Translate this page into:
Identification of anticancer bioactive compounds derived from Ficus sp. by targeting Poly[ADP-ribose]polymerase 1 (PARP-1)
⁎Corresponding author at: King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia. shamstabrez1@gmail.com (Shams Tabrez)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The nuclear enzyme Poly [ADP-ribose] polymerase 1 (PARP-1) facilitates DNA damage repair which is essential for maintaining genome integrity. The PARP-1 has been touted as an important target for the anticancer therapeutic development as it is overexpressed in various forms of cancer. Several drugs have been approved, and many are under investigation to potentially target PARP-1 and inhibit cancer growth. Of late, anticancer drug development has witnessed a tremendous shift toward natural compounds because of the side effects of conventional therapies. In this study, an attempt has been made to identify the bioactive compounds of Ficus sp. that can potentially bind and inhibit PARP-1 activity by using molecular docking approach. For this purpose, 46 compounds have been retrieved from the IMPPAT database and filtered for their pharmacokinetic properties using Lipinski's Rule of five. The screened compounds were then subjected to docking-based virtual screening using PyRx software, and finally, a total of eight compounds were selected based on their binding affinity towards PARP-1. The interactions of these ligands with PARP-1 were further studied using site-specific molecular docking. These ligands showed strong interaction via several hydrogen bonds and hydrophobic interactions with PARP-1 and had low inhibitory constant (Ki). Based on these findings, it is evident that the bioactive compounds derived from Ficus sp. can be considered as potential anticancer drug candidates. However, further studies in different in vitro and in vivo models would corroborate the inhibitory activity of the ligands targeting PARP-1.
Keywords
Cancer therapy
Ficus sp.
PARP-1
Phytomedicine
Molecular docking
1 Introduction
The maintenance of genome integrity is crucial for cell sustenance and functioning, and prevention of diseases such as cancer. This intricate process is elegantly carried out by various DNA repair factors to strictly scrutinize, detect and repair DNA damage. Impairment of this repair mechanism causes genome instability, a known hallmark of cancer, leading to cancer initiation, development, and progression (Singh et al., 2020; Yusoh et al., 2020). Poly (ADP-ribose) polymerases (PARPs) are nuclear enzymes that catalyze poly-ADP-ribosylation (PARylation), that is, the transfer of ADP-ribose moieties to target proteins (Rose et al., 2020). The PARP-1, a vital member of the human PARP enzymes, regulates several key cellular processes via PARylation, such as chromatin remodeling, DNA damage repair, transcription, replication, inflammation, and metabolism (Dias et al., 2021). The PARP-1 senses the occurrence of any DNA damage and facilitates its repair. It primarily promotes the restoration of single-stranded DNA breaks via the base-excision repair pathway by recruiting other repair factors (Singh et al., 2020).
It has been reported in several studies that the loss-of-function mutation of breast and ovarian cancer-associated tumor suppressor genes BRCA1/2 promote PARP-1 hyperactivation in cancer cells (Dias et al., 2021; Singh et al., 2020). However, several other BRCA mutation-independent cancers, including gastric, pancreatic, and lung cancer, have also been reported to have overexpressed PARP-1 activity. Therefore, PARP-1 has been touted as an important target for novel anticancer therapeutic development. Several PARP-1 inhibitors have been approved for clinical use, and many potential drugs are being investigated on human subjects and at the preclinical stages. For instance, to date, the US Food and Drug Administration (FDA) has approved four PARP inhibitors: Olaparib, niraparib, rucaparib, and talazoparib for the treatment of ovarian and breast cancer (Mirza et al., 2020; Wang et al., 2021).
The conventional therapeutic regimes for cancer are often associated with various side effects and toxicities. Therefore, efforts are being made to develop new cancer therapeutics from natural sources with minimum side effects (Alam et al., 2022; Uddin and Hoque, 2021). Plant extracts are being used to treat various diseases, including cancer, for centuries. Approximately 25% of all anticancer drugs approved between 1981 and 2019 were derived from natural sources (Huang et al., 2021). The fig plants of genus Ficus belonging to the Moraceae family have been reported to possess tremendous medicinal properties. These plants are traditionally used to treat various diseases such as respiratory, gastrointestinal, cardiovascular, diabetes, and cancer (Gurung et al, 2021). The anticancer properties of crude extracts of the Ficus sp. plants have been reported in several studies (Choudhari et al., 2013; Gurung et al., 2021; Soltana et al., 2019; Sumi et al., 2021). However, the PARP-1 inhibitory activity of Ficus sp. derived compounds has not been reported so far. This study attempts to identify the potent bioactive compounds derived from various Ficus sp. that potentially bind and inhibit PARP-1 using molecular docking approach and explore the possible underlying anticancer mechanisms.
