Translate this page into:
Antibacterial and cytotoxicity activities of bioactive compounds from Micrococcus species OUS9 isolated from sea water
⁎Corresponding author. krishna.ganduri@kluniversity.in (V.S. Ramakrishna Ganduri)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Bacterial screenings were collected from Nellore coastal regions of Bapatla and Vishakhapatnam from south India. Altogether, 29 bacterial isolates were attained from seawater and sediment samples consuming Zobel-culture media (pH = 7.3 ± 0.2) and incubated at 27 °C of temperature. Among these 29 bacterial isolates, 3 bacterial strains were shown the highest zone of inhibition producing the molecular characterization. The strains of Micrococcus species OUS9 was demonstrated to be potential. To characterize such activity and recognize the promising isolates, further research is required. This new natural biological product is also used in enormous sectors, pharmaceutical and food industries for preservation of natural environment. This study has confirmed that the bacteria isolates environment for halophilic bacterial growth, where there are numerous bacterial communities and many industrial applications are possible.
Keywords
Industrial applications
Zobel
Micrococcus sp. OUS9
1 Introduction
Marine bacteria are documented as an invaluable resource in delivering bioactive compounds and the marine natural products plays a vital role in biomedical research and pharmaceutical industry (Karbalaei-Heidari et al., 2020). For thousands of years, nature was the origin of medicinal products. There are an imposing number of contemporary drugs, primarily based on traditional medicine use, isolate from microorganisms. Microorganisms have played a growing part in antibiotic manufacturing and in another drug manufacturing in the previous century. As sources of precious bioactive metabolites, terrestrial fungi and bacteria have been extremely important for more than half a century. There has been a continuing search for fresh drug products from the ocean for over two centuries. Prior researchers have revealed as marine invertebrates are significant source for fresh biochemical compounds and well-known fact as established through the large number of compounds present in the clinical trials, proper and reliable supplies of these compounds have proved notoriously hard to acquire from the nature. Due to these issues, (Faulkner, 2000) studies have well-documented about the avenue centered on marine microorganisms for considerable attention. Therefore, the anticipated huge biodiversity of sea organisms could have been at first sight for one of reasons (Burch-Brown and Archer, 2017); though marine derived microorganisms were not taxonomically sounded. The groundwork studies showed the wealth of microbial diversity in the oceans of the world making the capable of frontiers for discovering fresh drugs (Blunt et al., 2004).
Marine derived bacteria are defined through marine water demands in particular with sodium for growth (Fenical, 1997). Among the cases of marine associated with fungi are without ion demands and mandatory marine species are lives only sea water (Jensen and Fenical, 2000). This issue is compounded in samples near shore/estuarine, where the large proportion of resident microbes were modified for variable marine levels of exposure. All microbes may be isolated from the environmental marine for the purpose of microbial drug discovery (Gerwick and Moore, 2012). Maximum of fresh complexes have recorded from marine microorganisms which have been acquired from various species, isolated either from land, sea or both. Coherent investigation for all the microbes has been isolated from extreme sea water environment for the purpose of microbial drug discovery. Based on different studies on species, the majority of new bioactive metabolites from sea associate microorganisms reports were derived from species which can be isolated. While, these optional species of marine constitute obvious sources of new metabolites, their environmental functions and adaptation levels for marine environment are not known (Garcia and Rosenberg, 2010). Survey carried on marine bacteria were separated from marine algae and invertebrates which showed as antimicrobial metabolites are produced in a large proportion (Burgess et al., 1999). The production of non-bacterial bioactive metabolites which inhibit the setting of potential competitors has also been shown to cause by epiphytic bacteria present in marine combines with rich algal surface and invertebrates (Petersen et al., 2020).
The novel cyclical decapeptide antibiotic Loloatin B, which prevent development by M−resistant S. aureus MRSA (Staphylococcus aureus methicillin resistant), and VRE (Vancomycin Enterococcus resistant) was made by species Bacillus in Papoa-Nova Guinea (Calfee, 2012). Thiomarinol, eminent antibiotics has discovered to produce marine bacterium Alteromonasrava. Marine microorganisms, including Bacillus loloatins, have been recorded as antibiotics. Ascomycin through ascomyceticus var. Streptomyces hygroscopicus, Rhamnose from Saccharopolyspora spinosa (Madduri et al., 2001), pyrones from Pseudomonas (Singh et al., 2003), Kanglemycin from Nocardia mediterranei var. kanglensis, Zorbamycin from Streptomyces flavovirdis. Therefore, current study aims to explore to isolate bioactive potential bacteria with seawater for an antibacterial activity.
