Translate this page into:
The nanomolar affinity of C-phycocyanin from virtual screening of microalgal bioactive as potential ACE2 inhibitor for COVID-19 therapy
⁎Corresponding authors at: Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Padjadjaran, Jl. Raya Bandung Sumedang KM. 21, 45363 Jatinangor, Indonesia (Dr. Muhammad Yusuf). fiddy.semba.prasetiya@brin.go.id (Fiddy S. Prasetiya), m.yusuf@unpad.ac.id (Muhammad Yusuf)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The global pandemic of COVID-19 caused by SARS-CoV-2 has caused more than 400 million infections with more than 5.7 million deaths worldwide, and the number of validated therapies from natural products for treating coronavirus infections needs to be increased. Therefore, the virtual screening of bioactive compounds from natural products based on computational methods could be an interesting strategy. Among many sources of bioactive natural products, compounds from marine organisms, particularly microalgae and cyanobacteria, can be potential antiviral agents. The present study investigates bioactive antiviral compounds from microalgae and cyanobacteria as a potential inhibitor of SARS-CoV-2 by targeting Angiotensin-Converting Enzyme II (ACE2) using integrated in silico and in vitro approaches. Our in silico analysis demonstrates that C-Phycocyanin (CPC) can potentially inhibit the binding of ACE2 receptor and SARS-CoV-2 with the docking score of −9.7 kcal mol−1. This score is relatively more favorable than the native ligand on ACE2 receptor. Molecular dynamics simulation also reveals the stability interaction between both CPC and ACE2 receptor with a root mean square deviation (RMSD) value of 1.5 Å. Additionally, our in vitro analysis using the surface plasmon resonance (SPR) method shows that CPC has a high affinity for ACE2 with a binding affinity range from 5 to 125 µM, with KD 3.37 nM. This study could serve as a reference to design microalgae- or cyanobacteria-based antiviral drugs for prophylaxis in SARS-CoV-2 infections.
Keywords
Bioactive compounds
Microalgae
Molecular docking
Molecular dynamics
SARS-CoV-2
Surface plasmon resonance
1 Introduction
Among the various diseases caused by viruses, the 2019 coronavirus disease (COVID-19) caused by a new coronavirus (SARS-CoV-2) was first detected in December 2019 and has since become a global pandemic (Chan et al., 2020; Chen et al., 2020). This virus has been reported as a new member of the β-coronavirus genus. It is closely related to severe acute coronavirus respiratory syndrome (SARS-CoV) and several bat coronaviruses (Zhou et al., 2020). Compared to coronavirus types previously found (SARS-CoV and MERS-CoV), the SARS-CoV-2 virus shows faster human-to-human transmission. Thus, the World Health Organization (WHO) has established public health emergencies worldwide (Chan et al. 2020; Chen et al., 2020). Additionally, therapeutic compounds and effective treatment options targeting this virus are still minimal. Therefore, efforts are needed to design new drugs that can be used as SARS-CoV-2 antivirus candidates with virtual drug screening methods. To date, no effective antiviral therapy has been found. However, several broad-spectrum antivirals have been recommended, such as the Nucleoside analogs and HIV-Protease Inhibitors (lopinavir, ritonavir), as an alternative to temporary therapy until specific antivirals are found (Zhou et al., 2020). Additionally, aside from vaccine development, repurposing approved antiviral drugs (e.g., remdesivir) is a practical clinical approach to overcome the SARS-CoV-2 global pandemic (Wu et al., 2020). Nevertheless, designing broad-spectrum antiviral agents that are effective against a wide range of SARS-CoV-2 and other emergent classes of viruses could be a sound strategy (Cho and Glenn, 2020; Hall and Ji, 2020).
The broad spectrum of antiviral drugs can be divided into two mechanisms of action: 1) by inhibiting the interactions between virus particles outside of cells and the receptors on the cell surface, thus preventing infection (Hangartner et al., 2006; Kim et al., 2012) and 2) by stopping viral genome replication to minimize the production of new virus particles (Clercq, 2004; de Wilde et al., 2018). Previous studies revealed that the Angiotensin Converting Enzyme-2 (ACE2) receptor has a significant role in SARS-CoV-2 infection (Wan et al., 2020; Zhang et al., 2020). SARS-CoV-2 recognizes this receptor to facilitate the entry of the virus into the target cell. In addition, the binding site between the ACE2 receptor and SARS-CoV has been identified at the molecular level (Zhang et al., 2020). Therefore, inhibiting this interaction with ligands on the ACE2 receptor site can be a potential approach in the preliminary drug discovery for COVID-19 therapy (Zhang et al., 2020).
Microalgae and cyanobacteria are essential organisms at the basis of most aquatic ecosystems. They can produce various high-value bioactive compounds (Cuellar-Bermudez et al., 2015; de Morais et al., 2015; Hayes, 2012a), as high-value lipophilic and hydrophilic-like compounds (e.g., tocopherols, polyphenols, tannins, and flavonoids) and pigments (e.g., carotenoids, astaxanthin, lutein, and phycobiliprotein) (Cuellar-Bermudez et al., 2015; Gastineau et al., 2014; Hayes, 2012b; Prasetiya et al., 2020a, 2020b). The diversity of these bioactive compounds makes microalgae and cyanobacteria ideal candidates for extensive applications, including the pharmaceutical industry, as a source of new drugs (Olaizola, 2003; Vo et al., 2011; Yim et al., 2004).
One of the applications of bioactive compounds produced by microalgae and cyanobacteria for the pharmaceutical and health sectors concerns the development of drugs with antiviral activity. Indeed, several microalgal species have been identified as potential sources of antiviral compounds. For example, spirulan polysaccharides and cyanovirin-N derived from Arthrospira sp. and Nostoc ellipsosporum, respectively, are reported to inhibit the spread of the virus HIV-1, HIV-2, HSV, influenza (de Morais et al., 2015; El-Baz et al., 2013; Smee et al., 2008). In addition, marennine-like pigments produced by the blue diatoms from the genus Haslea have been shown to display antiviral activity against the HSV-1 virus (Gastineau et al., 2014, 2012). Furthermore, several other compounds such as lutein, astaxanthin, and terpenoids produced by the genus Chlorella microalgae can also inhibit HPV virus, hepatitis B and C types, including viruses with severe acute respiratory syndrome SARS-CoV (de Morais et al., 2015; Silva et al., 2018).
