug-likeness properties and ADMET parameters of the candidate compounds were predicted using the Canonical SMILES files, which were submitted to SwissADME (https://www.swissadme.ch) and pkCSM online server (https://biosig.lab.uq.edu.au/pkcsm/prediction).
2.5 Based-virtual screening for potential ligand
The ligand was applied for based-virtual screening in the ChEMBL database using the SwissSimilarity online tool and based on the combined method for the search algorithm (Zoete et al., 2016). The hits were selected by combining the results of molecular docking screening and similarity scoring values greater than 0.6. Molecular docking screening using AutoDock Vina (Trott and Olson, 2010) and SwissADME prediction (https://www.swissadme.ch) were performed to obtain the most suitable hit.
2.6 Evaluation of the docking methodology for 1PA4 and 6TLZ proteins
Decoys were generated using the Directory of Useful Decoys-Enhanced (DUD-E) online tool (https://dude.docking.org/) to evaluate the docking methodology for protein RBfA, and the active ligands were designed in the ChEMBL database using the SwissSimilarity online tool based on ZINC04259381. These ligands were randomly selected, and molecular docking with RBfA (PDB ID: 1PA4) (Table S1) was performed using AutoDock Vina (Trott and Olson, 2010).
To determine the reliability of the docking methodology for the P40/P90 complex, two decoys (laninamivir and oseltamivir) were selected. The two compounds exhibited comparable pharmacophores, featuring the N-acetylneuraminyl moiety of 3′-sialyllactose and a robust interaction with the NA of the influenza virus. However, no clinical studies have demonstrated these NA inhibitors' ability to reduce the growth of M. pneumoniae in children with influenza (Hume-Nixon et al., 2022). Meanwhile, the active ligands were generated from 3′-sialyllactose using the SwissSimilarity online tool based on the ChEMBL database. These compounds were randomly chosen, and molecular docking was performed with the P40/P90 complex at the active site (Table S1) using AutoDock Vina (Trott and Olson, 2010).
2.7 Molecular docking study of candidates with 1PA4 and 6TLZ proteins
The potential candidates were docked into RBfA (PDB ID: 1PA4) and P40/P90 complex (PDB ID: 6TLZ) at the active site (Table S1) using AutoDock Vina parameters assisted with the PyRx software (Trott and Olson, 2010). The interaction results were visualized and analyzed using Discovery Studio Visualizer v.2.5 and Chimera v.1.16 software.
2.8 Molecular dynamics simulations
The ligand–protein complexes of ChEMBL1779463 with PDB ID 3CL0 or 6TLZ were performed in the molecular dynamics simulations based on the NAnoscale Molecular Dynamics (NAMD) software (Phillips et al., 2005). Topologies and parameter files of ligands and proteins were parameterized using CHARMM36m force field. The solvated and ionized complexes were carried out as in the previous study (Ha et al., 2024). The simulation steps were set at 500 ns using the NPT ensemble and a constant temperature of 310 K. The number of hydrogen bonds (HBs) and the backbone root mean square deviation (RMSD) of these complexes were analyzed using Visual Molecular Dynamics (VMD) v.1.93 software.
3 Results and discussion
3.1 Molecular docking screening for 3CL0 protein
The molecular docking method's reliability was evaluated in a previous study (Ha et al., 2024). The workflow of this study is illustrated in Fig. 1. Since the NA-resistant oseltamivir protein (PDB ID: 3CL0) was used as the target protein, the binding energy of oseltamivir with the catalytic active site (−6.7 kcal/mol) exhibited a significantly higher value than its laninamivir (−8.1 kcal/mol) (Table S2). Therefore, our recent study carried out the molecular docking screening for over 900 flavan-3-ols and flavanones collected from the COCONUT database, and the candidates were selected when binding energies were lower than the threshold of −8.1 kcal/mol (Ki value < 1.145 µM). After this step, the remaining 9 flavan-3-ols and 122 flavanones (Tables S3 and S4) showed suitable properties for further visualization and analysis.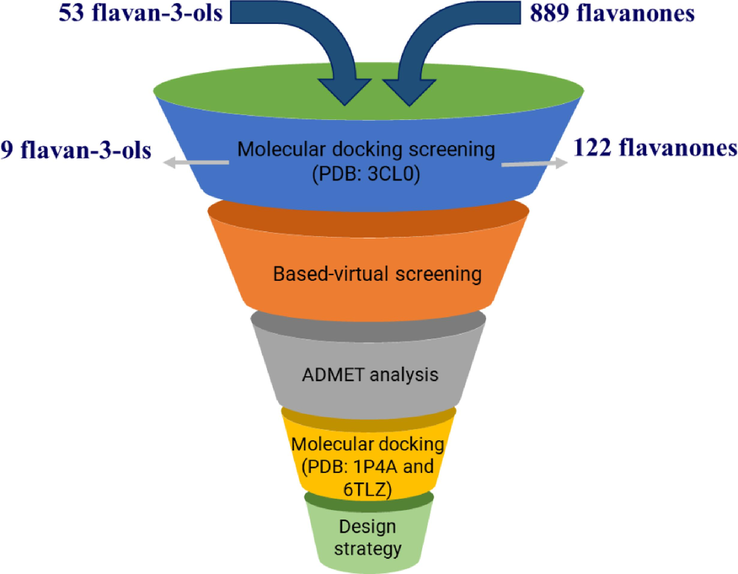
Procedure of molecular docking screening, based-virtual screening, ADMET, and synthesis prediction of flavan-3-ols and flavanones from the COCONUT database.
