Translate this page into:
Study on the prediction of congenital cardiac abnormalities using various Machine learning models
⁎Corresponding author. alzubiahmadali@gmail.com (Ahmad Ali AlZubi),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
Congenital heart disease (CHD) involves structural heart defects present from birth. Ventricular septal defects (VSDs) are among the most common types. Early diagnosis is important and can be done using fetal echocardiography at 12–14 weeks of gestation. However, detection rates depend on the quality of diagnostic tools and expertise. Machine learning (ML) can enhance detection through various diagnostic modalities, including electrocardiogram (ECG) and ultrasonography (US).
Aim and Objectives
This study aims to improve CHD detection by integrating fetal echocardiography with machine learning techniques.
Method
The study explores methods for detecting CHD using an online dataset, employing preprocessing, feature extraction, and deep learning classification.
Results
There was notable variability in model performance metrics. The Decision Support System for Early Prediction (DSSEP) had the highest sensitivity (80.11%) but a lower positive predictive value (PPV) and specificity compared to the Heart Deep Learning model (CDLM), which showed the highest specificity (88.25%) and PPV (91.31%). The Predictive Analysis of Congenital Heart Defects (PACHD) model had the lowest sensitivity (59.78%) and PPV (56.45%), while the Machine Learning-Based Discharge Prediction (MLBDP) model had the lowest specificity (59.78%) and the highest miss rate (40.22%). These findings highlight the importance of selecting appropriate models based on performance metrics.
Conclusion
The DSSEP model demonstrated higher sensitivity and lower miss rates, making it strong for early detection, whereas the CDLM model offered higher specificity and PPV, reducing false positives.
Keywords
Congenital Heart Disease (CHD)
Ventricular septal defects (VSDs)
Heart Deep Learning Model
Electrocardiogram (ECG)
1 Introduction
Congenital Heart Disease (CHD) represents a significant structural anomaly present from birth, affecting various components of the heart, such as valves, walls, and major vessels. (Pachiyannan et al., 2023; Sun et al., 2020). Each year, millions of infants are diagnosed with CHD, making it the leading cause of death among newborns, accounting for over one-third of infant fatalities (Qu et al., 2022). Among the different types of CHD, the ventricular septal defect (VSD) is the most common, varying in severity and impacting the heart's efficiency (Mullen et al., 2021; Shivadekar et al., 2024). Despite advancements in prenatal diagnostics, many cases remain undetected until after birth due to subtle early signs and limitations in current diagnostic techniques. Fetal echocardiography is the primary method for diagnosing congenital heart disease (CHD) and is recommended to be performed by the supervising physician between 12 and 14 weeks of pregnancy for all women.
However, the effectiveness of this technique is often compromised by inconsistent interpretation skills among healthcare providers. As a result, current detection rates for CHD range from 30 % to 51 %, revealing a significant gap in early diagnosis (Nidhi et al., 2021). Machine learning (ML) techniques offer a promising solution to enhance these detection rates by improving the accuracy of fetal echocardiographic assessments. ML algorithms have demonstrated superior predictive performance compared to traditional methods, enabling better identification and classification of heart defects from diverse data sources, including imaging and electrocardiograms (Kumar and Singh, 2022; Khan et al., 2023). Addressing the challenges of CHD detection is important. These conditions often coexist with other health issues in newborns, complicating treatment options. Integrating machine learning with fetal echocardiography can significantly improve diagnostic accuracy. This integration enables timely interventions and reduces the disease burden on affected infants.
This study aims to detect congenital heart disease (CHD) by integrating fetal echocardiography data with machine learning models. This integration seeks to improve CHD detection through advanced algorithms. The effectiveness of various machine learning models is evaluated using key metrics such as sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
2 Materials and methods
2.1 Computational resources
The algorithm is built in a Jupyter Notebook using Anaconda and trained on a PC with Python v3.11 in a TPU-v3:8 environment with 16 GB of RAM. It utilizes TensorFlow and Keras, along with an 8-core Tensor Processing Unit (TPU), to speed up machine learning tasks.