2 Materials and methods
2.1 Sampling site and collection
Both seawater and sample soils were collected from Nellore coastal region premises in south India. The water samples were collected in 500 mL of sterilized and autoclavable collection bottles. The sedimentation samples were collected into sterile plastic polythene bags and sealed. The samples were collected under aseptic conditions and were placed on sterile icepacks until further process. These samples were inoculated with 1–2 h after collection (Jayaprakashvel et al., 2010).
2.2 Isolation of marine bacteria
All the collected samples were marked according to the locations collected, from each sample 60 µL of water were shredded over the Zobells agar plates purchased from Hi-Media and incubated at 28 °C for 24 and 48 h in incubator. After the incubation, different colonies were transferred to the fresh sterile slants for further use (Jayadev and Lekshmi, 2016).
2.3 Screening of for bioactive compounds producing bacterial
2.3.1 Antagonism assay
Strahl et al (2002) has standardized the invitro antagonism assay method against bacterial strains like Escherichia coli and Staphylococcus aureus. Lawn culture was performed through utilizing sterilized cotton swab and incubates for 1 min. Ten micro-liter of bacterial culture was transferred into wells and petri dishes were incubated at 30 °C for overnight. Antagonistic interactions were scored based on presence and appearance of inhibition zones. One of the isolates strains which scored higher inhibition zone was selected for further characterization.
2.3.2 Molecular-based characterization
The 16S rRNA sequence was used in bacterial strains which showed best inhibition against the opted pathogens for molecular identification. The bacterial culture DNA sample, which was amplified using the 16S rRNA primers mainly forward (5′-TCACGGAGTTT-GATCCTG-3) and reverse (5′-GCGGCTGCACGTA GTT-3′) primers were used to sequence 16S rRNA gene-sequence. Genotyping was performed as per the documented studies (Khan et al., 2019). A phylogenetic tree was acquired with maximum probability demonstrating the evolutionary association with the selected sequence.
2.3.3 Fermentation process
Using 250 mL of Erlenmeyer flasks, fermentation was performed with the selected active bacterial strains consists of 100 mL of Zobell-broth medium. Pure selected bacteria strains were inoculated with 1 mL of culture suspension for the sterilized fermented broth. Using rotary shaking incubator at 250 rpm, inoculated flasks were incubated at 28 °C for 5 days. The fermented media was centrifuged for crude extract preparation at 10,000 rpm or 20 mins after incubation.
2.3.4 Solvent stabilization
Selection of appropriate solvent to extract the bacterial pigment, initially the solvent capable of supporting maximum pigment output was standardized using various solvents such as ethanol, acetone, methanol, ethyl acetate, petroleum ether, chloroform, hexane, distilled water and diethyl ether (Mahato et al., 2019).
2.3.5 Antibacterial activity
Agar technique was implemented for the antibacterial activity of extracts with crude pigments and evaluated against numerous bacterial pathogens such as Salmonella species (PM-08), Staphylococcus aureus (PM-14), E.coli (PM-04), B. subtilis, procured from M/s. Pure microbes, Pune. Plates were incubated at 37 °C of room temperature for 18–24 h and the diameter of inhibition area (mm) were assessed in the final of the experiment and the activity index calculation was performed. The measurement took three distinct, set instructions and recorded average values (Fatima et al., 2019).
2.3.6 Antioxidant activity
DPPH scavenging activity of crude extracts was evaluated. In short, 10 µL of each sample was mixed in methanol (1.0 mL) with 90 µmolL-1 DPPH; prepared with methanol at final quantity of 4 mL. These mixtures were vigorously shaken at 37 °C and incubated in the dark mode for 1hr. Resulting solution absorbance was appraised at 517 nm. Total reactions were carried out in triplicates with Butylated hydroxy anisole as positive control.
2.3.7 Extraction of secondary metabolites
Cells were separated from broth by centrifuging at 6000 rpm at 10 °C for 30 mins. The metabolites were extracted from the supernatant with liquid–liquid extraction technique with equal amount of ethyl acetate and concentration of crude extracts through rotary evaporation (Ming et al., 2018). Cells were washed and separated with methanol. Next, methanolic fraction was obtained and concentrated with yield of crude extracts.
2.3.8 Isolation and purification of bioactive compounds
Davies-Bolorunduro et al (2019) method was adopted from Kwon et al (2009) studies and we have applied in this study to isolate and purify the bioactive compounds. Flash column chromatography was performed.
2.3.9 Anticancer activity
The MTT assay is used to determine the cellular viability or metabolic activity in microcapsules which determines the cell proliferation and cytotoxicity with colorimetric assay based on Yellow-colored, water-soluble tetrazolium dye; MTT which lowers in formazan crystals. Mitochondrial lactate dehydrogenase is produced through liver cells lowers in MTT to insoluble formazan crystals which exhibit purple color when dissolved into the suitable solvent and intensity of number of viable cells can be recorded at 570 nm spectrophotometrically (Moorthy and Rangappan, 2016).