In the flavan-3-ols group (abbreviated as FL) (Fig. 2 and Tables S3, S5), only 3 compounds (FL13, FL22, and FL26) interact with both the active site and the adjacent 430-cavity. FL22 showed the lowest binding energy (−9.9 kcal/mol) with a Ki value of approximately 54.8 nM (Table 1). Several key interactions between FL22 with the catalytic active site and adjacent 430-cavity were obtained, such as Asp151, Arg224, Glu227, Glu277, Arg371, Ile427, Arg430, Pro431, and Lys432 through various interaction types (conventional hydrogen bond, π-alkyl, π-anion,…). Besides, FL22 also has two types of interactions (Van der Waals and conventional hydrogen bond) with the triad of arginine residues Arg118-Arg292-Arg371.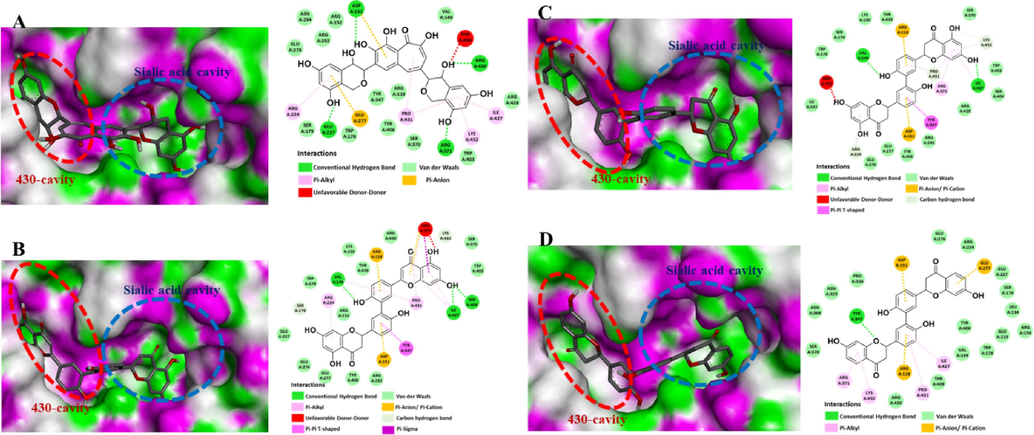
3D molecular docking binding into the sialic acid binding cavity and the 430-cavity, and 2D interaction diagrams of N1-H274Y-oseltamivir protein (PDB ID: 3CL0) with a flavan-3-ol FL22 (Fig. A) and three flavanones FN166, FN316, and ChEMBL 1,779,463 (Fig. B-D, respectively).
ID code
Coconut's ID
Compound name
Formula
Bingding
energy (kcal/mol)
Ki value (nM)
π-Anion/ π-cation
H-bond
Flavan-3-ol
FL-13
CNP0319747
2-(3,4-dihydroxyphenyl)-6-[2-(3,4-dihydroxyphenyl)-3,7,8-trihydroxy-3,4-dihydro-2H-1-benzopyran-5-yl]-3,4-dihydro-2H-1-benzopyran-3,7,8-triol
C30H26O12
−8.5
583.0
Arg118
Asp151
Arg292Arg118
ser246
tyr406arg430
FL-22
CNP0247600
3,4,6-trihydroxy-2,8-bis(3,5,7-trihydroxy-3,4-dihydro-2H-1-benzopyran-2-yl)-5H-benzo[7]annulen-5-one
C29H24O12
−9.9
54.8
Asp151
Glu277Asp151
glu227
arg371arg430
FL-26
CNP0372621
11-[(1,3-dihydroxypropan-2-yl)oxy]-2-[4-hydroxy-3-(2-hydroxyethoxy)phenyl]-6-[(1H-indol-5-yl)methyl]–2H,3H,4H,5H,6H-phenanthro[2,1-b]pyran-3,8-diol
C37H37NO9
−8.4
690.3
Arg118
Asp151
Glu277
Arg371Glu119
arg152
arg156glu227
Flavanone
FN-166
CNP0184920
2,3-Dihydro-3′,3′''-Biapigenin
C30H20O10
−11.4
4.4
Arg118
Asp151
Arg371Val149
ser404ile427
FN-316
CNP0203327
3′,3′''-Binaringenin
C30H22O10
−11.7
2.6
Arg118
Asp151Val149ile427
ChEMBL 1,779,463
2,2′-(6,6′-dihydroxy-[1,1′-biphenyl]-3,3′-diyl)bis(7-hydroxychroman-4-one)
C30H22O8
−11.4
4.4
Arg118
Asp151
Glu277Tyr347
Other 2 flavan-3-ols, FL13 and FL26, exhibited weak binding energies (−8.5 kcal/mol and −8.4 kcal/mol, respectively) with Ki values around 583.0–690.3 nM (Table 1). Similar to FL22, these compounds also have several key interactions such as conventional HBs, π-alkyl, π-anion/cation, carbon-hydrogen bond, and Van der Waals, with some essential amino acids Arg118, Glu119, Asp151, Arg152, Glu227, Ser246, Glu277, Arg292, Arg371, Ile427, Arg430, and Pro431 (Tables S3, S5). Our docking screening results for 53 flavan-3-ols from the COCONUT database suggested that FL22 was the most suitable candidate compared to FL13 and FL26. Even though only some previous studies suggested that FL22, theaflavin, and their derivatives had the in vitro and in silico anti-NA of different influenza virus strains (Mohamed et al., 2022), there is no report about the role of theaflavin as a potential for interaction with both the active site and the adjacent 430-cavity through the triad of arginine residues. Our results suggested the new mechanism of FL22 and proposed it as a potential bifunctional NA inhibitor (Fig. 2).