2.2 Study population selection and data acquisition process
In this study, a dataset from an open-source database is utilized. It includes ultrasound evaluations of female patients at 12–14 weeks of gestational age (Stoean et al., 2022). This dataset has been referenced in other studies as well (Hutchinson et al., 2017; Ungureanu et al., 2023). The dataset includes 6,720 images taken from original video files showing four main views of the heart.: the atrioventricular flows in the four-chamber view, the aorta in the left ventricular outflow tract plane, the intersection of the great vessels in the right ventricular outflow tract plane (shaped like an “X”), and the arterial arches in the three-vessel plane (shaped like a “V”). An additional “other” class was also included, containing random frames that do not fit the specified categories. To diagnose heart defects, the study employed techniques like rhythmic tapping, palpation, auscultation, and physical examination, which are important for the early evaluation of congenital heart defects (CHDs) and help direct further imaging (Fig. 1).
Diagnosis of heart abnormalities.
Fig. 2 shows a holistic block diagram of the proposed CHD detection, classification, and therapy paradigm.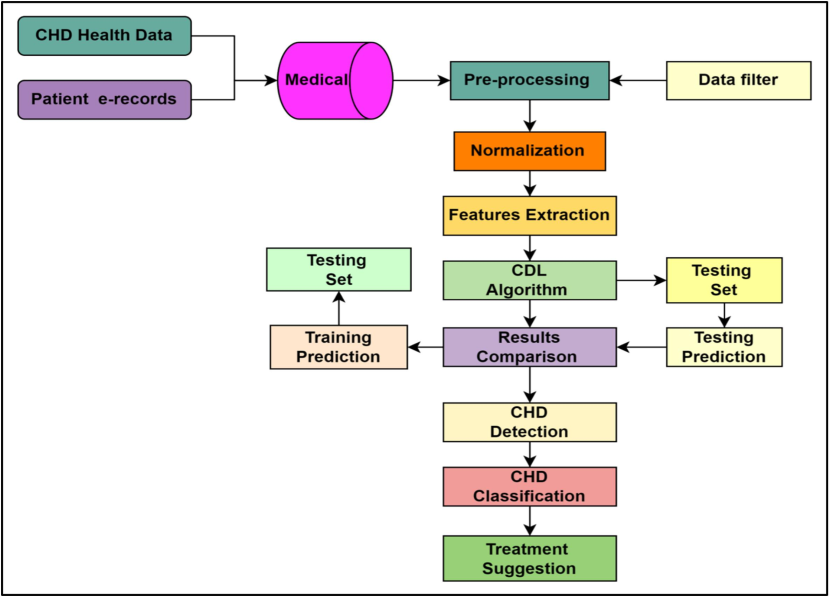
Flowchart showing the proposed design of the model.
2.3 Preprocessing
Pre-processing was done by 3 techniques, namely, K-Nearest Neighbors (KNN) estimation, “Min_Max” Normalization and One Hot encoding. KNN estimates the similarity of two data points and can identify similar cases within the obtained dataset. Then the similar data point is replaced by a missing value. In simpler language, it finds the closest point of the reference points, marked as “K”, which is based on the nearby reference points.
Min-max normalization is a popular data preprocessing technique employed to scale numerical features within a fixed range, typically between 0 and 1. The process involves two main steps. Firstly, the minimum and maximum values of the feature of interest are determined within the dataset. Subsequently, each data point in the feature is scaled using the following formula: X = original value of the feature
Xmin = the minimum value of the feature in the dataset.
Xmax = the maximum value of the feature in the dataset, and.
Xnormalized = the normalized value of the feature.
This normalization method proves beneficial when dealing with features that exhibit varying scales, enabling them to be compared on a level playing field without compromising the original data distribution. It finds widespread application in machine learning algorithms, particularly those reliant on gradient descent techniques, where the scale of features can significantly influence model performance. The last technique that is used during the pre-processing stage is “One hot encoding”, in which data analysis is conducted to convert categorical variables into a binary representation, involving each category within a variable as a binary vector, where only one element is marked as “1” while all others are “0”. This technique is particularly useful when dealing with categorical data in machine learning algorithms that require numerical input. Transforming categorical variables into this binary format, allows algorithms to interpret and process the data more effectively. For instance, if we have a variable like “color” with categories red, green, and blue, one hot encoding would represent each color as a distinct binary vector, facilitating analysis and modeling tasks.