2 Materials and methods
2.1 Preparation of ligand
The 3D structures of bioactive compounds derived from eight Ficus sp. (Ficus religiosa, Ficus racemosa, Ficus palmata, Ficus lacor, Ficus hispida, Ficus carica, Ficus benghalensis and Ficus benjamina) were retrieved from the IMPPAT: Indian Medicinal Plants, Phytochemistry And Therapeutics, a curated database (https://cb.imsc.res.in/imppat/home) in .sdf format (Mohanraj et al., 2018). Using Open Babel GUI, all atomic coordinates were modified to .pdbqt set-up. We employed the PyRx integrated universal force field for optimization and ligand preparation. Using Data Warrior Program, version 5.5.0, the bioactive compounds of Ficus sp. were filtered out based on the Lipinski rule of five (Sander et al., 2015). The ligands with zero violations of the rule were considered for further studies to identify potential inhibitors against PARP-1.
2.2 Preparation of target protein
The 3D structure of the target protein PARP-1 was retrieved from the Protein Data Bank (https://www.rcsb.org/) (PDB ID: 6I8M). The protein was then prepared for molecular docking studies by removing the heteroatoms (water and ions), adding polar hydrogen, and assigning Kollman charges. Grid boxes of appropriate sizes were placed around the bound cocrystal ligand to demarcate the active areas.
2.3 Virtual screening
For the best ideal hit against the PARP-1 target, the natural compound library was prepared from the IMPPAT database after filtering based on the Lipinski rule of five. The virtual screening was performed to identify potential PARP-1 inhibitors using PyRx with default settings, and the best hits were selected based on their set binding affinity threshold of ≤−8.0 kcal/mol. For further analysis, these selected compounds were subjected to site-specific docking.
2.4 Molecular docking
The selected bioactive components of Ficus sp. were subjected to site specific molecular docking against the PARP-1 molecular target using the AutoDock 4.2 program. The Lamarckian Genetic Algorithm was employed with the following parameters: a starting population of 150 randomly inserted individuals; a maximum number of energy assessments of 2,500,000; a mutation rate of 0.02; and a crossover rate of 0.8. For each ligand, a total of 50 dockings were done independently. The grid box had the following centre points: X: 6.610, Y: 23.444, Z: 20.261, and the following dimensions: X:70, Y:70, and Z:70. The grid point spacing was 0.375. The conformation with the lowest binding free energy (ΔG) and inhibition constant (Ki) was considered to be the best. The BIOVIA Discovery Studio was used to examine the molecular interactions of the selected ligands with the receptor.
2.5 ADMET prediction
The physicochemical and pharmacokinetic features of the potential compounds were evaluated by analyzing their ADMET properties predicted using the online tool pkCSM (Pires et al., 2015).
3 Results and discussion
The PARP-1 has been an excellent target for anticancer therapeutic development. Previously, PARP-1 targeting bioactive compounds derived from Rauwolfia serpentina were identified by employing molecular docking technique (Abuzenadah et al., 2022a). The molecular docking is an in silico technique that can be conveniently used to identify potential drug candidates against certain disease specific target proteins (Abuzenadah et al., 2022b; Jabir et al., 2021a, 2021b, 2021c). In this study, to identify the potential bioactive compounds derived from various Ficus sp., the ligands were retrieved from the IMPPAT database and filtered based on the Lipinski rule of five using the DataWarrior program, version 5.5.0 (Sander et al., 2015). The compounds that obeyed the rule were selected for further investigation. The Lipinski rule of five provides reliable guidelines, used as a thumb rule to identify 'drugability' and pharmacological activities of novel compounds suggesting possible clinical use (Lipinski et al., 1997). Forty-six bioactive compounds of Ficus sp. were retrieved from the IMPPAT database (Supplementary Table S1). Out of these, only 19 compounds were found to obey the Lipinski rule of five with zero violation of the rule and hence selected for further studies. Other 27 compounds showed a rule violation at least in one parameter and therefore not considered for possible therapeutic evaluation against PARP-1. The Lipinski rule of five parameters for all the ligands are shown in Supplementary Table S2, where the selected compounds are highlighted.