Regarding the flavanone group (abbreviated as FN), the binding energies of this skeleton were significantly lower than those of flavan-3-ols. During the analysis, 46 compounds showed binding interactions with both active site and adjacent 430-cavity through several critical amino acids (Tables S4, S6). Among those, two compounds, FN166 and FN316, exhibited the lowest binding energies following −11.4 kcal/mol and −11.7 kcal/mol, respectively, with Ki values around 2.6–4.4 nM (Fig. 2 and Tables 1, S4, S6). Besides the most suitable skeleton (biflavonoid) for bifunctional NA inhibitory effect, the results also revealed that flavanone diglycosides (FN150, FN193, and FN330) showed binding energies better than flavanone glycoside (FN106, FN270, and FN334). Because of their high similarity pharmacophore, FN166 and FN316 were focused for further analysis. As shown in Fig. 2, FN316 and FN166 exhibited key interactions with the catalytic active site and adjacent 430-cavity through the triad of arginine residues.
3.2 Based-virtual screening for ligand FN316
During the above screening procedures for over 900 flavan-3-ols and flavanones, three anti-NA candidates (FL22, FN166, and FN316) were selected for further evaluation for the “drug-likeness” properties based on the SwissADME online tool. As illustrated in Fig. 3 and Table S7, FN166 and FN316 showed fewer violations of Lipinski's rule than FL22. However, FN166 and FN316 still presented unsuitable values, including the number of hydrogen bond donors of more than 5 and molecular weights of over 500 Da. Hence, this study examined further similarities with these flavanones that mitigate the violations of Lipinski's rule. Previous reports intimated that the ChEMBL database contains NPs and NP derivatives, NP mimetics, and NP pharmacophores. Hence, this database was used for based-virtual screening for ligand FN316 using the SwissSimilarity online tool and combined method for the search algorithm with similarity scoring values greater than 0.6. Five similarities were selected, with at least one flavanone moiety not present in the previous list of 889 flavanones. Among those five, four compounds (ChEMBL3763296, ChEMBL3765207, ChEMBL4075132, and ChEMBL4086291) contain alkylamines, while ChEMBL1779463 is a biflavanone structure (Fig.S1). The “drug-likeness” properties of five similarities showed that ChEMBL3763296, ChEMBL4086291, and ChEMBL1779463 had only a violation of Lipinski's rule, while other compounds (ChEMBL3765207 and ChEMBL4075132) did not exhibit any violation of this rule.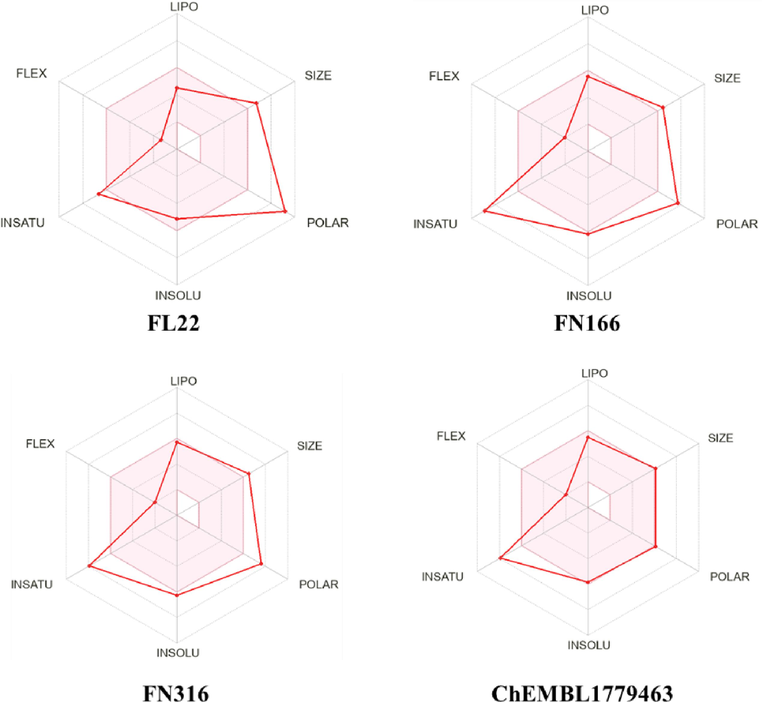
The “drug-likeness” properties based on the SwissADME online tool for four potential anti-NA candidates (FL22, FN166, FN316, and ChEMBL1779463).
After performing molecular docking screening of the five similarities for 3CL0 protein using the AutoDock Vina tool, ChEMBL1779463 showed the lowest binding energy (−11.4 kcal/mol) with a Ki value of around 4.4 nM (Table S8). In addition, ChEMBL1779463 showed several key interactions with both the catalytic active site and adjacent 430-cavity (Fig. 2). It was suggested that the biflavanone structure could become a more suitable skeleton than flavanone linked with alkylamines.