3 Results and discussion
The composition of the marine ecosystem and biogeochemistry of the oceans play a crucial role. The great variety of microbes in our waters continues therefore vital to be understood. In order to capture rare, less abundantly organisms, efforts are required to investigate regions containing the biggest biodiversity, such as aquatic settings (Salihoglu et al., 2013). The notable variety of marine micro-organisms is due to their initial development, speed of generations and micro-environmental heterogeneity (Staley, 2006). The ocean is heterogenous owing to its nutrient patches and microscale incline, which can help into various inches (Kassen and Rainey, 2004).
In the present investigation the total 29 different bacterial cultures were isolated from different locations of Nellore district regions the bacterial colonies present on agar plates with morphologically different pigment producing has been identified. The selected colonies were sub-cultured in individual testtubes and observed for morphological characterization shown in the (Table 1). The chosen colonies were screened by well diffusion for antagonistic activity. Process against two pathogenic bacteria out of 29 selected bacterial cultures OUS9 have shown efficient (Table 2) antagonistic activity, the results showed that the selected isolate have good growth and ability of high pigment producing capacity as shown in the (Fig. 1). In one of studies of two human-pathogenic bacteria have discovered that marine isolates are more efficacious than bacteria which live freely living bacteria (Nair and Simidu, 1987). Studies investigating the frequencies of marine bacterial isolates; antagonistic interactions discovered between 5 and 8 percent of isolates expressing activity level (Krasilnikova, 1961). Although these trials concentrated on marine isolates as prospective antibiotic producers, the target species were non-marine bacteria, often human pathogens. Similarly, more than our understanding of antibiotics producing and understanding of environmental antibiotic sensitivity is restricted. Kanamycin and penicillin antibiotics have diverse form of concentration for inhibition of marine bacteria.
NAME OF THE INDIVIDUAL BACTERIA
COLONY CHARACTERISTICS OF ISOLATED BACTERIA
GRAMS STAINING
MOTILITY
OUS1
Yellow pigmented irregular, Opaque, Rough,
Gram positive cocci in bunch
Non-motile
OUS2
Yellow pigmented, Circular, Opaque, Rough, Slightly
Gram positive cocci
Non-motile
OUS3
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci in bunch
Non-motile
OUS4
Lemon yellow pigmented, Circular, Opaque, Rough, Convex
Gram positive cocci in bunch
Non-motile
OUS5
Pale yellow pigmented Circular, Translucent, Smooth,
Gram positive rods
Non-motile
OUS6
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram negative rods
Motile
OUS7
Yellow pigmented, Circular, Translucent, Smooth, Convex
Gram positive short rods
Motile
OUS8
Yellow pigmented, Circular, Opaque, Smooth, Convex
Gram negative cocco bacilli
Motile
OUS9
Bright yellowish pigmented, Circular, Opaque.
Gram positive in cocci
Non-Motile
OUS10
Pale yellow pigmented, Circular, Opaque, Smooth, Flat
Gram positive cocci in bunch
Non-Motile
OUS11
Thick Yellow pigmented, Elliptical, Opaque, Smooth, Convex
Gram positive cocci in bunch
Non-motile
OUS12
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive short rods
Motile
OUS13
Light yellow pigmented, Circular, Translucent, Smooth,
Gram positive cocci in bunch
Non-motile
OUS14
Golden yellow pigmented, Congregated, Opaque, Smooth,
Gram positive cocci in bunch
Non-motile
OUS15
Yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci
Non-motile
OUS16
Lemon yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci in bunch
Non-Motile
OUS17
Light yellow pigmented, Irregular, Translucent, Smooth, Flat
Gram negative rods
Motile
OUS18
Lemon yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci
Non-motile
OUS19
Yellow pigmented, Circular, Opaque, Rough, Convex
Gram positive cocci
Non-motile
OUS20
Lemon yellow pigmented, Circular, Opaque, Rough, Convex
Gram positive cocci
Non-motile
OUS21
Bright yellow pigmented, Circular, Opaque, Smooth,
Gram positive cocci
Non-motile
OUS22
Yellow pigmented, Circular, Translucent, Smooth, Convex
Gram positive cocci
Non-motile
OUS23
Light yellow pigmented, Circular, Translucent, Smooth,
Gram positive cocci
Non-motile
OUS24
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci
Non-motile
OUS25
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram negative rods
Motile
OUS26
Yellow pigmented, Circular, Opaque, Rough,
Gram positive cocci
Non-motile
OUS27
Pale yellow pigmented, Circular, Opaque, Smooth, Convex
Gram positive cocci in bunch
Non-motile
OUS28
Lemon yellow pigmented, Circular, Opaque, Rough, Convex
Gram positive cocci in bunch
Non-motile
OUS29
Pale yellow pigmented Circular, Translucent, Smooth,
Gram positive rods
Non-motile
Isolates
Gram positive
Gram negative
OUS1
ND
ND
OUS2
ND
ND
OUS3
ND
ND
OUS4
ND
ND
OUS5
ND
ND
OUS6
ND
ND
OUS7
ND
ND
OUS8
ND
ND
OUS9
6
9
OUS10
ND
ND
OUS11
ND
ND
OUS12
ND
ND
OUS13
ND
ND
OUS14
ND
ND
OUS15
1
ND
OUS16
ND
3
OUS17
ND
ND
OUS18
ND
ND
OUS19
ND
ND
OUS20
ND
ND
OUS21
ND
ND
OUS22
ND
ND
OUS23
2
ND
OUS24
ND
ND
OUS25
ND
ND
OUS26
ND
ND
OUS27
ND
ND
OUS28
ND
ND
OUS29
ND
ND

Pigmentation of isolated bacteria from marine intertidal regions.