Potential protease inhibitors of metabolites produced by microalgae and cyanobacteria are very promising. Hence, preliminary studies show that some active compounds such as scytovirin, sulfoglycolipid, lutein, palmitic or linoleic acid can be used as inhibitors of different viruses (De Morais et al., 2015; Gastineau et al., 2012; Santoyo et al., 2012; Silva et al., 2018; Vitale et al., 2015). Some of them have been hypothesized as potential inhibitors of viruses related to SARS (Pendyala et al., 2021; Smee et al., 2008). Recent in silico studies from (Al-Khafaji et al., 2020) and (Naidoo et al., 2021) demonstrated that some cyanobacterial proteins could serve as potential inhibitors against SARS-CoV-2. Nevertheless, both studies have yet to confirm their results with in vitro experiments. These promising results should be confirmed, and further study is needed regarding the potential of marine bioactive compounds from microalgae and cyanobacteria as SARS-CoV-2 virus inhibitors.
The present study aims at obtaining structural insight into the ACE2 receptor of SARS-CoV-2 and identifying which microalgal and cyanobacterial bioactive compounds could be used as potential SARS-CoV-2 inhibitors. Therefore, a list of bioactive compounds was retrieved from the established database, targeting the ACE2 receptor to specifically develop anti-COVID-19 drugs. Furthermore, a compound selected by virtual screening using molecular docking analysis (AutoDock VINA software) was further validated by molecular dynamics simulation and tested in vitro to identify possible molecular binding interactions with the ACE2 receptor, using the promising surface plasmon resonance (SPR) method (Zhu et al., 2021).
2 Materials and methods
2.1 Software and data collection
Molecular docking simulations were carried out using AutoDock VINA software, downloaded from the website of The Scripps Research Institute (https://vina.scripps.edu/). Visualization of the docking results was performed with BIOVIA Discovery Studio Visualizer 2020. The protein’s crystal structure was downloaded from the Protein Data Bank website (https://www.rcsb.org). ChemDraw Ultra 8.0 (PerkinElmer Inc.) was used to draw 2D and 3D structures and optimize ligand geometry. Data collection strategies and structure of compounds from potential microalgae and cyanobacteria as antivirals were carried out through MarinLit database searches (https://pubs.rsc.org/marinlit/) and publications related to active compounds from marine organisms from 1970 to 2020.
2.2 Ligand preparation and target receptors for molecular docking
The 2D structure of the compound was prepared with ChemDraw Ultra 8.0 and then converted to a 3D structure using the Chem 3D program. All ligands were saved in PDB format. The PDB format of the crystal structure of ACE2 receptor in complex with their inhibitors was downloaded from the RCSB Protein Data Bank (https://www.rcsb.org). The complex for ACE2 and its inhibitor, MLN-4760, was available under PDB ID 1R4L. The MLN-4760 is a cell-permeable, bioavailable, highly potent inhibitor of ACE2 (IC50 = 440 pM against soluble human ACE2) that exhibits good selectivity over bovine carboxypeptidase A and porcine ACE. Afterward, the Biovia Discovery Studio 2020 (BDS) was used to separate the protein from the inhibitor. Concomitantly, undesired molecules were also eliminated. Both protein and inhibitor molecules were then saved into two different PDB files (target receptor and control ligand). Finally, the inhibitor molecule file was put aside, whereas the protein file was loaded into Autodock Tools 1.5.6 software for further preparation steps. These steps cover the addition of polar hydrogen and Kollman charges, removing non-polar hydrogen atoms, and converting into PDBQT format following (Afriza et al., 2018; Khayrani et al., 2021). The file conversion into PDBQT format allowed the file to be loaded in Autodock Vina for molecular docking simulation (Huey et al., 2012).
2.3 Preparation of grid and molecular docking parameters
The grid parameter files were prepared with the help of AutoDockTools 1.5.6. As for its input and output, Vina uses a similar PDBQT molecular structure file format used by AutoDock. The pdbqt files were generated interactively and viewed using MGLTools. The lowest binding energy and Root Mean Square Deviation (RMSD) were further analyzed.
2.4 Molecular docking simulations
Positive control docking was performed using the Autodock Vina program to find the coordinates and parameters that are most appropriate on the active sites. The observed docking parameters were amino acid residues and hydrogen bonds, and the RMSD value should not be more than 2 Å. As for target receptors that are not complex with ligands, blind docking was performed to determine the binding site of the ligand with the receptor.
2.5 Virtual screening of potential compounds and molecular interaction analysis
Virtual screening was performed using PaDEL-ADV software (https://www.yapcwsoft.com/dd/padeladv/) following the protocol from (Hardianto et al., 2018). This program reads each ligand file in pdbqt format, previously prepared using AutodockTools (see sections 2.2 and 2.3). Autodock Vina was then used to dock the ligand with the receptor. Individual binding modes were extracted from the output PDBQT file using vina_split. Results for each binding mode were extracted from the log file and saved into the csv file. The log file and all the related PDB and PDBQT files were then compressed into a zip file. The docking results were sorted by their free energy binding of compounds and by the most histogram groups. Afterward, the compound with the lowest binding energy or maximum multitarget receptors was chosen for further analysis of the interaction between the ligand compound and its receptor.
Autodock tools integrated with the Autodock Vina generated grid maps for each ligand atom. The grid boxes were made to include one site at a time and perform docking. Each coordinate used in the grid box is described in Table 1, with 1 Å spacing on the grid box. Autodock Vina produces energy binding for each molecule and ten models with different conformations and energy. The lowest binding energy was considered as the ligand with the maximum binding affinity. Additionally, Visualization from docking analysis was performed using Biovia Discovery Studio by examining the number and type of interactions between the ligand and the target protein and its native ligand that was used as the control.
Protein (PDBID)
Center coordinate
Size box
X
Y
Z
X
Y
Z
ACE2 (1R4L)
40.199
6.024
29.006
18
22
24
2.6 Molecular dynamic simulations
The molecular dynamics simulation was carried out on the ACE2 protein system with potential inhibitors from screening results on the ACE2 binding site. This simulation used the GPU with the program pmemd.cuda on AMBER20 (Case et al., 2021). Initial minimization was performed using 1000 steps of steepest descent. Afterward, 2000 steps of conjugate gradient minimization were carried out by applying 5 kcal mol−1 harmonic force resistance. The force resistance was gradually released from all heavy atoms (including side chains) to only atoms in the mainframe with the same minimization protocol. Additionally, 5000 steps of unimpeded conjugate gradient minimization were performed to eliminate spatial collisions.
Afterward, the system was set to 310 K in increments (0–100 K; 100–200 K; 200–310 K for 20 ps each) for 60 ps. Moreover, the system density and pressure were equilibrated for 1000 ps. The resistance force was released gradually during the equilibrium phase. Then the production stage was carried out for 200 ns. The time-step in the production phase was set to 2 fs. The MD simulation trajectories were then analyzed using the cpptraj module on AmberTools 20.