3.3 ADMET study
Four potential anti-NA candidates (FL22, FN166, FN316, and ChEMBL1779463) were evaluated for ADMET parameters (Table 2). Several ADMET parameters were analyzed in detail, as follows: (1) aqueous solubility of testing compounds was in accepted ranges from −6.5 to 0.5; (2) absorption in the human intestine of ChEMBL1779463 was highest than others; (3) the blood–brain barrier (BBB) permeability of four compounds are lower than 0.3, which considered to un-cross the BBB (Muehlbacher et al., 2011); (4) ChEMBL1779463 showed a better bioavailability score than the remaining other candidates; (5) other indicators (such as P-glycoprotein substrate, total clearance,…) suggested that those testing compounds could be used as safety drugs with less toxicity for human, especially ChEMBL1779463 may become the most potential than others. *: using SwissADME web server; other predicted parameters using pkCSM web server.
No.
ADMET properties
FL22
FN166
FN316
ChEMBL 1779463
1
Human intestinal absorption (%)
55.652
79.101
81.574
99.391
2
Blood-brain barrier permeability (logBB)
−2.436
−2.176
−2.083
−1.456
3
P-glycoprotein substrate
Yes
Yes
Yes
No
4
Total clearance [log ml/(min.kg)]
0.43
0.254
0.102
0.083
5
AMES toxicity
No
No
No
No
6
Max tolerated dose [log mg/(kg.d)]
0.366
0.399
0.338
0.377
7
CYP1A2 inhibitor
No
No
No
No
8
CYP2C19 inhibitor
No
No
No
No
9
CYP2C9 inhibitor
Yes
No
No
Yes
10
CYP2D6 inhibitor
No
No
No
No
11
CYP3A4 inhibitor
No
No
No
No
12
hERG I inhibitor
No
No
No
No
13
hERG II inhibitor
Yes
Yes
Yes
Yes
14
Oral rat acute toxicity, LD50 (mol/kg)
2.483
2.424
2.382
2.335
15
Oral rat chronic toxicity (log mg/kg_bw/day)
4.694
2.980
3.386
1.818
16
Skin sensitization
No
No
No
No
17
T. pyriformis toxicity (log µg/L)
0.285
0.285
0.285
0.285
18
Minnow toxicity (log mM)
4.17
1.687
1.467
−0.593
19
Bioavailability score*
0.17
0.17
0.17
0.55
3.4 Molecular docking between potential candidates with m. Pneumoniae proteins
RBfA is a cold shock response protein required for bacterial growth at low temperatures and efficient processing of the 5′ end of 16S rRNA during assembly of the 30S subunit (Maksimova et al., 2021). Previous studies revealed that aminoglycosides and ZINC04259381 could bind to the 30S subunit, leading to transcriptional errors that interfere with its structural function and inhibit the protein synthesis of bacteria (Rodrigues et al., 2019). Therefore, in this study, ZINC04259381 was used as a positive control for screening the potential of the four above candidates against 30S RBfA. In comparison with a previous report, several key interactions of ZINC04259381 with target protein were similar in binding to the active site through some essential amino acids such as Arg15, Asn18, Val35 (Fig.S2) (Rodrigues et al., 2019). In addition, this protein's docking method was successfully evaluated for 10 random decoys and 05 random active ligands (Table S9). These results suggested that our docking methodology was suitable for further investigation of testing candidates. As summarized in Table 3, the binding energies of the four candidates were from −8.6 kcal/mol to −9.2 kcal/mol, similar to its positive control (−9.2 kcal/mol). Several key interactions between four candidates with the active site were also analyzed. Fig. 4A demonstrated the interaction of FL22 with RBfA via several amino acids such as Lys6, Ile17, Asn18, His34, and Val35 by various interaction types. Compounds FN166, FN316, and ChEMBL1779463 did not have π-cation likely FL22, but they still bound into the active site through several critical amino acids (Fig. 4B-D).
ID code
1PA4
6TLZ
Bingding
energy (kcal/mol)
Ki value (nM)
π-Anion/ π-cation/ π-sigma
H-bond
π-π T-shaped
Bingding
energy (kcal/mol)
Ki value (µM)
π-Anion/ π-cation
H-bond
π-π T-shaped
FL-22
−9.2
178.76
Ile17
His34
Val35Lys6
−
−7.7
2.25
−
Asp687
Phe831
FN-166
−9
250.58
Ile17
Val35Val30
Val21
−8.7
0.42
Asp210
Asp687Asn223
Pro235
Leu630
Asp687Phe631
FN-316
−8.6
492.42
Ile17
Glu10
Thr36Val35
−8.5
0.58
Asp210
Asp687Pro235
Leu630Phe631
CHEMBL 1,779,463
−8.9
296.69
Ile17
Val35−
Val49
−8.7
0.42
Arg226
Arg226
Thr632Phe631
ZINC-04259381
−9.2
178.76
−
Asn18
His34−
−
−
−
−
−
3′-Sialyllactose
−
−
−
−
−6.1
33.57
−
Glu219
Ser220
Asn223
Pro235
Lys238
Leu630−
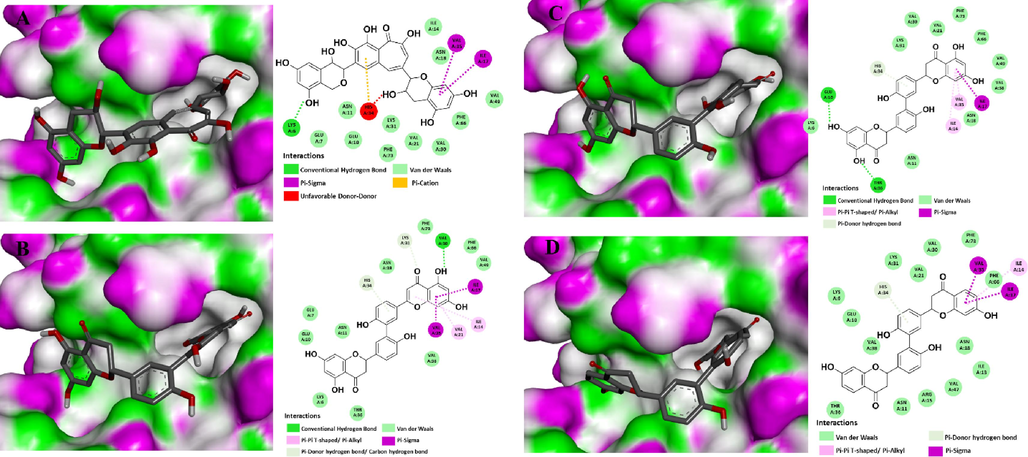
3D molecular docking binding into the active site and 2D interaction diagrams of 30S RBfA (PDB ID: 1PA4) with a flavan-3-ol FL22 (A) and three flavanones FN166, FN316, and ChEMBL 1779463 (B-D, respectively).