2.4 Proposed model
The proposed machine learning approach to the detection of CHD involves using a supervised learning algorithm. The structure of this algorithm is outlined in Algorithm 1. It is trained using historical data of newborns with CHD, along with factors such as gestational age, birth weight, and maternal health history.
Algorithm
// Get ultrasound image samples and set the range;
Input: Ain; Output: Aout;
// Segment the images;
For each cluster pair (Ax_in, Ay_in)
If min(Ax_in, Ay_in) < |Qx_in − Qy_in|
Then merge Ax_in, Ay_in into Az;
// Feature extraction;
AZi = AZx_in + AZy_in;
Qi = (AZx_in * Qx_in + AZy_in * Qy_in) / (AZx_in + AZy_in);
// Preprocessing of samples;
Where Ain is the x × y matrix;
For i = 1:x
For j = 1:y
If (Ain(x,y) < 0)
Ain_s = 1;
Else
Ain_s = 0;
End;
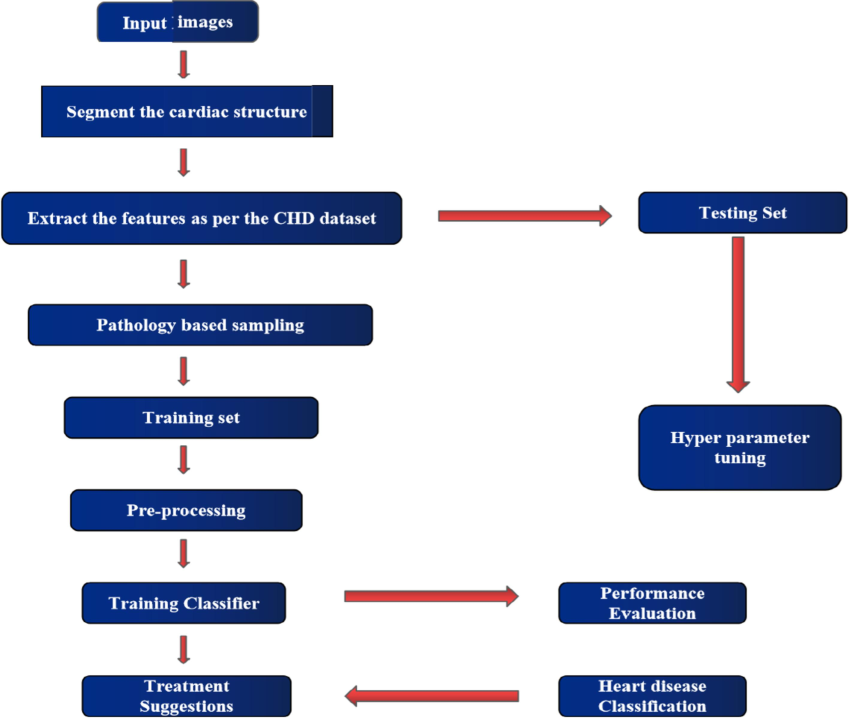
Proposed flow diagram showing the detection stage.
2.5 Comparative analysis
A comparison of CHD prediction models in a certain population includes accuracy, sensitivity, specificity, positive predictive value, and negative predictive value. The Heart DL Model (CDLM) is compared to the Novel Healthcare Framework (NHF), Decision Support System for Early Prediction (DSSEP), Machine Learning-Based Discharge Prediction (MLBDP), and Predictive Analysis of Congenital Heart Defects (PACHD) (Nurmaini et al., 2022).