Carlos et al (2013) has evaluated the 16S rDNA for the bacterial diversity for the similar samples of coral settings and for large abundance of proteobacteria. For different weavy softs, stony corals, sponges and sea water of this phylum was recorded earlier in an elevated abundance (Porporato et al., 2013). These strains were further reconfirming through ribotyping molecular technique using 16S rRNA gene sequence and this partial sequence of 16S rRNA gene was obtained with the blast which was documented in Micrococcus species OUS9 (Fig. 2) and further the sequence was deposited in GenBank (Accession number MN108086). Microorganisms living in the rhizospheres of diversity of crops are probable due to the abundance of substrate shown from the roots when compared with non-rhizosphere soils to synthesize and release auxin as secondary metabolites (Ahmad et al., 2005). Nair and Simidu (1987) studies have documented this antibacterial compound are not only inhibits terrestrial bacteria also, for ecological importance to native bacterial strains. Yellow pigment extracted from isolated bacteria for high content of pigments was investigated. High contents of pigment were obtained from methanol solvent when compared with ethanol, hexane and petroleum benzene solvents.
Phylogenetic tree of Micrococcus OUS9 bacterial isolate from marine intertidal regions.
Many new species are acknowledged as promising source for fresh bio-active compounds in taxonomic terms (Huang et al., 2012). Wealthy cause for secondary metabolism is with the novel structures at outstanding biological activities has been found in marine bacteria extracted from deep sea sediments. A strain AB 18–032 termed as actinomycete Verrucosispora have been extracted from the sediment sample obtained at 289 m of depth from Japanese Sea (Bister et al., 2004). Abyssomicin C (8) has shown methicillin-resistant antibiotic activity and S Aureus (MRSA) and S-resistant to vancomycin had 4 and 13 µg/mL of MIC values. A representative class-I spirotetron is regarded as Abyssomicin C. This biosynthesis is initially recognized and obtained over bacterial source which inhibits para aminobenzoic acid (PABA) biosynthesis. Abyssomicin C and its atropoisomeric inhibits PABA respectively. Further, discovering the marine environment is to be promising strategy for active compounds and for development of resistant therapies for many microorganisms, which indicates the requirement of new drugs are required. Precisely, for anti-fungal, anti-bacterial, anti-viral and anti-protozoal activities.