The compounds filtered based on the Lipinski rule of five were then subjected to molecular docking-based virtual screening to identify possible inhibitors of PARP-1 using PyRx software. The docking scores of these compounds are shown in Supplementary Table S3. The best ligands were selected based on the set threshold binding energy of −8.0 kcal/mol. There were a total of eight compounds found to be docked against PARP-1 with a stronger binding energy of ≤−8.0 kcal/mol. The molecular structures of these selected compounds are shown in Table 1. These eight bioactive compounds of Ficus sp. showing greater affinity towards PARP-1 in molecular docking-based virtual screening were further analyzed in site-specific molecular docking using AutoDock 4.2 to find prospective therapeutic candidates that can effectively bind and inhibit PARP-1. A redocking experiment with a cocrystal ligand was performed to validate the docking technique and algorithm before the molecular docking studies with the ligands. The root means square deviation (RMSD) between the docked and native cocrystal positions was less than 2 Å in all cases, indicating that our docking parameters were correct. Table 1 shows the molecular docking results of the chosen ligands against PARP-1, including binding energy (ΔG), minimal inhibition constant (Ki), and interacting amino acid residues.
Sl No
Ligand Name
Plant Source
Structure
ΔG (kcal/mol)
Ki
Interactive Residues
1
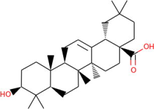
Oleanolic acid
Ficus hispida, Ficus carica
−9.84
60.77 nM
Tyr710, Gln763, Asp766, His862, Ile872, Gly876, Leu877, Arg878, Ile879, Ala880, Pro881, Tyr889, Gly894, Ile895, Tyr896, Tyr907
2
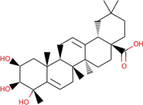
Triterpenoids
Ficus lacor
−9.39
130.40 nM
Gln763, Asp766, Asp770, His862, Gly863, Ile872, Gly876, Ley877, Arg878, Ile879, Ala880, Tyr889, Lys893, Gly894, Ile895, Tyr896, Tyr907
3
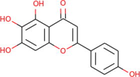
Scutellarein
Ficus lacor
−8.97
265.55 nM
Trp861, His862, Gly863, Ser864, Tyr896, Phe897, Ala898, Gly888, Tyr889, Met890, Lys903, Ser904, Tyr907, Glu988
4
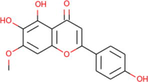
Sorbifolin
Ficus lacor
−8.49
602.84 nM
Trp861, His862, Gly863, Gly888, Tyr889, Met890, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Glu988
5
Guaiol acetate
Ficus hispida
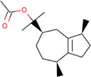
−8.4
695.75 nM
His862, Gly863, Gly894, Ile895, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Glu988
6
Nodakenetin
Ficus carica

−7.66
2.43 μM
Trp861, His862, Gly863, Gly888, Tyr889, Met890, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Glu988
7
Hispidin
Ficus hispida
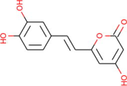
−7.52
3.08 μM
Trp861, His862, Gly863, Gly888, Tyr889, Met890, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Leu984, Glu988
8
Imperatorin
Ficus benjamina
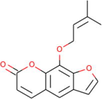
−7.15
5.77 μM
Trp861, His862, Gly863, Tyr889, Tyr896, Phe897, Gly894, Ile895, Ala898, Lys903, Tyr907, Glu988
Oleanolic acid derived from Ficus hispida and Ficus carica, was shown to be the best docked to PARP-1 with ΔG of −9.84 kcal/mol and Ki of 60.77 nM, introducing two hydrogen bonds through Arg878 and other important and hydrophobic interactions via residues Tyr710, Gln763, Asp766, His862, Ile872, Gly876, Leu877, Ile879, Ala880, Pro881, Tyr889, Gly894, Ile895, Tyr896, and Tyr907 (Fig. 1). Previous studies also suggest the possible anticancer role of oleanolic acid against various forms of cancer by targeting multiple signaling pathways (Tang et al., 2022; Žiberna et al., 2017). The strong binding affinity and small inhibition constant of oleanolic acid against PARP-1 suggest its possible therapeutic use against cancer by specifically targeting PARP-1. However, further studies in animal models and human trials will ensure its use for clinical purposes.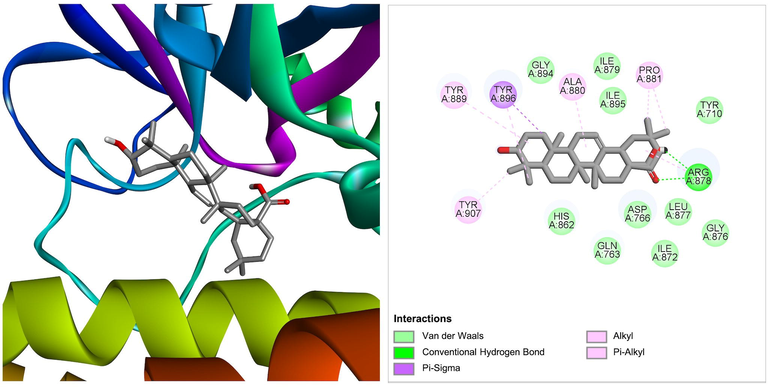
The molecular interaction of oleanolic acid derived from Ficus hispida and Ficus carica with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and oleanolic acid.