Simulation is done with Matlab r2022a. The ratio of true positives (TP) to total diseased patients, presented as a percentage, measures a test's sensitivity (Se) in CHD prediction. A more sensitive test or treatment is more accurate. Specificity measures the ability to correctly identify true negative (TN) cases. It was calculated using The Positive Predictive Value (PPV) reflects the accuracy of a test in identifying individuals with CHD and was computed as NPV assesses the proportion of true negatives among negative predictions and was calculated using
3 Results
In this study, the predictive performance of various deep learning models was evaluated to diagnose congenital heart disease (CHD) using ultrasound images of fetuses at 12–14 weeks gestational age. The models assessed include NHF, DSSEP, MLBDP, PACHD, and CDLM, with a total dataset organized into training (80 %), validation (10 %), and test (10 %) sets. The TP, TN, FP, FN, accuracy, confusion matrix and ROC (AUC) curves for 100 images are presented in Table S1 and Figures (S1 and S2) (Supporting Information).
3.1 Sensitivity Measurements
Table 1 presents the evaluation of sensitivity percentages for each model in predicting heart disease. The results of the sensitivity evaluation indicate that CDLM significantly outperformed the other models, achieving a sensitivity range of 87.05 % to 91.55 %.
No. of Images
MLBDP
PACHD
CDLM
NHF
DSSEP
100
73.25
64.95
91.55
70.29
80.01
200
72.95
62.36
90.44
70.2
79.81
300
71.68
61.1
89.96
69.18
81.09
400
68.2
59.85
89.78
59.82
80.04
500
67.21
57.26
88.92
58.69
80.12
600
66.75
55.96
88.60
59.01
80.81
700
66.1
56.95
87.05
59.81
81.02
In contrast, NHF showed a sensitivity of 59.81 % to 70.29 %, while DSSEP maintained a more stable sensitivity around 79.81 % to 81.02 %. MLBDP and PACHD exhibited lower sensitivity, with MLBDP ranging from 66.10 % to 73.25 % and PACHD from 56.95 % to 64.95 %. The average sensitivity for all the models is plotted in Fig. 4.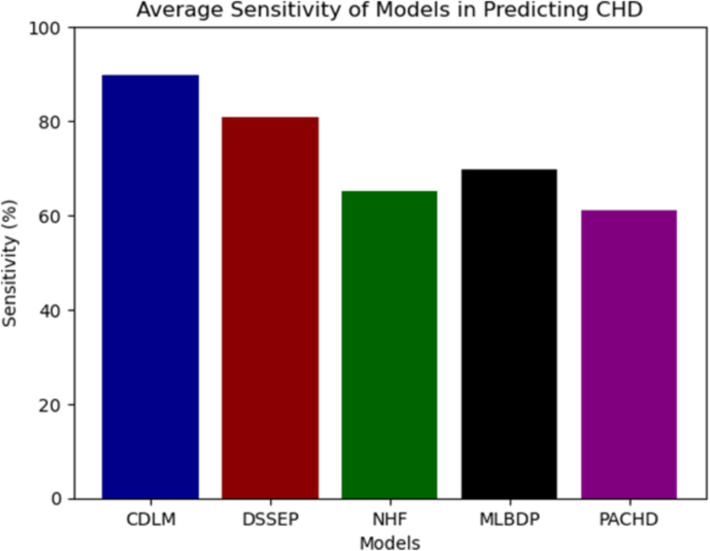
Average sensitivity of models.
3.2 Specificity evaluation
Table 2 summarizes the specificity percentages for the models, which measure the ability to correctly identify negative cases. Most models showed a decline in specificity as the number of images increased, indicating challenges in accurately identifying negative cases with larger datasets. CDLM, however, consistently maintained higher specificity across different image counts, demonstrating robust effectiveness in recognizing negative situations.