The effect of four anti-NA candidates were investigated the binding effect on the immunodominant protein P40/P90 complex against M. pneumoniae adhesion to host cells. A previous study reported the interaction of P40/P90 with 3′-sialyllactose through several important amino acids, such as Pro235, Pro237, Lys238, Phe631, and Thr632 (Vizarraga et al., 2020). Therefore, the site containing these amino acids was described as the binding site for sialic acid-based molecular mimicry. In comparison with the previous report, key interactions of sialyllactose with protein P40/P90 were bound to the sialic acid binding site (Fig.S3) (Vizarraga et al., 2020). In addition, results in Table S10 indicated that 02 decoy compounds showed binding energies from −4.9 to −5.3 kcal/mol, significantly higher than 3′-sialyllactose (−6.1 kcal/mol). However, 07 random active ligands exhibited binding energies similar to 3′-sialyllactose. These results revealed that this molecular docking methodology could be applied for further study. Four anti-NA candidates showed binding energies significantly lower than 3′-sialyllactose, with values from −7.7 to −8.7 kcal/mol (Table 3). In a detailed analysis, three compounds (FN166, FN316, and ChEMBL1779463) exhibited great interactions with protein P40/P90 via several amino acids, while FL22 exhibited fewer interactions than others (Fig. 5).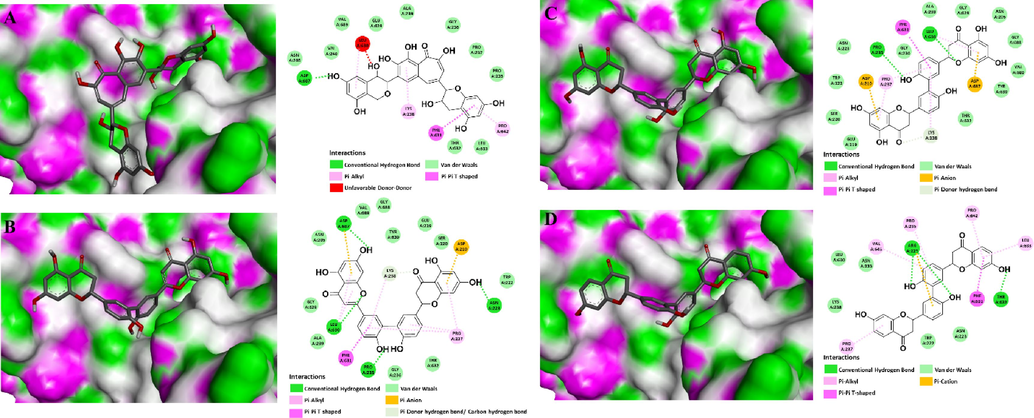
3D molecular docking binding into sialic acid cell receptor and 2D interaction diagrams of the immunodominant protein P40/P90 complex (PDB ID: 6TLZ) with a flavan-3-ol FL22 (Fig. A) and three flavanones FN166, FN316, and ChEMBL 1,779,463 (Fig. B-D, respectively).
Biflavanone could become a potential structure for inhibiting the M. pneumoniae via (1) binding to the 30S subunit, leading to transcriptional errors and interfering with the protein synthesis; (2) inhibiting the bacteria attachment to host cells through binding to the sialic acid cell receptor P40/P90. According to the above molecular docking results, molecular docking combined with other computer techniques allowed for inexpensive, highly effective, and easy-to-perform, leading to advantages for further drug development. However, several drawbacks need to be improved for this computer tool, including the conformational flexibility of target proteins and weak or transient binding of ligand–protein complexes. Therefore, further testing in vitro experiments is suggested for potential candidates.
3.5 Molecular dynamics simulations
3CL0 and 6TLZ were chosen to further validate the ligand–protein complexes' stability since they showed at least one hydrogen bond with ChEMBL1779463. As shown in Fig.S4, RMSD values of these complexes showed less than 2Å, suggesting that the predicting crystallographic binding orientation was a good solution (Ha et al., 2024). The molecular dynamics simulation of ChEMBL1779463 with protein 3CL0 showed the number of HBs from 2 to 4 bonds in the range from 0 to 500 ns, suggesting that this ligand has proper interaction with several types of amino acids (Arg118, Glu119, Val149, Arg156, Glu227, Arg292, Tyr347, and Arg371) (Table S11 and Fig.S4). The complexes of ligand–protein 6TLZ exhibited HBs from 2 to 4 during the molecular dynamics simulation period of 0–400 ns, significantly reduced after the 400–500 ns. Some amino acids with the HBs interactions with ChEMBL1779463 were listed as Asn223, Arg226, Asn233, Leu630, and Thr632 (Table S11 and Fig.S4 D).