The compound named under triterpenoids (derived from Ficus lacor) was found to interact with target PARP-1, with a ΔG of −9.39 kcal/mol and a Ki of 130.40 nM, introducing two hydrogen bonds through Gln763, and Arg878, as well as other significant and hydrophobic contacts via residues Asp766, Asp770, His862, Gly863, Ile872, Gly876, Ley877, Ile879, Ala880, Tyr889, Lys893, Gly894, Ile895, Tyr896, Tyr907 (Table 1). The molecular interaction between the molecule and PARP-1 has been shown in Fig. 2. The triterpenoids have been previously reported for their potential ability to target cancer cells by modulating their metabolism (Mamouni et al., 2021). Our findings would augment the research on triterpenoids-based anticancer drug development.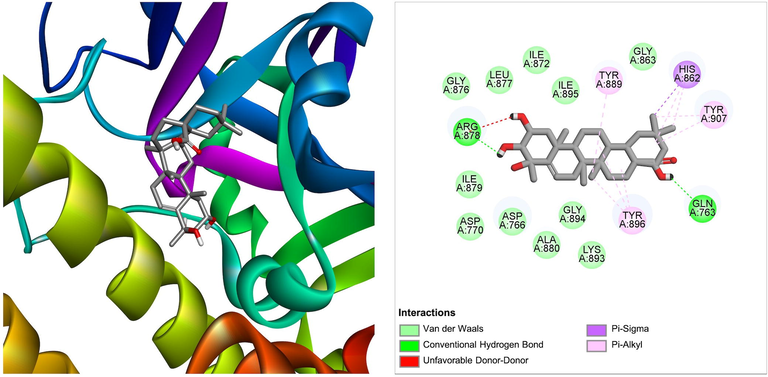
The molecular interaction of triterpenoids derived from Ficus lacor with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and triterpenoids.
Another compound scutellarein derived from Ficus lacor was found to best bind the PARP-1 with a ΔG of −8.97 kcal/mol and a Ki of 265.55 nM, introducing four hydrogen bonds through Gly863, Gly888, and Ser904, additional significant hydrophobic contacts via residues Trp861, His862, Ser864, Tyr896, Phe897, Ala898, Tyr889, Met890, Lys903, Tyr907, Glu988 (Table 1). Fig. 3 shows the molecular interactions between the scutellarein and PARP-1. Several studies reported the anticancer activity of the flavonoid scutellarein by inducing apoptosis and inhibiting metastasis (Abusaliya et al., 2021). However, its ability to bind and inhibit PARP-1 has not been explored so far. Our findings would pave the way to conduct further studies on the anticancer activity of scutellarein by targeting PARP-1.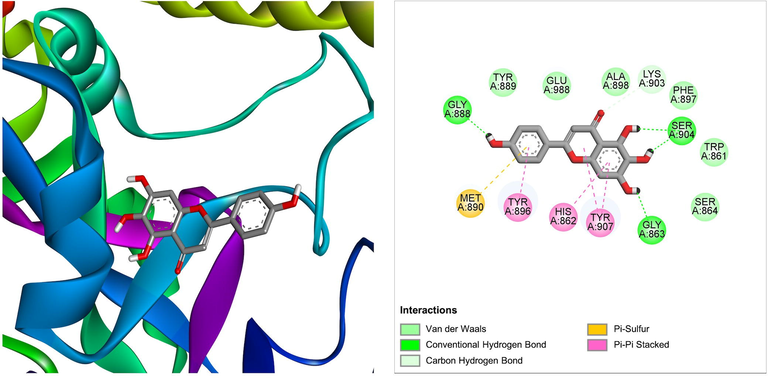
The molecular interaction of scutellarein derived from Ficus lacor with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and scutellarein.