No. of Images
MLBDP
PACHD
CDLM
NHF
DSSEP
100
60.75
62.36
89.25
71.69
81.57
200
60.02
61.85
88.72
71.52
81.15
300
59.94
61.25
88.25
71.21
80.48
400
59.6
60.55
87.93
70.85
80.01
500
59.5
59.74
86.54
70.74
79.87
600
59.4
59.30
86.25
70.22
79.43
700
59.31
58.1
85.5
69.40
79.02
3.3 Positive Predictive Value (PPV)
Table 3 shows the PPV for the models, which indicates the proportion of true positives among all positive predictions. CDLM demonstrated consistently high PPV values, ranging from 85.32 % to 89.25 %. NHF’s PPV declined from 70.58 % to 65.51 %, indicating challenges in maintaining predictive accuracy. DSSEP started strong at 85.40 % but dropped to 75.05 %. MLBDP and PACHD exhibited lower PPV, with MLBDP ranging from 66.30 % to 72.48 % and PACHD from 52.10 % to 60.26 %.
No. of Images
MLBDP
PACHD
CDLM
NHF
DSSEP
100
72.48
60.26
89.25
70.58
85.4
200
71.95
59.12
88.73
69.93
82.15
300
70.45
57.64
88.22
69.1
79.5
400
69.99
56.3
87.12
68.74
79.1
500
68.39
55.2
88.59
66.68
78.5
600
67.65
54.70
88.42
65.9
77.4
700
66.30
52.1
85.32
65.51
75.05
Negative prediction values(NPV%) for proposed models are summarized in Table 4. CDLM exhibited the highest NPV values among the proposed models. That ranges from 82.66 % to 96.66 %. NPV for NHF decreased from 66.95 % to 61.25 %. DSSEP maintained a reasonable NPV from 79.02 % to 88.12 %, while both PACHD and MLBDP displayed lower NPV values.
No. of Images
MLBDP
PACHD
CDLM
NHF
DSSEP
100
70.99
71.69
96.66
66.95
79.02
200
66.44
67.62
93.56
64.91
79.96
300
62.12
63.72
92.1
64.29
80.85
400
59.75
60.73
89.69
64.25
80.12
500
56.21
57.4
85.69
63.95
80.45
600
52.45
55.68
85.42
62.33
88.12
700
50.96
54.95
82.66
61.25
81.01
Table 5 presents the findings of miss rates (%) across various healthcare frameworks. Miss rates represent the percentage of positive cases that are incorrectly classified as negative, indicating the model's failure to identify actual cases of congenital heart disease (CHD). In this evaluation, the proposed cardiac deep learning model (CDLM) achieved the lowest miss rates, ranging from 10.15 % to 10.92 %. NHF had significantly higher miss rates, ranging from 35.63 % to 36.85 %. DSSEP’s miss rates varied between 19.21 % and 20.48 %, while PACHD had the highest miss rates, between 39.47 % and 41.05 %.
Number of Images
MLBDP
PACHD
CDLM
NHF
DSSEP
100
30.95
39.8
10.27
35.74
20.38
200
29.95
41.05
10.8
36.85
19.21
300
30.25
39.47
10.34
35.96
20.12
400
30.21
40.88
10.15
36.5
20.03
500
30.82
40.45
10.92
36.15
19.77
600
30.43
39.98
10.68
35.63
20.48
700
31.36
40.03
10.4
36.77
19.25
Fig. 5 compares five healthcare frameworks' performance characteristics. Recall indicates the percentage of real positive cases the model properly identifies out of all positive cases. NHF detects 63.79 of positive cases with sensitivity. Specificity quantifies how many true negative cases the model accurately identifies out of all negative cases. For instance, DSSEP has 80.25 specificity, identifying 80.25 of negative cases. PPV, or precision, estimates the proportion of real positive instances among all model-identified positive cases.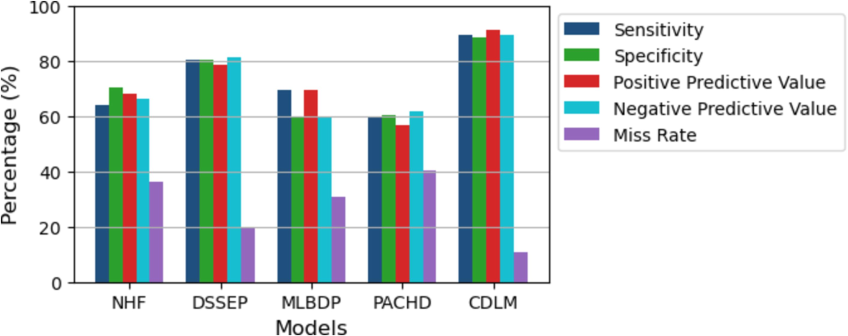
Comparison of the output of models.