2.7 CPC isolation from Arthrospira platensis
Dry powder of A. platensis was obtained from the Jepara Aquaculture Research Facility (BBPBAP Jepara), Indonesia. A total of 20 mg of A. platensis dry biomass with 5 mL of sodium phosphate buffer (0.1 M, pH 7) used and subjected to repeated maceration extraction (ME) and then vortexed (Vortex V1 plus, BOECO, Germany). The cell debris were removed by centrifugation (Tomy MX-307, Tomy Seiko Co.ltd., Japan) at 3140 × g for 5 min. The supernatant was then pooled (500 mL) and marked as crude extract and the extraction solvent was colorless. The phycocyanin crude extract was then purified following protocol from (Liao et al., 2011) and the purity of resulting CPC was calculated. The CPC was then freeze dried (BIOBASE BK-FD10PT, China) for in vitro experiments.
2.8 Surface plasmon Resonance-Based competition assay
2.8.1 ACE2 immobilization
ACE2 was immobilized onto the BA1080 standard SPR chip with a refractive index of 1.61, purchased from NanoSPR LLC, US. First, the chip must be cleaned by soaking it in piranha solution (v/v 3:1 (NH4OH: 30 wt% H2O2) at room temperature) for 10 s then streamed with tap water, soapy water, acetone, and distilled water, respectively. Afterward, the SPR chip was dried using an air blower. After the SPR chip was clean, the 150 μL 3-MPA 10 mM was dropped onto the SPR chip surface and let for 30 min. Then, it was rinsed with phosphate-buffer saline (PBS) solution. Afterward, the N-ethyl-N’-(3-(dimethylamino)propyl)carbodiimide/N-hydroxysuccinimide (EDC-NHS) solution was dropped and incubated for 30 min. Afterward, it was rinsed with PBS solution and dried using an air blower. Furthermore, the chip was installed in the NanoSPR 8 chip holder (NanoSPR LLC, US) for immobilizing the ACE2. Moreover, the ACE2 was flowed onto the SPR chip with a flow rate of 40 μL min−1 for 5 min, allowed to stand for 30 min and then flowed with PBS for 10 min. Afterward, 1 % BSA was flowed onto the SPR chip to block the residual active sites with the same parameters as ACE2 immobilization.
2.8.2 SPR screening measurements
In this study, SPR screening was performed using Nano SPR 8 (NanoSPR LLC, US), and its dynamic responses were observed by SPR intensity (RU) versus time parameters. The ACE2 - CPC interactions were performed with 5 min baseline, 40 min association process, and 30 min dissociation process using PBS as the media with 40 μL min-1 flow rate. Afterward, the prepared CPC solution used had a 5–125 μM in the PBS solution.
2.8.3 Kinetic binding analysis
The kinetic binding analysis was performed to obtain binding kinetic and affinity parameters of ACE2 – CPC. In this study, the association and dissociation dynamic responses of CPC by ACE2 were followed using the Pseudo-second-order and Avrami kinetic adsorption models. These models were represented by Eqs. (1) and (2), respectively, with slight modifications (Ahmad et al., 2015; Vargas et al., 2011). From this model, the observed rate (kobs) could be used to obtain the value of the association rate (kon) and dissociation rate (koff) through linear regression of the C vs kobs with the relationship as shown in Eq. (3) (Schasfoort, 2017; Swinney et al., 2014).
Where is the dynamic response at a specific time, t is the time, ho is the initial adsorption rate, k2 is the adsorption rate of the Pseudo-second-order, nAV is the Avrami’s constant, and kAV is the adsorption rate on the Avrami model (Ahmad et al., 2015; Vargas et al., 2011). Parameters k2 and kAV are the kobs values.
2.8.4 Isotherm analysis
In this study, isotherm analysis was performed to predict the binding interaction behavior between CPC and ACE2. This was obtained by making a curve C vs
fitted using a non-linear regression according to adsorption isotherm models. Additionally, the curve follows the Brouers-Sotolongo (BS) adsorption model with the modified equation shown below (Karoui et al., 2020; Vargas et al., 2011):
Where is the dynamic response of SPR (%), is the maximum adsorption capacity, C is the concentration of CPC, KBS is the Brouers–Sotolongo constant or isotherm constant, and α is the surface heterogeneity constant.
3 Results and discussion
3.1 Molecular docking analysis for ACE2 receptor
In this study, docking analysis between the control ligand and protein ACE2 showed five hydrogen bonds with several amino acids, such as Arg273, Thr371, Tyr515, Pro346, and Thr371 (Table 2). Apart from the main hydrogen bonds, other interactions were also found, two types of electrostatic bonds with Arg514 and Arg518, as well as hydrophobic interaction between ligand and amino acids Tyr510 and His345. From the docking analysis on several bioactive compounds from microalgae and cyanobacteria, and the various interactions that occur between these compounds and the ACE2 receptor, a total of three potential candidate compounds were selected based on the number of interactions that resemble the native ligand one, as well as the number of overall interactions involved. Phycocyanobilin, phycocyanin, and 7-dehydroporiferasterol peroxide have 5, 4, and 3 hydrogen bonds, respectively, with several hydrophobic interactions. Although not many interactions were formed compared to the native ligand, some of these compounds have approximately similar potential as the native ligand, especially for the phycocyanobilin compound with five hydrogen bonds and three hydrophobic interactions. Based on theoretical and experimental correlations between H-bond pairings and their effects on ligand binding affinity, H-bonds could enhance receptor-ligand interactions when both the donor and acceptor have either significantly stronger or significantly weaker H-bonding capabilities than the hydrogen and oxygen atoms in water (Chen et al., 2016). Thus, our virtual screening revealed that the binding affinity of the screened compounds was similar to or higher than that of the control ligand of ACE2 (MLN-4760).
No.