Another flavonoid from Ficus lacor, the sorbifolin, was shown to be the best docked to PARP-1 with ΔG of −8.49 kcal/mol and Ki of 602.84 nM, introducing three hydrogen bonds through Gly888, Met890, and Ser904, other important and hydrophobic interactions via residues Trp861, His862, Gly863, Tyr889, Tyr896, Phe897, Ala898, Lys903, Tyr907, Glu988 (Table 1). The molecular interaction between the PARP-1 receptor and flavonoid sorbifolin is shown in Fig. 4. Sorbifolin has previously been reported for its antiviral activity and protective role in myocarditis (Badshah et al., 2021; Enayati et al., 2021). Its role in cancer therapeutic development remains unexplored. To this end, our findings suggest its potential anticancer activity by targeting PARP-1.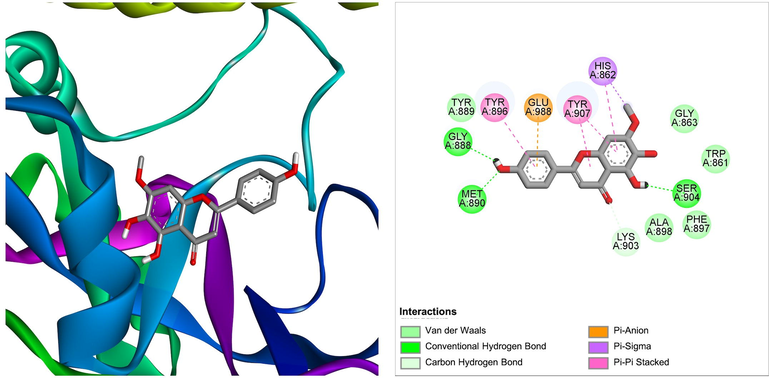
The molecular interaction of sorbifolin derived from Ficus lacor with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and sorbifolin.
The compound guaiol acetate derived from Ficus hispida was found to interact with target PARP-1 via residues His862, Gly863, Gly894, Ile895, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, and Glu988, with a ΔG of −8.4 kcal/mol and a Ki of 695.75 nM (Table 1). The molecular interactions between the target proteins and the ligand are shown in Fig. 5. The anticancer activity of this compound has not been reported earlier. The results obtained in this study show the promising ability of guaiol acetate as an anticancer agent, especially by targeting the PARP-1 receptor.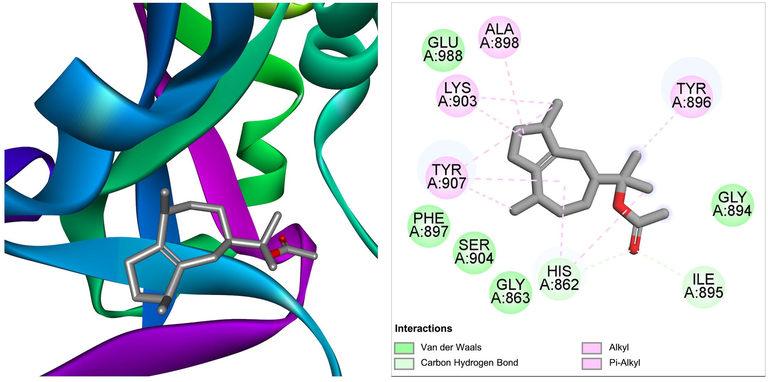
The molecular interaction of guaiol acetate derived from Ficus hispida with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and guaiol acetate.