3.6 Proposed the total synthesis strategy and biological pathways
Even though NPs have substantive roles in the research and development of medicine, the limited plant sources encourage researchers to investigate the synthesis strategy. So, this study also suggests a scheme for the total synthesis of the second metabolites of natural resources. Among four candidates, FL22 (theaflavin) is a major agent from black tea, a commercial product with an antioxidant effect, while compounds FN166 and FN316had been successfully proposed for the total synthesis (Sagrera and Seoane, 2010). Therefore, this study focuses on the ChEMBL1779463 to propose a total-synthesis strategy using commercial and affordable reagents with some key steps, such as using a green procedure and protecting the phenol group using tetrafluoropyridyl (TFP) ether moiety attachment (Brittain and Cobb, 2019; Nishimura et al., 2010). First, 4-hydroxybenzaldehyde was reacted with horseradish peroxidase enzyme in the presence of hydrogen peroxide (3% in water) at pH∼4 to form a dimerization structure (1) (Fig. 6) (Nishimura et al., 2010). Intermediate compound (1), or 2′,4′-dihydroxyacetophenone, were mixed with pentafluoropyridine (PFP) and K2CO3 in the acetonitrile (MeCN) at room temperature for 16h to obtain readily cleavable protecting group for phenols (2) or (3), respectively (Brittain and Cobb, 2019). The intermediate compounds (2) and (3) were then linked together through the reaction with NaH in the tetrahydrofuran (THF) at room temperature for 24h (Sagrera and Seoane, 2010). Compound (4) was then created with acetic acid with the assistance of the reflux method for 96h to form the compound (5). Finally, the tetrafluoropyridyl (TFP) ether groups of compound (5) were removed to obtain ChEMBL1779463 following the procedure: mixing the intermediate compound with potassium fluoride, 18-crown-6, and methyl thioglycolate at 50°C for 24h (Sagrera and Seoane, 2010).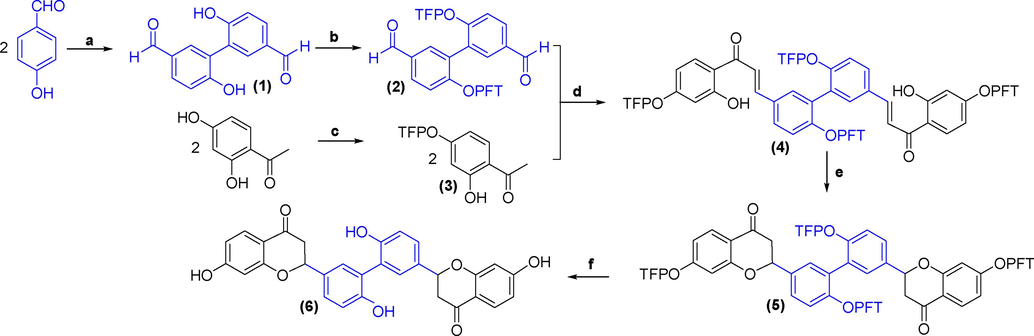
The proposed total synthesis of ChEMBL1779463. (a) Horseradish Peroxidase, H2O2, pH 4, 5–10 min. (b and c) PFP, K2CO3, MeCN, rt, 16 h. (d) NaH, THF, rt, 24 h. (e) AcOH, reflux, 96 h. (f) KF, 18-crown-6, methyl thioglycolate, CD3CN, D2O, 50 °C, 24 h.
The noteworthy points of this study were the suggestion of the biological pathways of biflavanone, the most potential candidate for simultaneously inhibiting bifunctional NA protein and anti-M. pneumoniae. Biflavanone (1) inhibited the final step of influenza A viral life cycle (virion release); (2) could bind to the 30S subunit, leading to transcription mistakes and bacteria's protein synthesis interference (Maksimova et al., 2021); and (3) could interact with the sialic acid receptor P40/P90, preventing bacteria from attaching to host cells (Vizarraga et al., 2020) (Fig. 7).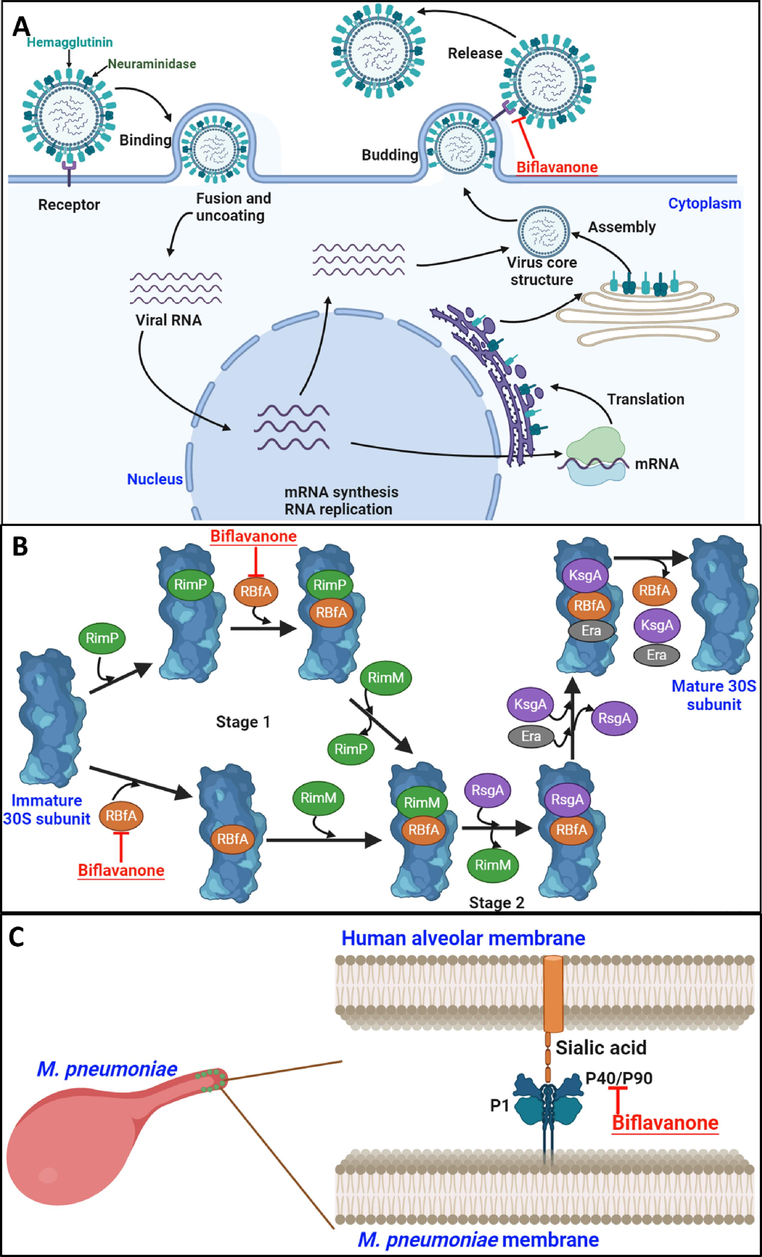
The proposed biological pathways of biflavanone for inhibiting the final step (virion release) of influenza A virus via NA receptor (A), binding to the 30S subunit (B) and the sialic acid cell receptor P40/P90 (C) of M. pneumoniae.