Ligands
Interactions
Binding Affinity (kcal/mol)
Hydrogen Bonds
Hydrophobic Bonds
Other Bonds
1
Native ligand 1R4L as control (MLN-4760)
Arg273, Thr371, Tyr515, Pro346, Thr371
Tyr510 (pi-sigma), His345 (Pi-Pi)
Arg514, Arg518 (electrostatic)
−9.0
2
7-Dehydroporiferasterol peroxide
Arg273, Glu402, Tyr515
His374 (Pi-sigma), Ala153, Tyr515
−10.1
3
24-Methylcholestan-3beta-ol
–
Phe274 (Pi-sigma)
−9.7
4
24-Methylcholesterol
Asp382
His378 (Pi-pi stacked), Phe274, His374, His401
−10.4
5
Brassicasterol
–
Phe274 (Pi-sigma)
−9.6
6
Crinosterol
–
His374, Phe274(Pi-sigma), Pro346, Phe274, His345
−10.0
7
Cryptophycin
Asn149, Thr371, Tyr127, Tyr515
Glu375, Glu402 (Pi-anion), Phe274 (Pi-sigma),Leu 144, Val343, Tyr127, His345, Phe504
−9.5
8
Ergosterol
Glu406
Phe274 (Pi-sigma)
−9.7
9
Ergosterol peroxide
Arg273, His374, Glu402
Phe274 (Pi-sigma), Ala153, His374
−10.5
10
Phycocyanin
Asn131,
Arg255,
Glu388,
Asp251
Arg255,
Phe256,
Ala135,
Phe486,
His487,
Tyr492,
Tyr497
−9.7
11
Phycocyanobilin
Lys363, Thr371, Arg518, His345, Glu406, Glu145
Arg273 (Pi-cation), Tyr510, Phe274 (Pi-sigma), His378, Pro346
−9.5
12
Scytovirin N
Tyr127, Thr371
Lys363 (Pi-cation), Leu144 (Pi-sigma), Phe274 (Pi-pi stacked), Tyr127, His345 (Pi-pi T-shaped) Pro346, Met360,
−12.4
13
Stigmasterol
Arg518
Pro346, Lys363, Cys344, Phe274
−9.7
The docking prediction using Autodock VINA software revealed that among the different molecules targeted, phycocyanobilin could have various interactions with the ACE2 receptor. In this receptor, phycocyanin has a docking score of −9.7 kcal mol−1, which is lower than the ACE2′ native ligand with four hydrogen bonds formed in their amino acids’ residues (Asn131, Arg255, Glu388, and Asp251; Fig. 1a). Additionally, a total of seven hydrophobic bonds were also observed in the amino acid residues (Arg255, Phe256, Ala135, Phe486, His487, Tyr492, and Tyr497; Fig. 1a). On the other hand, phycocyanobilin has a slightly higher docking score (−9.5 kcal mol−1) compared to that of phycocyanin by forming six hydrogen bonds with the residues of amino acids Lys363, Thr371, Arg518, His345, Glu406, and Glu145 (Fig. 1b). Apart from the hydrogen bonds, phycocyanobilin also forms electrostatic interactions with the ACE2 receptor at the residue Arg273 as well as hydrophobic interactions phi – sigma (Tyr510 and Phe274) and phi – alkyl (His378 and Pro346). From docking prediction, it can thus be assumed that the binding affinity of phycocyanin and phycocyanobilin is lower than those of the native ligand. This result suggests that these compounds can potentially be used as ACE2 inhibitors.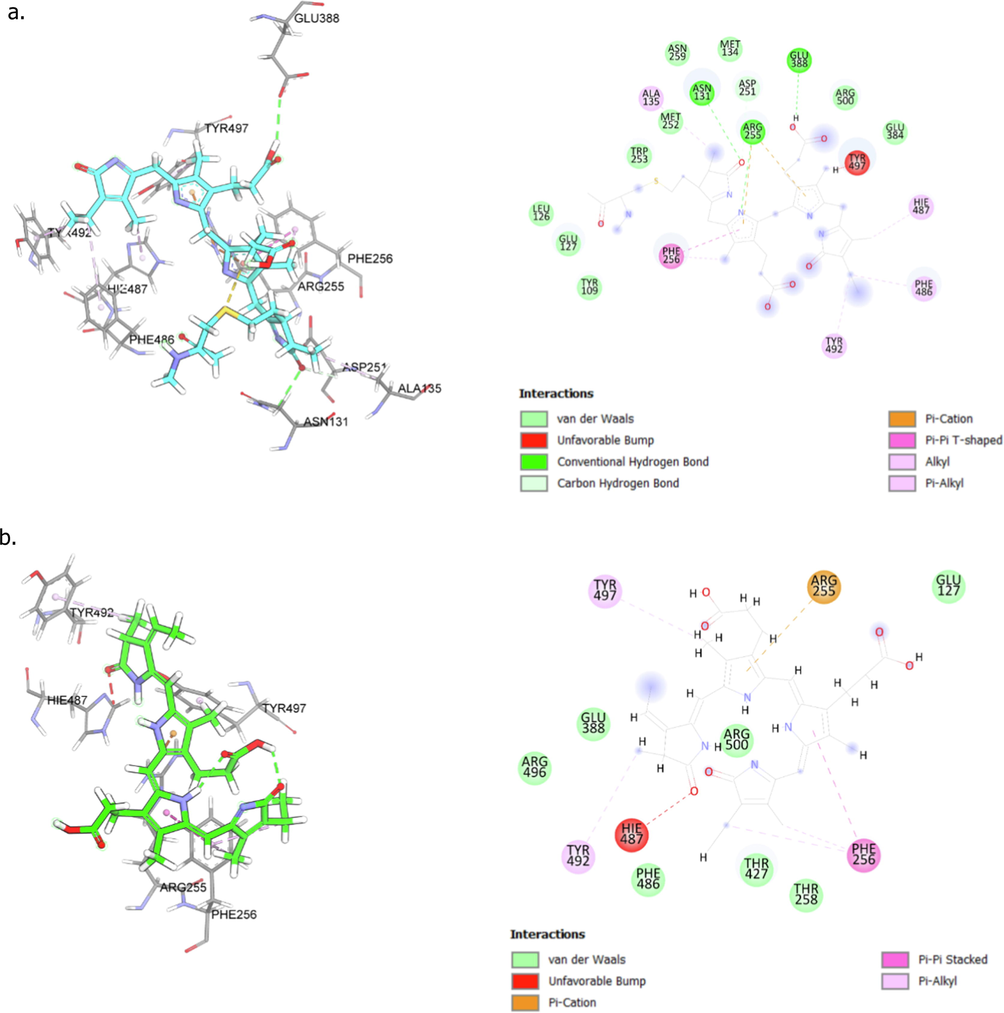
A detailed view of low-energy binding conformations of phycocyanin (a) and phycocyanobilin (b) bound to the active site of ACE2 generated by molecular docking (Autodock VINA software). Image was created using BIOVIA Discovery Studio 2020. The green dashed line indicates the hydrogen bonds with amino acid residues around the ligand, while the pink dashed line indicates hydrophobic interaction between the ligand and the amino acid residues. The orange dashed line indicates electrostatic interaction.
Phycocyanobilin (PCB, C33H38N4O6) and phycocyanin (CPC, C165H185N20O30) could thus be good candidates for developing anti-COVID drugs, and more tests were conducted on these compounds. PCB is a blue phycobilin commonly found in cyanobacteria and the chloroplasts of red algae, glaucophytes, and some cryptomonads (Hayes, 2012b; Santoyo et al., 2012). This molecule has a molecular weight of 586.68 g mol−1. It is present only in the phycobiliproteins, allophycocyanin (APC), and phycocyanin (CPC), of which it is the terminal acceptor of energy (de Morais et al., 2015). A previous study by (Radibratovic et al., 2016) revealed that PCB could bind to human serum albumin (HSA), a protein found mainly in the blood of humans, increasing its ability to prevent the proteolytic activity of other proteins.