Nodakenetin derived from Ficus carica was shown to be the best docked against PARP-1 with ΔG of −7.66 kcal/mol and Ki of 2.43 μM, introducing one hydrogen bond through Met890, other important and hydrophobic interactions via residues Trp861, His862, Gly863, Gly888, Tyr889, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Glu988 (Table 1). Molecular interactions between PARP-1 and the ligand at the best binding pose are shown in Fig. 6. With a ΔG of −7.52 kcal/mol and a Ki of 3.08 μM, another bioactive compound hispidin derived from Ficus hispida was found to be the best docked to PARP-1, introducing four hydrogen bonds through Trp861, Gly863, Gly888, and Met890, additional significant hydrophobic contacts via residues His862, Tyr889, Tyr896, Phe897, Ala898, Lys903, Ser904, Tyr907, Leu984, Glu988 (Table 1). The molecular interactions between the PARP-1 and hispidin at the best binding pose are shown in Fig. 7. The hispidin found in many fungal and plant species has been reported to possess several medicinal properties, including antioxidant, anti-inflammatory, anti-apoptotic, antiviral, and anticancer activity (Palkina et al., 2021).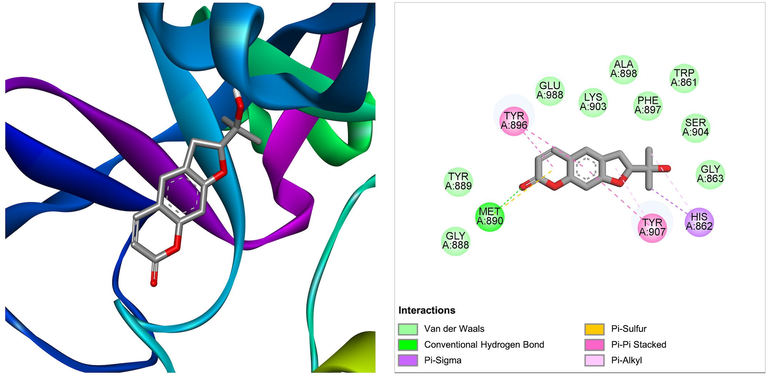
The molecular interaction of nodakenetin derived from Ficus carica with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and nodakenetin.
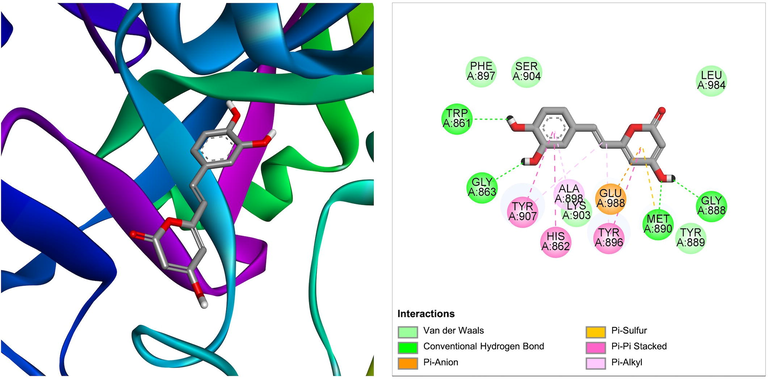
The molecular interaction of hispidin derived from Ficus hispida with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and hispidin.
Similarly, a compound derived from Ficus benjamina, imperatorin was found to bind at the best pose to PARP-1 target, with a ΔG of −7.15 kcal/mol and a Ki of 5.77 μM, involving one hydrogen bond through Gly863, as well as additional significant and hydrophobic contacts through residues Trp861, His862, Tyr889, Tyr896, Phe897, Gly894, Ile895, Ala898, Lys903, Tyr907, Glu988 (Table 1). The molecular interactions between the PARP-1 target and imperatorin at the best binding pose are shown in Fig. 8. The imperatorin is a naturally occurring furanocoumarin, that shows several pharmacological activities, including anticancer, anti-inflammatory, anti-hypertension, antibacterial, and neuroprotective roles (Deng et al., 2020). Our studies surely expand the scope of exploring the anticancer activity by specifically targeting PARP-1 molecules.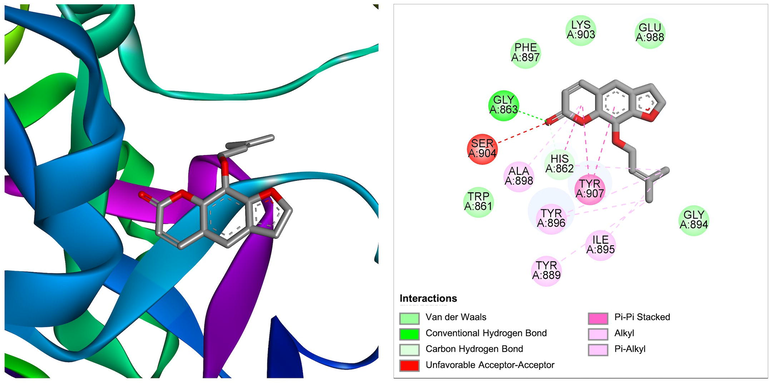
The molecular interaction of imperatorin derived from Ficus benjamina with PARP-1. The 3D and 2D images were generated by using BIOVIA Discovery Studio showing amino acid residues involved in various interactions between PARP-1 and imperatorin.