4 Conclusion
This is the first report in silico evaluating the role of flavan-3-ol and flavanone as potential candidates for treating pneumonia coinfected by the influenza A virus and M. pneumoniae. 03 flavan-3-ols and 46 flavanones showed the potential interaction with both the active site and the adjacent 430-cavity. Based on the Swiss tool for based-virtual screening, the four most suitable candidates (FL22, FN166, FN316, and ChEMBL1779463) were selected and further investigated the anti-M. pneumoniae through some critical targets (inhibiting 30S RBfA and immunodominant protein P40/P90 complex). Our docking data indicated that among various skeletons such as flavan-3-ol, biflavan-3-ol, flavanone, biflavanone, and alkylamine derivatives,… biflavanone exhibits potential structure for future treatment of oseltamivir-resistant influenza A virus and M. pneumoniae, simultaneously. Furthermore, ChEMBL1779463 was predicted safety for humans and successfully proposed the total-synthesis strategy using commercial reagents and modern techniques.
CRediT authorship contribution statement
Nguyen-Huan Pham-Khanh: Data curation, Formal analysis, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing. Thi-Kim-Quy Ha: Data curation, Formal analysis, Methodology, Supervision, Visualization, Writing – review & editing. Khe-Vinh Duong: Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Flavonoids are promising safe therapy against COVID-19. Phytochem. Rev.. 2021;1–22
- [CrossRef] [Google Scholar]
- Tetrafluoropyridyl (TFP): a general phenol protecting group readily cleaved under mild conditions. Org. Biomol. Chem.. 2019;17:2110-2115.
- [CrossRef] [Google Scholar]
- Overview of antimicrobial options for M ycoplasma pneumoniae pneumonia: focus on macrolide resistance. Clin. Respir. J.. 2017;11:419-429.
- [CrossRef] [Google Scholar]
- Total Flavonoids from Camellia oleifera Alleviated Mycoplasma pneumoniae-Induced Lung Injury via Inhibition of the TLR2-Mediated NF-κB and MAPK Pathways. Molecules. 2023;28:7077.
- [CrossRef] [Google Scholar]
- In silico design of novel tetra-substituted pyridinylimidazoles derivatives as c-jun N-terminal kinase-3 inhibitors, using 2D/3D-QSAR studies, molecular docking and ADMET prediction. Int. J. Pept. Res. Ther.. 2020;26:1335-1351.
- [CrossRef] [Google Scholar]
- Bifunctional inhibitors of influenza virus neuraminidase: molecular design of a sulfonamide linker. Int. J. Mol. Sci.. 2021;22:13112.
- [CrossRef] [Google Scholar]
- Coinfections with influenza virus and atypical bacteria: Implications for severe outcomes? Clin. Respir. J.. 2016;12:366-367.
- [CrossRef] [Google Scholar]
- Antivirals targeting the neuraminidase. Cold Spring Harb. Perspect. Med.. 2022;12:a038455
- [CrossRef] [Google Scholar]
- Molecular docking screening, dynamics simulations, ADMET, and semi-synthesis prediction of flavones and flavonols from the COCONUT database as potent bifunctional neuraminidase inhibitors. Pharmacia. 2024;71:1-10.