The present study was carried out with crude pigments were verified for antimicrobial activity with pathogenic bacteria has also been observed that out of Chloroform, Ethanol, Methanol and Acetone extracts, with standard drug reference and Methanol crude pigment showed zone of inhibition greater against Bacillus subtilis 15 mm, followed by S. aureus with 12 mm, Salmonella 11 mm, Escherichia coli 09 mm, P. aeruginosa 06 mm and lowest zone was observed to K. pneumoniae 02 mm (Fig. 3, Table 3). This confirms as crude Yellow pigment of Micrococcus species isolates were well and active against both gram positive and negative bacteria. For their natural product contents such as algae, ascidians, corals, microorganisms and sponges which is known to be total forms of marines were investigated (Arif et al., 2004). Throughout thousands of years, ecological pressures like space competition, symbiosis, predation and tide variations have led to the biosynthesis of complicated of secondary metabolite species, which in turn enable them for adopting the competitive and hostile environment (Da Silva et al., 2006). Different marine organisms such as algae and invertebrates in sea surface or cavum has been found to be more nutritive than inanimate and marine water and may have large number of bacteria among them (Bonugli-Santos et al., 2015). These few species of bacteria are usually non-symbiotic of host which can be allied bacteria (Bultel-Ponce et al., 1999) and are compelled to create antibiotic resistance secreted by their host.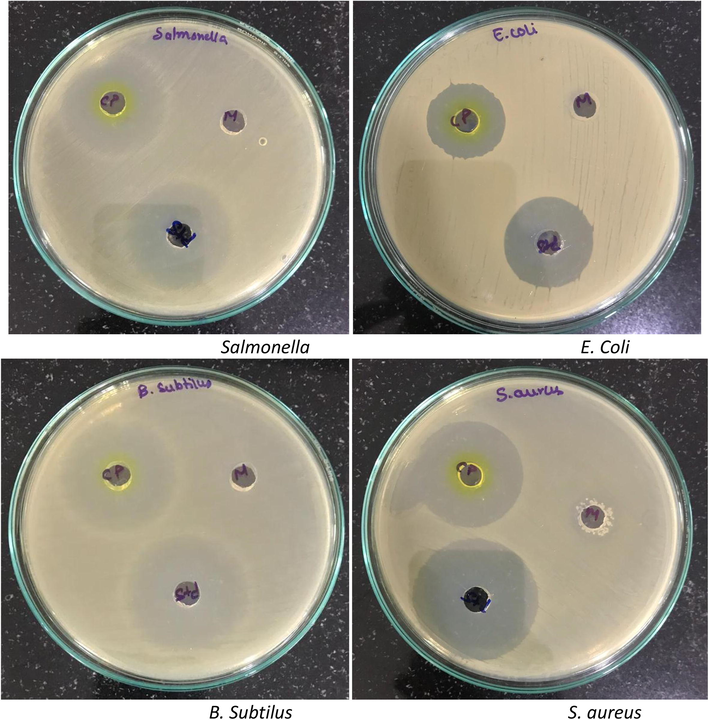
Antibacterial activity of Methanol crude extracts from Micrococcus species OUS9.
Gram Staining
Gr+
Shape and growth
Cocci
Motility stab
Non-motile
Growth and macconkey plates
+
Tellurite reduction
Oxidase Production
–
Catalase Production
+
Indole Production
–
O/F test
ONPG Hydrolysis
H2S Production
–
Growth at 4 °c
Growth at 40 °c
Production of extra-cellular enzymes
Amylase
Gelatinase
+
Cellulase
urease
–
Caesinase
Acid from
D-Arabinose
–
D-Inositol
–
D-Lactose
+
D-Maltose
–
D-Rhamnose
–
D-Sorbitol
D-Glucose
–
D-Mannose
+
D-Trehalose
−/+
D-Xylose
+
Maltotriose
+
Accetate
+
Lactic acid
+
Malic acid
+
Propionic acid
+
Pyruvic acid
+
Glycerol
+
Alpha Hydroxybutyrate
+
Beta Hydroxybutyrate
+
Serine
+
Glutamate
–
Sorbitol
+
Glucose
Growth in NaCl
0%
+
0.5%
+
1%
+
Tween 40
+
Tween 80
+
0%
+
0.5%
+
1%
+
In the present investigational study, carried out the free radical scavenging activity of Micrococcus species OUS9 extract with concentration range from 25, 75 to 100 mg/ml and ascorbic acid standard used as a standard. The result of DPPH, ABTS and NO scavenging activity indicates Micrococcus species OUS9 extract are potentially active. As shown clearly in the Table 4 that the extract shows 48.44 ± 6.2% inhibition as compared to standard 56.21 ± 4.9 in DPPH activity followed by 44.14 ± 2.9 in ABTS and 42.13 ± 3.4% inhibition in NO with 100 (mg/ml) (Table 5). With promising antioxidant activity, the inhibition effect increased in higher levels. It has a maximum of 176 of 95 per cent. Species of reactive oxygen plays important role to protect human body from damage (Govindarajan et al., 2005). In addition, past epidemiological surveys have shown a reduction in consumption of natural antioxidants in cancer and other oxidatively damaged illnesses (Rietveld and Wiseman, 2003).