3.2 Molecular dynamics simulation of PCB and CPC on ACE2 receptor
The molecular dynamics simulations of the ACE2 receptor that binds to PCB, CPC, and native ligands showed a significant change in distance (Å) from the initial conformation when compared to the structure of ACE2 Apo (Fig. 2). The dynamics of the receptor that binds to the ligand was more stable than the Apo structure. This result is indicated by the root mean square deviation (RMSD) value, which increased to 3.5 Å after 150 ns of simulation (Fig. 2). In the receptor bound to the control, the RMSD value ranged from 2 to 2.5 Å during the simulation. On the other hand, the receptor bound to the phycocyanin and phycocyanobilin compounds, the RMSD profile showed a very stable value of 1.5 Å. This stable conformation indicates favorable interaction between the ligand and the receptor, thereby changing the flexible nature of the ACE2 structure to become more rigid.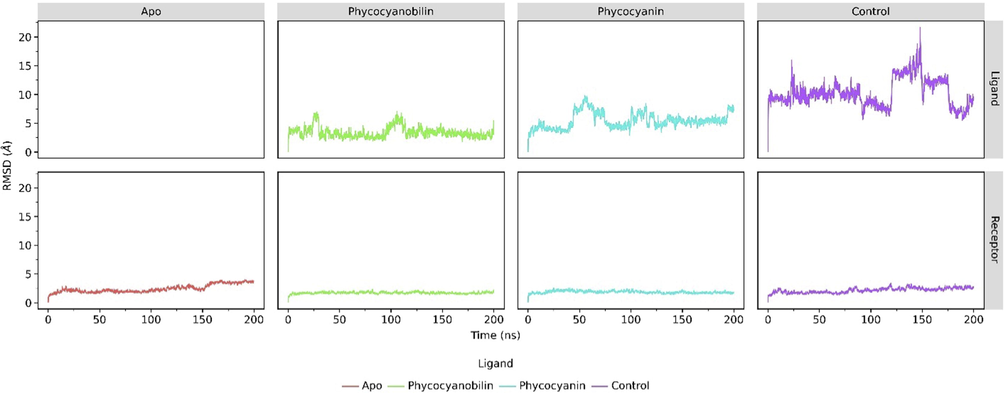
The RMSD analysis of 200 ns-length MD simulation results for ACE2 receptor and each of its ligands (i.e control, phycocyanin, and phycocyanobilin).
The RMSD value of the control ligand experienced a relatively high progression compared to the phycocyanin and phycocyanobilin compounds, where the shift occurred up to 15 Å at 150 ns of the simulation (Fig. 3a-c). After this period, the native ligand again dropped its RMSD value to the similar level as the phycocyanin ligand (Fig. 3). This result indicates lower bond stability in the native ligand, which causes the RMSD value to increase to 15 Å.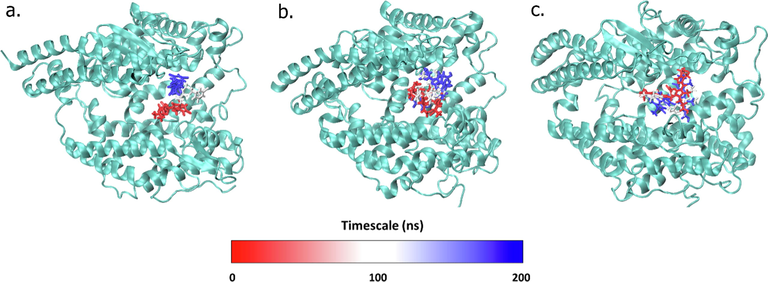
The conformational changes of each ligand which is control (a) phycocyanin (b), and phycocyanobilin (c) at 100 ns interval throughout the simulations.
The ACE2 protein has 597 amino acids. From the root mean square fluctuation (RMSF) profile, the ACE 2 receptor that is not bound to the ligand has a higher fluctuation than the system bound to the ligand. In contrast, as for phycocyanin and phycocyanobilin ligands, RMSF values produced a more stable profile than the native system, suggesting that these two compounds have a potential interest as inhibitors for ACE2 protein (Fig. 4).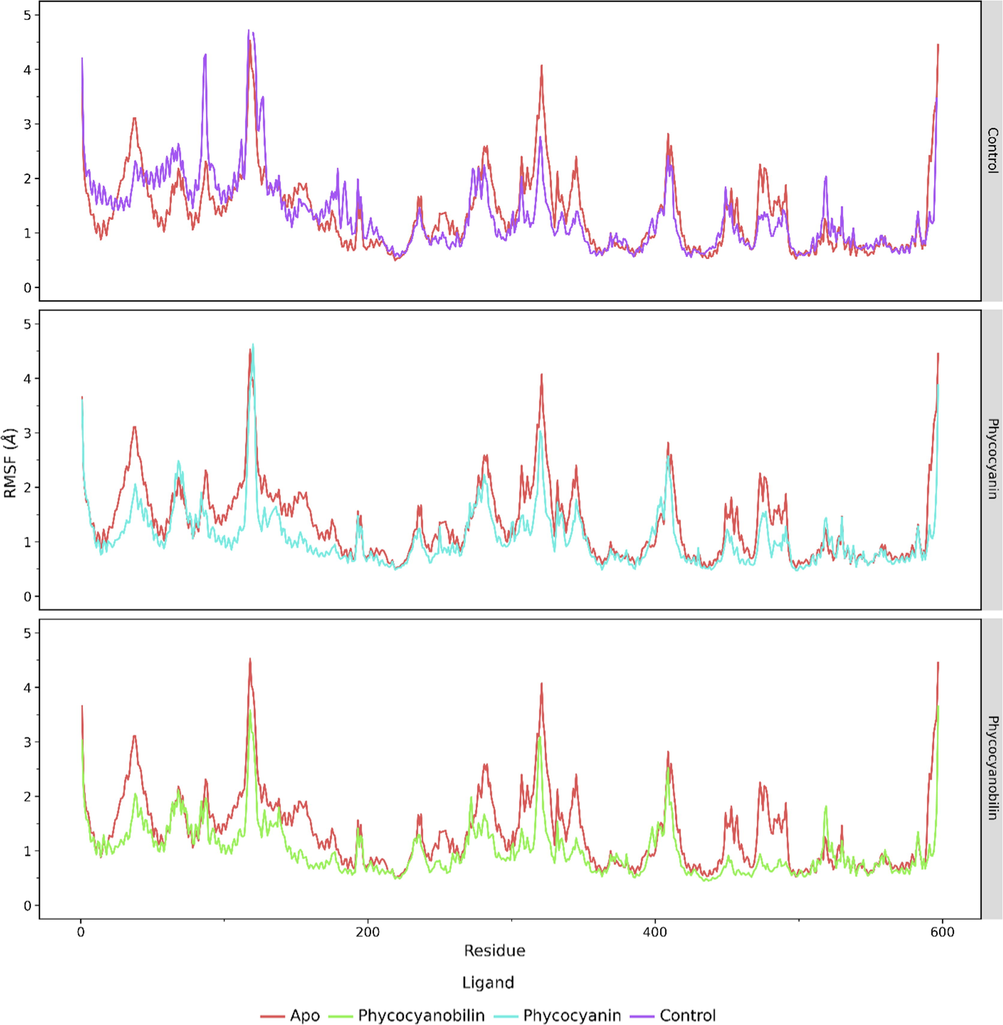
The RMSF analysis of the 597 amino acid residues of ACE2 receptor during the 200 ns simulation.