Overall, out of total 46 bioactive compounds derived from the eight species of Ficus plant as obtained from IMPPAT database, only 8 compounds were identified to potentially target PARP-1. These selected compounds are mainly derived from only four species of Ficus plant. No compounds derived from Ficus religiosa, Ficus racemosa, Ficus benghalensis, and Ficus palmata were found to possess strong binding affinity for PARP-1. All the selected compounds that exhibit strong affinity for PARP-1 are derived from Ficus hispida, Ficus carica, Ficus lacor, and Ficus benjamina. The ligand oleanolic acid that showed best binding affinity for PARP-1 is found in both Ficus hispida and Ficus carica. Moreover, the ligands guaiol acetate and hispidin are also derived from Ficus hispida. The Ficus carica derived another compound nodakenetin also showed strong binding against PARP-1. Similarly, there are three ligands derived from Ficus lacor (triterpenoids, scutellarein, and sorbifolin) found to display strong binding affinity for PARP-1 indicating their anticancer therapeutic potential. On the other hand, only one compound derived from Ficus benjamina was found to possess binding affinity and inhibitory potential against PARP-1.
The selected molecules derived from Ficus sp. were further investigated to predict their pharmacokinetic features of using a set of parameters in ADMET (Table 2). Of the selected eight compounds, only three, triterpenoids, nodakenetin, and imperatorin, showed toxicity patterns. All compounds have an intestine absorption rate of greater than 30%, indicating that they are well-absorbed molecules (Pires et al., 2015). Further modification of the compounds and modulating their concentration would ensure their safety for clinical purposes.
Sl. No.
Ligand Name
Absorption
Distribution
Metabolism
Excretion
Toxicity
Intestinal absorption (human)
Water solubility
BBB permeability
CYP2D6 substrate
CYP2D6 inhibition
Renal OCT2 substrate
AMES toxicity
Hepatotoxicity
1
Oleanolic acid
98.956
−3.399
−0.158
No
No
No
No
No
2
Triterpenoids
67.761
−3.162
−0.628
No
No
No
No
Yes
3
Scutellarein
73.581
−3.156
−1.263
No
No
No
No
No
4
Sorbifolin
77.413
−3.584
−1.173
No
No
No
No
No
5
Guaiol acetate
94.836
−4.617
0.511
No
No
No
No
No
6
Nodakenetin
96.502
−2.505
0.024
No
No
No
Yes
Yes
7
Hispidin
91.731
−2.519
−0.981
No
No
No
No
No
8
Imperatorin
95.987
−3.786
0.221
No
No
Yes
Yes
No
4 Conclusion
The PARP-1 enzyme has been found to be overexpressed in various cancers, and therefore it is considered as an excellent target for anticancer therapeutic development. This study reports eight bioactive compounds (oleanolic acid, triterpenoids, scutellarein, sorbifolin, guaiol acetate, nodakenetin, hispidin, and imperatonin) derived from four species of figs (Ficus hispida, Ficus carica, Ficus lacor, and Ficus benjamina) with strong binding affinity and inhibitory activity against PARP-1, indicating their potential as anticancer therapeutics. Based on these findings, it is evident that the bioactive compounds derived from Ficus sp. can potentially be used as cancer therapeutics by targeting PARP-1. However, further studies on different in vitro and in vivo models would be required to corroborate their therapeutic and pharmacological features.
Acknowledgements
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number “IFPRC-146-141-2020” and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Competing financial interests
The authors declare no competing financial interests.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Glycosidic flavonoids and their potential applications in cancer research: a review. Mol. Cell. Toxicol. 2021
- [CrossRef] [Google Scholar]
- Identification of potential Poly (ADP-Ribose) polymerase-1 inhibitors derived from rauwolfia serpentina: possible implication in cancer therapy. Evid.-Based Compl. Alternative Med.. 2022;2022:1-9.
- [Google Scholar]
- Elucidating anti-angiogenic potential of Rauwolfia serpentina: VEGFR-2 targeting based molecular docking study. Evid.-Based Compl. Alternative Med.. 2022;2022:1-10.
- [Google Scholar]
- Therapeutic and pharmacological potential of Tanshinones against lung cancer: a systematic review. Phytomed. Plus. 2022;2(1):100202.