Bacteria Pathogens
Zone of inhibition in mm
Chloroform extract
Ethanol extract
Methanol extract
Acetone extract
Standard
Bacillus subtilis
–
–
15
–
19
Salmonella
–
–
11
–
16
Escherichia coli
–
–
09
02
12
P. aeruginosa
05
–
06
–
08
K. pneumoniae
–
–
02
–
14
S. aureus
–
01
12
04
15
Concentration
(mg/ml)Antioxidant activity of methanol extracted crude pigment isolated from micrococcus sp
Standard
ABTS
NO
DPPH
100
56.21 ± 4.9
44.14 ± 2.9
42.13 ± 3.4
48.44 ± 6.2
75
42.27 ± 1.9
32.95 ± 3.9
39.26 ± 4.2
43.19 ± 5.7
25
32.19 ± 1.3
20.24 ± 2.9
25.75 ± 1.4
31.47 ± 2.9
Extraction of crude through solvent extraction is a significant phenomenon for opting the appropriate solvent for extracting high yields of bioactive compounds. Previous studies have confirmed the strong antimicrobial potential for ethyl acetate extraction against the pathogens of fungi and bacteria. Here from the eluted fractions with column chromatography is tested for antimicrobial study and the best fraction was further identified. KLUF-10 and KLUF-13 with molecular weight 568.9 and KLUF-13 the molecular weight with 263.38. On the basis of these observations the purified compound is proposed to be KLUF-10 as 3-Hydroxy-β, ε-caroten-3′-one, KLUF-13 1-(1-(4-methoxphyenyl)-2-(methyl amino) ethyl) cyclohexan-1-ol and which was isolated from Micrococcus species OUS9 for the first time and is to appear that novel compound with good antimicrobial activity as shown in the Figs. 4 and 5 supplementary table 1). The most promising results for anticancer activity were obtained with KLUF-10 and KLUF-13 fractions of the Micrococcus sp. OUS9 strain. The inhibitory effect of pure compounds treated cells was observed after 24 h of incubation. The results of cytotoxicity study performed by MTT assay suggest that the given test compound, KLUF-10 and KLUF-13 are anticancer in nature against the both HeLa and MCF-7 Cell lines and the IC50 concentrations are 34.5ug/ml and 34.95ug/ml respectively IC50 concentrations are HeLa 28.9 ug/ml and MCF-7 26.6ug/ml (Supplementary Table 2; Supplementary Figs. 1-2). In multiple microbial metabolites, the role of pigment is to act as coloring agent in food processing and cosmetic industries and also own anti-oxidant, anti-inflammatory, anti-cancer, and anti-microbial activities. The current results of this study suggest that the coastal region's ability as an origin of marine bacteria to generate antibacterial compounds which can and must be explored for many pharmaceutical applications.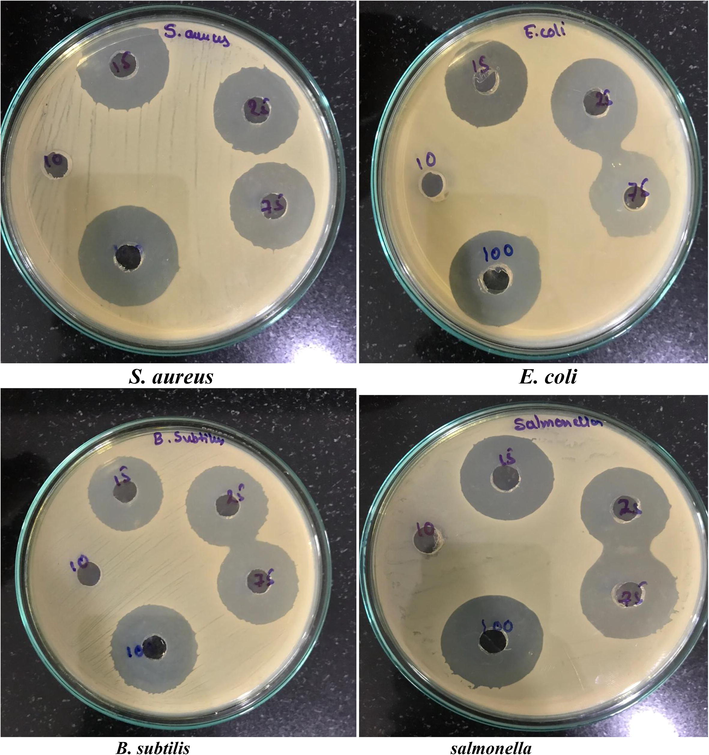
Antibacterial activity (MIC) of pure compound (13th Yellow fraction) isolated from Micrococcus species OUS9.
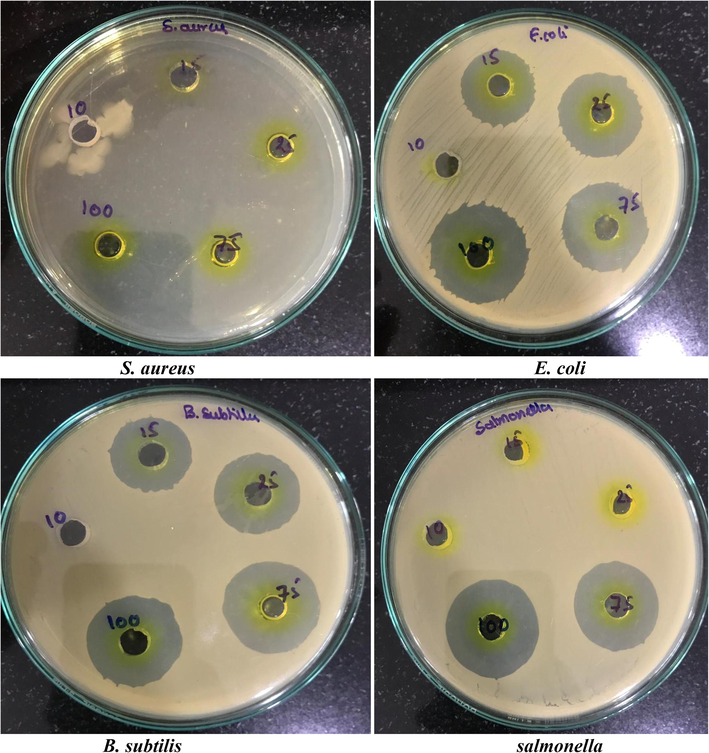
Antibacterial activity (MIC) of pure compound (10th Yellow fraction) isolated from Micrococcus species OUS9.