The molecular mechanics/generalized Born surface area (MM/GBSA) method evidenced that during the MD simulation, the phycocyanin had the lowest total binding free energy (–32.8158 kJ mol−1) compared to other ligands, such as phycocyanobilin (-17.162 kJ mol−1) and the native ligands (-5.0989 kJ mol−1, Table 3). Further hydrogen bond analysis results are also consistent with these findings, where the average number of hydrogen bonds of phycocyanin was the highest among all, followed by phycocyanobilin and the control, respectively (Fig. 5). It is also indicating that the hydrogen bond contributes to most of the interactions between ligand and ACE2 receptor. Therefore, this finding suggests that the interaction of phycocyanin and phycocyanobilin compounds with the ACE2 receptor is relatively strong, and they have stable binding free energy. To our knowledge, little is known about the potential of phycocyanin and phycocyanobilin as potential SARS-CoV-2 inhibitors by targeting the ACE2 receptor. Recent studies from Al-Khafaji et al. (2020) demonstrated that the molecular docking calculations and molecular dynamic analysis support the fact that the phycocyanin could be a potential candidate inhibitor for the ACE2 receptor towards SARS-CoV-2. On the other hand, Pendyala et al. (2021) demonstrated that phycocyanobilin potentially inhibits the binding of other SARS-CoV-2 receptors, such as the main protease (Mpro) and papain-like protease (PLpro). The in silico study revealed that the binding affinity score of phycocyanobilin with Mpro and PLpro were −8.6 and −9.8 kcal mol−1, respectively.
Energy Calculations
Control
(kcal mol−1)Phycocyanin
(kcal mol−1)Phycocyanobilin
(kcal mol−1)
E Van der Waals
–23.9605
−45.8258
−39.7384
E Electrostatic
711.9812
−67.3703
−6.8781
E GB
−690.1198
86.8252
34.9951
E Surface
−2.9998
−6.4449
−5.5409
ΔG Gas
688.0207
−113.1961
−46.6166
ΔG Solv
−693.1195
80.3803
29.4542
ΔG Total
−5.0989
–32.8158
−17.1623
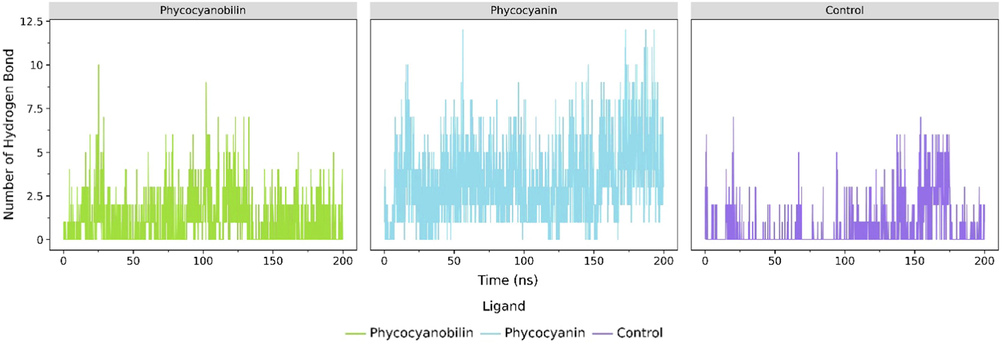
The average number of hydrogen bonds between each ligand throughout 200 ns simulations. The angle of 135° and 3.0 Å cutoff were applied to the calculations.
3.3 Surface plasmon Resonance-Based competition assay
The SPR method is a powerful tool for discovering protein–protein interaction (PPI) inhibitors. Moreover, this method can quantitively explain highly potent PPIs and weak fragment or low molecular weight (LMW) ligand–protein interactions. In this study, the function of phycocyanin (CPC) as an inhibitor of SARS-CoV-2 was evaluated to further validate the molecular docking and molecular dynamics studies using the SPR technique. With this method, first, we confirmed that after the dissociation of ACE2 and BSA 1 %, the dynamic response did not return to the baseline (Fig. 6). This result indicates that ACE2 and BSA 1 % well bonded on the surface of the SPR chip so that the measurement of CPC binding is ready to be carried out. This SPR technique can provide information on the affinity and kinetics of CPC binding by ACE2. The ACE2 − CPC binding SPR response data at different concentrations (5–125 μM) are shown in Fig. 7a-b.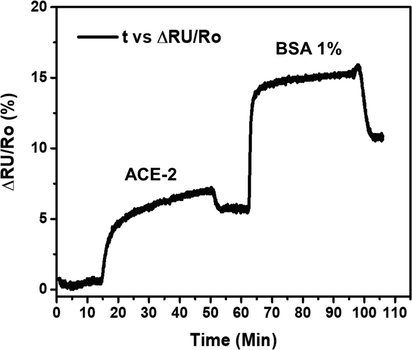
The SPR dynamic responses of ACE2 and BSA1% immobilization processes.