- [Google Scholar]
- Antiviral activities of flavonoids. Biomed. Pharmacother.. 2021;140:111596
- [CrossRef] [Google Scholar]
- The aqueous extract of ficus religiosa induces cell cycle arrest in human cervical cancer cell lines SiHa (HPV-16 positive) and apoptosis in HeLa (HPV-18 positive) PLoS ONE. 2013;8:e70127
- [CrossRef] [Google Scholar]
- Imperatorin: a review of its pharmacology, toxicity and pharmacokinetics. Eur. J. Pharmacol.. 2020;879:173124
- [CrossRef] [Google Scholar]
- Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol.. 2021;18:773-791.
- [CrossRef] [Google Scholar]
- Protective role of nutraceuticals against myocarditis. Biomed. Pharmacother. 2021:112242.
- [Google Scholar]
- Molecular docking and dynamics simulation study of bioactive compounds from Ficus carica L. with important anticancer drug targets. PLOS ONE. 2021;16:e0254035
- [CrossRef] [Google Scholar]
- Natural products in cancer therapy: past, present and future. Nat. Prod. Bioprospecting. 2021;11:5-13.
- [CrossRef] [Google Scholar]
- Concatenation of molecular docking and molecular simulation of BACE-1, γ-secretase targeted ligands: in pursuit of Alzheimer’s treatment. Ann. Med.. 2021;53(1):2332-2344.
- [Google Scholar]
- Identification of butyrylcholinesterase and monoamine oxidase B Targeted ligands and their putative application in Alzheimer’s treatment: a computational strategy. Curr. Pharm. Des.. 2021;27:2425-2434.
- [CrossRef] [Google Scholar]
- In silico screening of glycogen synthase kinase-3β targeted ligands against acetylcholinesterase and its probable relevance to Alzheimer’s disease. J. Biomol. Struct. Dyn.. 2021;39:5083-5092.
- [CrossRef] [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 1997;23:3-25.
- [CrossRef] [Google Scholar]
- Targeting mitochondrial metabolism in prostate cancer with triterpenoids. Int. J. Mol. Sci.. 2021;22:2466.
- [CrossRef] [Google Scholar]
- The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann. Oncol.. 2020;31:1148-1159.
- [CrossRef] [Google Scholar]
- IMPPAT: A curated database of Indian medicinal plants. Phytochem. Therap. Sci. Rep.. 2018;8:4329.
- [CrossRef] [Google Scholar]
- Therapeutic potential of hispidin—fungal and plant polyketide. J. Fungi. 2021;7:323.
- [CrossRef] [Google Scholar]
- pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem.. 2015;58:4066-4072.
- [CrossRef] [Google Scholar]
- PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front. Cell Dev. Biol.. 2020;8:564601
- [CrossRef] [Google Scholar]
- DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model.. 2015;55:460-473.
- [CrossRef] [Google Scholar]
- Therapeutic strategies and biomarkers to modulate PARP activity for targeted cancer therapy. Cancers. 2020;12:972.
- [CrossRef] [Google Scholar]
- Antitumoral activity of Ficus carica L. on colorectal cancer cell lines. Cell. Mol. Biol.. 2019;65:6.
- [CrossRef] [Google Scholar]
- Pharmacological activities of Ficus racemosa and analysis of its major bioactive polyphenols by HPLC-DAD. In: Rizvi A., ed. Highlights on Medicine and Medical Science Vol. 5. Book Publisher International (a part of SCIENCEDOMAIN International). 2021. p. :68-84.
- [CrossRef] [Google Scholar]
- Anticancer activity of oleanolic acid and its derivatives: recent advances in evidence, target profiling and mechanisms of action. Biomed. Pharmacother.. 2022;145:112397
- [CrossRef] [Google Scholar]
- Non-flavonoids targeting cancer stem cells: a promising therapeutic avenue for cancer treatment. In: Tabrez S., Imran Khan M., eds. Polyphenols-based Nanotherapeutics for Cancer Management. Singapore: Springer Singapore; 2021. p. :289-334.
- [Google Scholar]
- PARP inhibitors in gastric cancer: beacon of hope. J. Exp. Clin. Cancer Res.. 2021;40:211.
- [CrossRef] [Google Scholar]
- Combining PARP inhibition with platinum, ruthenium or gold complexes for cancer therapy. ChemMedChem. 2020;15:2121-2135.
- [CrossRef] [Google Scholar]
- Oleanolic acid alters multiple cell signaling pathways: implication in cancer prevention and therapy. Int. J. Mol. Sci.. 2017;18:643.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102079.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