4 Conclusion
In the marine environment, bioactive compounds that produce microorganisms are widely distributed. Bioactive metabolites from marine bacteria which acts as an anchor for survival between pathogenic bacteria and their environment. This research is a step towards understanding and testing bioactive metabolites generating marine bacteria in the region around the shore of Nellore, Andhra Pradesh, India. Screening of these bacteria by screening techniques for the manufacturing of bioactive metabolites. The strain of Micrococcus species OUS9 demonstrated potential. To characterize such activity and recognize the promising isolates, further research is required. This new natural biological product is also used in various sectors such as the pharmaceutical industry, the food industry and the preservation of the natural environment
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmad, F., Ahmad, I., Khan, M. S. J. T. J. O. B., 2005. Indole acetic acid production by the indigenous isolates of Azotobacter and fluorescent Pseudomonas in the presence and absence of tryptophan. 29, 29–34.
- Novel marine compounds: anticancer or genotoxic? J. Biomed. Biotechnol.. 2004;2004:93-98.
- [Google Scholar]
- Abyssomicin C—a polycyclic antibiotic from a marine verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate. Biosynthesis Pathway.. 2004;43:2574-2576.
- [Google Scholar]
- Marine Natural Products.. 2004;21:1-49.
- Bonugli-Santos, R.C., Dos Santos Vasconcelos, M.R., Passarini, M.R., Vieira, G.A., Lopes, V.C., Mainardi, P.H., Dos Santos, J.A., de Azevedo Duarte, L., Otero, I.V., dA Silva Yoshida, A.M., Feitosa, V.A., Pessoa, A., Jr., Sette, L.D., 2015. Marine-derived fungi: diversity of enzymes and biotechnological applications. Front. Microbiol., 6, 269.
- Metabolites from the Sponge-Associated Bacterium Pseudomonas Species. Mar Biotechnol (NY). 1999;1:384-390.
- [Google Scholar]
- Burch-Brown, J., Archer, A.J.B., Philosophy, 2017. In defence of biodiversity. 32, 969–997.
- Burgess, J.G., Jordan, E.M., Bregu, M., Mearns-Spragg, A., Boyd, K.G., Microbial antagonism: a neglected avenue of natural products research. Progress in Industrial Microbiology. Elsevier.
- Calfee, D.P.J.C.O.I.I.D., 2012. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci, and other Gram-positives in healthcare. 25, 385–394.
- Carlos, C., Torres, T.T., Ottoboni, L.M.J.S.R., 2013. Bacterial communities and species-specific associations with the mucus of Brazilian coral species. 3, 1624.
- In vitro antiviral activity of marine sponges collected off Brazilian coast. Biol. Pharm. Bull.. 2006;29:135-140.
- [Google Scholar]
- Davies-Bolorunduro, O.F., Adeleye, I.A., Akinleye, M.O., Wang, P.G.J.J.O.P.A., 2019. Anticancer potential of metabolic compounds from marine actinomycetes isolated from Lagos Lagoon sediment. 9, 201–208.
- Fatima, N., Khan, M.M., Khan, I.A.J.S.J.O.B.S., 2019. L-asparaginase produced from soil isolates of Pseudomonas aeruginosa shows potent anti-cancer activity on HeLa cells. 26, 1146–1153.
- Faulkner, D.J.J.A.V.L., 2000. Marine pharmacology. 77, 135–145.
- Fenical, W.J.T.I.B., 1997. New pharmaceuticals from marine organisms. 15, 339–341.
- Garcia, S.M., Rosenberg, A.A.J.P.T.O.T.R.S.B.B.S., 2010. Food security and marine capture fisheries: characteristics, trends, drivers and future perspectives. 365, 2869–2880.
- Gerwick, W.H., Moore, B.S., 2012. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. 19, 85–98.
- Antioxidant approach to disease management and the role of 'Rasayana' herbs of Ayurveda. J. Ethnopharmacol.. 2005;99:165-178.