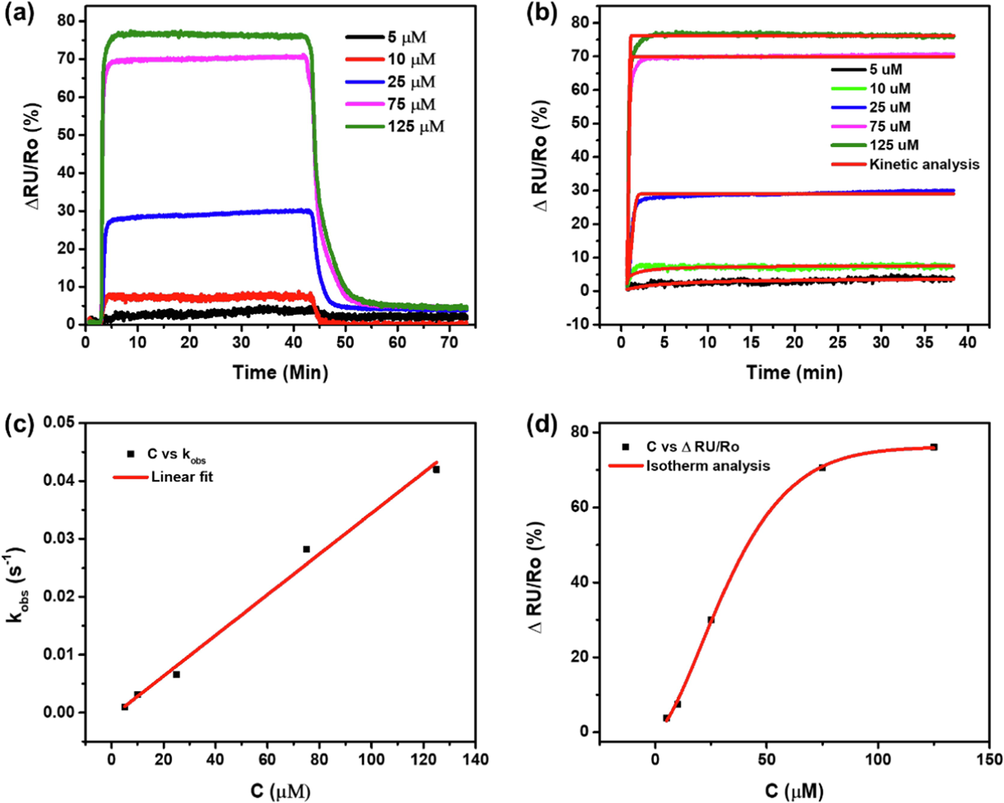
(a) The SPR dynamic response resulting from the interaction of ACE2 – CPC at a concentration of 5 – 125 μM, (b) kinetic adsorption analysis on each CPC association response by ACE2 with different concentrations to obtain the kobs values, and (c) the linear regression of C vs kobs to obtain the kon value, and (d) Non-linear regression curve of the BS isotherm adsorption model on the concentration effect to the dynamic response data.
To determine the efficacy of the developed inhibitor, it is necessary to evaluate the residence time (τ), where the longer the time, the more potent the drug developed (Zhu et al., 2021). The τ was obtained through the value of the dissociation rate (koff) (Copeland et al., 2006). Fig. 7b shows the SPR curve of the association of CPC with ACE2, which was then analyzed using an adsorption kinetic model. It was demonstrated that the association curve of SPR concentration of 5–125 μM follows the pattern of different adsorption kinetic models. Concentrations of 5 and 10 μM followed the Pseudo second-order model. In contrast, 25 – 125 μM concentrations follow Avrami's kinetic model with relative coefficient values (R2) shown in Table S1. The obtained linear regression parameters are summarized in Table S1.
The linear regression between C and kobs revealed the linearity relationship with R2 = 0.99214 (Fig. 7c-d). From the linear equation, the computed values kon and koff were 0.00035145 s−1 μM−1 and 0.000701142 s−1, respectively. On the other hand, the affinity constant (KA) and dissociation constant (KD) for ACE2 – CPC interactions were obtained using KD = and KA = , thus the value of KD and KA were 1.995 μM and 0.501 μM−1, respectively (Swinney et al., 2014). Additionally, the value of τ = 1/koff obtained was 0.396 h, while the half-life value (t1/2 = 0.693/koff) was 0.275 h (Copeland et al., 2006). It appeared that the range of residence time values obtained in this study was relatively narrow. This result is likely caused by the lack of strength of ACE2 in binding CPC. Nevertheless, this argument needs to be validated. Therefore, it is necessary to calculate the binding energy ( G0 = R.T. Ln KD) involved during the adsorption process (Karoui et al., 2020). As a hypothesis, if the gas constant (R) is 1.987 cal K−1 mol−1 and the temperature (T) is 298 K, the value of G0 obtained is − 7.77 kcal mol−1.
To predict the binding mechanism of CPC by ACE2, the data were evaluated using a non-linear regression isotherm adsorption model. The curve pattern follows the Brouers-Sotolongo (BS) model with a relative coefficient value (R2) of 0.9996. The parameters obtained from the C vs non − linear regression are = 76.087, KBS of 0.00337, and α of 1.548. In the present study, the value of α was greater than 1, suggesting that the CPC binding process by ACE2 takes place through slow kinetic biosorption, and each ACE2 binds to CPC molecules with unequal energy (Karoui et al., 2020).
3.4 Potential microalgae candidates for developing SARS-CoV-2 inhibitor
In the present study, our docking and molecular dynamics analysis to search for potential SARS-CoV-2 inhibitor from natural products demonstrated that phycocyanobilin, with CPC in particular, could be used as a potential inhibitor, as this compound has an excellent binding affinity and significant number type of interaction, both characteristics that resemble those of its natural ligands. Moreover, the results of these in silico experiments were also confirmed in vitro with the SPR experiment. CPC can be obtained from Arthrospira platensis (previously known as Spirulina platensis), a cyanobacteria species that is extensively cultured (Olaizola, 2003; Singh et al., 2011), mainly as a source of food, feed supplement, and natural blue pigment (Belay et al., 1996; McClane et al., 2006; Singh et al., 2011). It is worth noting that CPC from several Arthrospira species has also therapeutic potential but without any adverse effect to the living organisms (Chamorro et al., 1997, 1996), in contrast to other natural blue pigments and bioactive compounds such as marennine-like pigments from the marine diatoms Haslea ostrearia and H. nusantara (Prasetiya et al., 2017, 2019; Prasetiya et al., 2020a; Prasetiya et al., 2020b).
In this study, CPC demonstrated potential activity as a candidate for the SARS-CoV-2 inhibitor. A previous study from (McCarty, 2007) demonstrated that CPC constitutes up to 14 % of the total dry weight of A. platensis, in which phycocyanobilin represents 4.7 % of the mass of CPC. Regarding the production of phycocyanin from A. platensis biomass, several studies revealed that extraction and purification is relatively simple as it can be completed with four major steps that cover; crude extract preparation, ammonium sulfate precipitation, dialyses, and anion exchange chromatography (Kumar et al., 2014) or through a hexane extraction process combined with high pressure (Seo et al., 2013).