- [CrossRef] [Google Scholar]
- Hume-Nixon, M., Graham, H., Russell, F., Mulholland, K., Gwee, A., Group, A.R.I.R., 2022. Review of the role of additional treatments including oseltamivir, oral steroids, macrolides, and vitamin supplementation for children with severe pneumonia in low-and middle-income countries. J. Glob. Health 12. doi: 10.7189/jogh.12.10005.
- Drug resistance in influenza A virus: the epidemiology and management. Infect. Drug Resist.. 2017;121–134
- [CrossRef] [Google Scholar]
- Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol.. 1996;20:247-253.
- [CrossRef] [Google Scholar]
- Clinical and epidemiological characteristics in children with community-acquired mycoplasma pneumonia in Taiwan: A nationwide surveillance. J. Microbiol. Immunol. Infect.. 2015;48:632-638.
- [CrossRef] [Google Scholar]
- RbfA is involved in two important stages of 30S subunit assembly: formation of the central pseudoknot and docking of Helix 44 to the decoding center. Int. J. Mol. Sci.. 2021;22:6140.
- [CrossRef] [Google Scholar]
- Proadrenomedullin in Mycoplasma pneumoniae community-acquired pneumonia in children. Clin. Infect. Dis.. 2021;73:e1769-e1771.
- [CrossRef] [Google Scholar]
- In vitro virucidal activity of the theaflavin-concentrated tea extract TY-1 against influenza A virus. J. Nat. Med.. 2022;1–9
- [CrossRef] [Google Scholar]
- Qualitative prediction of blood–brain barrier permeability on a large and refined dataset. J. Comput. Aided Mol. Des.. 2011;25:1095-1106.
- [CrossRef] [Google Scholar]
- Green, enzymatic syntheses of divanillin and diapocynin for the organic, biochemistry, or advanced general chemistry laboratory. J. Chem. Educ.. 2010;87:526-527.
- [CrossRef] [Google Scholar]
- Addressing the mysterious respiratory illness outbreak in China. New Microbes New Infect.. 2024;56
- [CrossRef] [Google Scholar]
- Antibacterial potential of 2-(-(2-Hydroxyphenyl)-methylidene)-amino) nicotinic Acid: Experimental, DFT Studies, and molecular docking approach. Appl. Biochem. Biotechnol.. 2022;194:5680-5701.
- [CrossRef] [Google Scholar]
- Surveillance of animal influenza for pandemic preparedness. Science (80-.. 2012;). 335:1173-1174.
- [CrossRef] [Google Scholar]
- Scalable molecular dynamics with NAMD. J. Comput. Chem.. 2005;26:1781-1802.
- [CrossRef] [Google Scholar]
- Reverse vaccinology and subtractive genomics reveal new therapeutic targets against Mycoplasma pneumoniae: a causative agent of pneumonia. R. Soc. Open Sci.. 2019;6
- [CrossRef] [Google Scholar]
- Solution structure of a putative ribosome binding protein from Mycoplasma pneumoniae and comparison to a distant homolog. J. Struct. Funct. Genomics. 2003;4:235-243.
- [CrossRef] [Google Scholar]
- Total Synthesis of 3′, 3′′′-Binaringenin and Related Biflavonoids. Synthesis (stuttg).. 2010;2010:2776-2786.
- [CrossRef] [Google Scholar]
- Mycoplasma pneumoniae infection: Basics. J. Gen. Fam. Med.. 2017;18:118-125.
- [CrossRef] [Google Scholar]
- Novel aspects on the pathogenesis of Mycoplasma pneumoniae pneumonia and therapeutic implications. Front. Microbiol.. 2014;5:410.
- [CrossRef] [Google Scholar]
- Mycoplasma pneumoniae detections before and during the COVID-19 pandemic: results of a global survey, 2017 to 2021. Eurosurveillance. 2022;27:2100746.
- [CrossRef] [Google Scholar]
- Clinical effectiveness of neuraminidase inhibitors—oseltamivir, zanamivir, laninamivir, and peramivir—for treatment of influenza A (H3N2) and A (H1N1) pdm09 infection: an observational study in the 2010–2011 influenza season in Japan. J. Infect. Chemother.. 2012;18:858-864.
- [CrossRef] [Google Scholar]
- COCONUT online: collection of open natural products database. J. Cheminform.. 2021;13:1-13.
- [CrossRef] [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [CrossRef] [Google Scholar]
- Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae. Nat. Commun.. 2020;11:5188.
- [CrossRef] [Google Scholar]
- Epidemiology, microbiology, and treatment patterns of pediatric patients hospitalized with pneumonia at two hospitals in China: a patient chart review study. Ther. Clin. Risk Manag.. 2018;501–510
- [CrossRef] [Google Scholar]
- Analysis and assay of oseltamivir-resistant mutants of influenza neuraminidase via direct observation of drug unbinding and rebinding in simulation. Biochemistry. 2013;52:8150-8164.
- [CrossRef] [Google Scholar]
- World Health Organization, 2023. Disease Outbreak News: Upsurge of respiratory illnesses among children in northern China. https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON494 (accessed 23 November 2023) [WWW Document]. 2023.
- The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res.. 2024;52:D1180-D1192.
- [CrossRef] [Google Scholar]
- Macrolide-resistant mycoplasma pneumoniae. United States. Emerg. Infect. Dis.. 2015;21:1470.
- [CrossRef] [Google Scholar]
- Zoete, V., Daina, A., Bovigny, C., Michielin, O., 2016. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. doi: 10.1021/acs.jcim.6b00174.
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103242.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