- [Google Scholar]
- Huang, H., Yang, T., Ren, X., Liu, J., Song, Y., Sun, A., Ma, J., Wang, B., Zhang, Y., Huang, C.J.J.O.N.P., 2012. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. 75, 202–208.
- Jayadev, A., Lekshmi, M.J.E.L.S.R., 2016. Screening and Isolation of Protease producing Marine Bacteria. 2, 73–76.
- Jayaprakashvel, M., Muthezhilan, R., Srinivasan, R., Hussain, A.J., Gobalakrishnan, S., Bhagat, J., Kaarthikeyan, C., Muthulakshmi, R.J.J.A.B., 2010. Hydrogen cyanide mediated biocontrol potential of Pseudomonas sp. AMET1055 isolated from the rhizosphere of coastal sand dune vegetation. 9, 38–42.
- Jensen, P.R., Fenical, W.J.D.F.T.S., 2000. Marine microorganisms and drug discovery: current status and future potential. 6–29.
- Karbalaei-Heidari, H.R., Partovifar, M., Memarpoor-Yazdi, M.J.A.J.O.M.B., 2020. Evaluation of the Bioactive Potential of Secondary Metabolites Produced by a New Marine Micrococcus Species Isolated from the Persian Gulf. 12, 61–65.
- Kassen, R., Rainey, P.B.J.A.R.M., 2004. The ecology and genetics of microbial diversity. 58, 207–231.
- Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. DiabetesMetab. Syndr.. 2019;13:688-694.
- [Google Scholar]
- Krasilnikova, E.J.M.-U., 1961. Antibiotic properties of microorganisms isolated from various depths of world oceans. 30, 545-&.
- Kwon, H.C., Kauffman, C.A., Jensen, P.R., Fenical, W.J.T.J.O.O.C., 2009. Marinisporolides, polyene-polyol macrolides from a marine actinomycete of the new genus Marinispora. 74, 675–684.
- Madduri, K., Waldron, C., Merlo, D.J.J.J.O.B., 2001. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. 183, 5632–5638.
- Mahato, N., Sinha, M., Sharma, K., Koteswararao, R., Cho, M.H.J.F., 2019. Modern Extraction and Purification Techniques for Obtaining High Purity Food-Grade Bioactive Compounds and Value-Added Co-Products from Citrus Wastes. 8, 523.
- Ming, T., Han, J., Li, Y., Lu, C., Qiu, D., Li, Y., Zhou, J., Su, X.J.B.M., 2018. A metabolomics and proteomics study of the Lactobacillus plantarum in the grass carp fermentation. 18, 216.
- Moorthy, M., Rangappan, R.J.S.S., 2016. Binuclear Cu (II) Schiff base complexes as prescursors for the synthesis of CuO nanoparticles: anticancer activity against MCF-7 cell line. 4, 95–107.
- Nair, S., Simidu, U.J.A.E.M., 1987. Distribution and significance of heterotrophic marine bacteria with antibacterial activity. 53, 2957–2962.
- Petersen, L.-E., Kellermann, M.Y., Schupp, P.J., 2020. Secondary Metabolites of Marine Microbes: From Natural Products Chemistry to Chemical Ecology. YOUMARES 9-The Oceans: Our Research, Our Future. Springer.
- Diversity and antibacterial activity of the bacterial communities associated with two Mediterranean sea pens, Pennatula phosphorea and Pteroeides spinosum. (Anthozoa: Octocorallia). 2013;66:701-714.
- [Google Scholar]
- Antioxidant effects of tea: evidence from human clinical trials. J. Nutr.. 2003;133:3285s-3292s.
- [Google Scholar]
- Salihoglu, B., Neuer, S., Painting, S., Murtugudde, R., Hofmann, E., Steele, J., Hood, R., Legendre, L., Lomas, M., Wiggert, J. D. J. J. O. M. S. 2013. Bridging marine ecosystem and biogeochemistry research: lessons and recommendations from comparative studies. 109, 161–175.
- Singh, M.P., Kong, F., Janso, J.E., Arias, D.A., Suarez, P.A., Bernan, V.S., Petersen, P.J., Weiss, W.J., Carter, G., Greenstein, M. J. T. J. O. A., 2003. Novel α-Pyrones Produced by a Marine Pseudomonas sp. F92S91. 56, 1033–1044.
- Staley, J. T. J. P. T. O. T. R. S. B. B. S., 2006. The bacterial species dilemma and the genomic–phylogenetic species concept. 361, 1899–1909.
- Strahl, E., Dobson, W., Lundie JR, L. J. C. M. 2002. Isolation and screening of brittlestar-associated bacteria for antibacterial activity. 44, 450–459.
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.07.003.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