MLBDP has a PPV of 59.87, meaning 59.87 of the positive results are real positives. NPV is the percentage of model-identified negative scenarios that are true. PACHD has an NPV of 89.25, meaning 89.25 of its negative situations are true negatives. The false negative rate measures the percentage of results that are false negatives. CDLM misses 89.25 of positive cases while misclassifying them as negative.
4 Discussion
The findings from this study provide valuable insights into the performance of various deep learning models in predicting congenital heart disease (CHD) using ultrasound images. The analysis reveals important trends and implications for clinical practice, emphasizing the need for a multifaceted evaluation of model performance.
4.1 Performance variability among models
The results show that sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) varied significantly across models. CDLM consistently outperformed the other models in sensitivity (89.45 %) and specificity (88.25 %), indicating its robustness in correctly identifying both positive and negative cases. This suggests that CDLM may be particularly effective for clinical applications where both accurate identification of disease and avoidance of false positives are crucial. Conversely, models like NHF and PACHD demonstrated lower sensitivity and higher miss rates, especially at larger image counts. This decline could suggest that these models may not generalize well with increased data variability or complexity. It raises concerns about their reliability in clinical settings, where accurate identification of CHD is critical for timely intervention.
4.2 Impact of dataset Size
The analysis revealed that larger dataset sizes often reduce model sensitivity and positive predictive value (PPV). This decline may stem from the challenges of training on diverse data, which can create prediction variability. In contrast, the CDLM model maintained more stable performance, suggesting better generalization for real-world use. Variability in specificity across models also points to difficulties in identifying negative cases as datasets expand. This highlights the need for ongoing evaluation and adjustment to sustain model performance. Codes from the ICD (International Classification of Diseases) in administrative information are frequently used for the detection of coronary heart disease (CHD) (Bhatt et al., 2023). However, these codes might incorrectly categorize individuals with true positive (TP) cases of heart disease. Improving CHD surveillance can be achieved by accurately detecting CHD in administrative documents with machine learning (ML) techniques. ML enhances the precision of identifying true positive cases compared to relying solely on ICD codes. This approach increases the relevance of findings from large datasets for the CHD patient group, thereby strengthening public health monitoring efforts (Shi et al., 2023).
Cardiovascular disease is the leading cause of death worldwide, and predicting it from clinical data is challenging. Machine learning (ML) is effective for diagnosing and predicting heart disease (Griffeth et al., 2023). Previous studies have applied ML techniques to forecast heart illness. In one study, eight ML classifiers were used to identify key features that enhance prediction accuracy (Mohsin et al., 2023). Neural network models like Naïve Bayes and Radial Basis Functions achieved accuracies of 95 % and 91 %, respectively. Vector Quantization demonstrated a 99 % reliability rate, outperforming other methods for predicting cardiovascular issues. The goal of predicting cardiovascular disease is to save lives, improve outcomes, and optimize healthcare resources. Key contributions include early intervention, personalized treatment, technological advancements, and ongoing research to mitigate the impact of coronary heart disease (Srinivasan et al., 2023).
5 Conclusion
This study has demonstrated the transformative potential of deep learning models in diagnosing congenital heart disease (CHD) from ultrasound images of fetuses at 12–14 weeks gestational age. The Cardiac Deep Learning Model (CDLM) emerged as the leading approach. That shows significant advantages in sensitivity and specificity compared to traditional models. Its ability to accurately identify both positive and negative cases makes it a promising tool for clinical settings, where timely and accurate diagnosis is crucial. The high positive predictive value (PPV) of CDLM indicates its effectiveness in minimizing false positives, which is essential for reducing unnecessary interventions and alleviating parental anxiety. Furthermore, the impressive negative predictive value (NPV) underscores its capacity to reliably identify healthy cases, ensuring that expectant parents can have confidence in their ultrasound results. However, the study also revealed challenges faced by other models, particularly NHF and PACHD, which exhibited declines in performance metrics as dataset sizes increased.
The dataset, while large, may not fully represent the diverse populations found in clinical practice. Variations in ultrasound image quality, gestational age, and prenatal care practices could affect model performance. Additionally, relying solely on one imaging method might limit effectiveness; incorporating multiple imaging techniques could enhance diagnostic accuracy.
Future research will aim to expand the dataset to include a broader range of groups and clinical scenarios. Utilizing additional imaging methods, such as MRI, could further improve accuracy. Lastly, long-term studies are essential to evaluate the impact of these models on patient outcomes in real-world settings.
CRediT authorship contribution statement
Ahmad Ali AlZubi: Writing – original draft, Funding acquisition, Conceptualization. Abdulrhman Alkhanifer: Writing – review & editing, Visualization, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Effective Heart Disease Prediction Using Machine Learning Techniques. Algorithms. 2023;16(2):88.
- [CrossRef] [Google Scholar]
- Impact of heart failure on reoperation in adult congenital heart disease: An innovative machine learning model. J. Thorac. Cardiovasc. Surg. 2023
- [Google Scholar]
- First-Trimester Fetal Echocardiography: Identification of Cardiac Structures for Screening from 6 to 13 Weeks’ Gestational Age. J. Am. Soc. Echocardiogr.. 2017;30(8):763-772.
- [CrossRef] [Google Scholar]
- Khan, A., Qureshi, M., Daniyal, M., & Tawiah, K. (2023). A Novel Study on Machine Learning Algorithm-Based Cardiovascular Disease Prediction. Health & Social Care in the Community, 2023.
- Kumar, G. K. L., & Singh, A. R. (2022). An Automated Heart Disease Diagnosis System Using Adaptive Cross-Layer Stacked Residual Convolutional Neural Networks.
- The role of artificial intelligence in prediction, risk stratification, and personalized treatment planning for congenital heart diseases. Cureus. 2023;15(8)
- [Google Scholar]
- Race and genetics in congenital heart disease: application of iPSCs, omics, and machine learning technologies. Front. Cardiovasc. Med.. 2021;8:635280
- [Google Scholar]
- Deep Learning for Improving the Effectiveness of Routine Prenatal Screening for Major Congenital Heart Diseases. J. Clin. Med... 2022;11(21):6454.
- [Google Scholar]
- A Heart Deep Learning Model (CDLM) to Predict and Identify the Risk Factor of Congenital Heart Disease. Diagnostics. 2023;13(13):2195.
- [Google Scholar]
- Using Innovative Machine Learning Methods to Screen and Identify Predictors of Congenital Heart Diseases. Front. Cardiovasc. Med.. 2022;8
- [CrossRef] [Google Scholar]
- A machine learning model for predicting congenital heart defects from administrative data. Birth Defects Res.. 2023;115(18):1693-1707.
- [Google Scholar]
- Evaluation of Machine Learning Methods for Predicting Heart Failure Readmissions: A Comparative Analysis. International Journal of Intelligent Systems and Applications in Engineering. 2024;12(6s):694-699.
- [Google Scholar]
- An active learning machine technique based prediction of cardiovascular heart disease from UCI-repository database. Sci. Rep.. 2023;13(1):13588.
- [Google Scholar]
- First Trimester Fetal Echocardiography Data Set for Classification. figshare. Figure. 2022
- [CrossRef] [Google Scholar]
- Nursing Care of Neonatal and Infant Congenital Heart Disease with 256-Slice Computed Tomography. Investigación Clínica. 2020;61(1):506-516.
- [Google Scholar]
- Learning deep architectures for the interpretation of first-trimester fetal echocardiography (LIFE) - a study protocol for developing an automated intelligent decision support system for early fetal echocardiography. BMC Pregnancy Childbirth. 2023;23(1)
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103555.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