To complete this search of compounds that could be potential candidates of SARS-CoV-2 inhibitors, our approach highlighted that the genus Chlorella also presents a significant interest. According to our docking analysis, several bioactive compounds from the database such as oxocholesterol, ergosterol, dehydroporiferasterol, and triterpenoid are produced by Chlorella vulgaris and they display inhibitory activity against SARS-CoV-2. To our knowledge, most Chlorella species mainly inhabit freshwater and are commonly found in very nutrient-rich waters. However, it is possible to find these microalgae in marine environment since a few marine species are also known (Levine and Fleurence, 2018). Chlorella strains or species are the most cultivated microalgae since they are widely used as nutraceutical and feed supplements, as well as in the pharmaceutical and cosmetics industry (Borowitzka, 1997; Li et al., 2014; Silva et al., 2018; Yasuhara-Bell and Lu, 2010). Additionally, previous studies demonstrated that several species of Chlorella were found to contain ergosterol as their major sterol (de Morais et al., 2015; Rahal et al., 2014; Yasuhara-Bell and Lu, 2010). This molecule also has been reported to have the inhibitory activity to zoonotic influenza viruses, such as H5N1, H7N9, and H9N2 (Silva et al., 2018).
4 Conclusions
Compounds from natural products extracted from microalgae and cyanobacteria are potentially used for the treatment of viral diseases. In this study, we screened potential microalgal bioactive compounds and the selected hits that may inhibit SARS-CoV-2 on ACE2 receptor for COVID-19 preliminary drugs discovery. Through the combination of in silico and in vitro studies, we demonstrated that phycocyanin (CPC), which can be produced from the cyanobacteria genus Arthrospira (previously known as Spirulina), can be proposed as potential inhibitor. The molecular docking and simulation of molecular dynamics revealed that this compound potentially inhibits the binding of the ACE2 receptor and SARS-CoV-2, with the docking scores of −9.7 kcal mol−1. Additionally, SPR technique also supports the fact that CPC at certain concentration strongly binds with ACE2 receptor. Therefore, the genus Arthrospira and possibly other phycocyanin-producer genera could be important candidates for bioprospection regarding antiviral research. However, further validation through in-vivo assessment is required to validate the results and to transform this potential inhibitor into clinical drugs. Additionally, our study provides valuable insights for exploring and developing novel natural anti-COVID-19 therapeutic agents from microalgae species.
Acknowledgements
The authors wish to thank the Directorate of Research and Community Service (DRPM) Universitas Padjadjaran and the Ministry of Education and Culture (KEMENDIKBUD) of the Indonesian Government for their financial support. This study was funded by the Indonesian Research Collaboration Programme (PPKI) from the Indonesian World Class University Research Scheme granted to FSP (Contract number: 1959/UN6.3.1/PT.00/2021). This publication also benefited from the Horizon 2020 Research and Innovation Programme GHaNA (The Genus Haslea, New marine resources for blue biotechnology and Aquaculture) under Grant Agreement No 734708/GHANA/H2020-MSCA-RISE-2016 (JLM).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin. J. Phys. Conf. Ser.. 2018;1073
- [CrossRef] [Google Scholar]
- Modified durian seed as adsorbent for the removal of methyl red dye from aqueous solutions. Appl. Water Sci.. 2015;5:407-423.
- [Google Scholar]
- Phycocyanine as a Potential Inhibitor of SARS-CoV-2-Spike/TMPRSS2 and SARS-CoV-2-RBD/ACE2 interactions: an in silico approach. Biochem. Mol. Biol. J.. 2020;6:1-6.
- [CrossRef] [Google Scholar]
- Spirulina (Arthrospira): potential application as an animal feed supplement. J. Appl. Phycol.. 1996;8:303-311.
- [Google Scholar]
- Microalgae for aquaculture: Opportunities and constraints. J. Appl. Phycol.. 1997;9:393-401.
- [CrossRef] [Google Scholar]
- Case, D.A., Aktulga, H.M., Belfon, K., Ben-Shalom, I.Y., Brozell, S.R., Cerutti, D.S., Cheatam, T.E., Cisneros, G.A., Cruzeiro, V.W.D., Darden, T.A., Duke, R.E., Giambasu, G., Gilson, M.K., Gohlke, H., Goetz, A.W., Harris, R., Izadi, S., Izmailov, S.A., Jin, C., Kasavajhala, K., Kaymak, M.C., King, E., Kovalenko, A., Kurtzman, T., Lee, T.S., LeGrand, S., Li, P., Lin, C., Liu, J., Luchko, T., Luo, R., Machado, M., Man, V., Manathunga, M., Merz, K.M., Miao, Y., Mikhailovskii, O., Monard, G., Nguyen, H., O’Hearn, K.A., Onufriev, A., Pan, F., Pantano, S., Qi, R., Rahnamoun, A., Roe, D.R., Roitberg, A., Sagui, C., Schott-Verdugo, S., Shen, J., Simmerling, C.L., Skrynnikov, N.R., Smith, J., Swails, J., Walker, R.C., Wang, J., Wei, H., Wolf, R.M., Wu, X., Xue, Y., York, D.M., Zhao, S., Kollman, P.A., 2021. Amber 2020.
- Dominant lethal study of Spirulina maxima in male and female rats after short-term feeding. Phytother. Res.. 1996;10:28-32.
- [CrossRef] [Google Scholar]
- Reproduction and peri- and postnatal evaluation of Spirulina maxima in mice. J. Appl. Phycol.. 1997;9:107-112.
- [CrossRef] [Google Scholar]
- A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-523.
- [CrossRef] [Google Scholar]
- Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv.. 2016;2
- [CrossRef] [Google Scholar]
- Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513.
- [CrossRef] [Google Scholar]
- Materials science approaches in the development of broad-spectrum antiviral therapies. Nat. Mater.. 2020;19:813-816.
- [CrossRef] [Google Scholar]
- Antivirals and antiviral strategies. Nat. Rev. Microbiol.. 2004;2:704-720.
- [CrossRef] [Google Scholar]
- Drug–target residence time and its implications for lead optimization. Nat. Rev. Drug Discov.. 2006;5:730-739.
- [Google Scholar]
- Extraction and purification of high-value metabolites from microalgae: Essential lipids, astaxanthin and phycobiliproteins. J. Microbial. Biotechnol.. 2015;8:190-209.
- [CrossRef] [Google Scholar]
- Review article biologically active metabolites synthesized by microalgae. Biomed Res. Int.. 2015;2015:1-15.
- [CrossRef] [Google Scholar]
- Biologically active metabolites synthesized by microalgae. Biomed Res. Int.. 2015;2015
- [CrossRef] [Google Scholar]
- de Wilde, A.H., Snijder, E.J., Kikkert, M., van Hemert, M.J., 2018. Host Factors in Coronavirus Replication BT - Roles of Host Gene and Non-coding RNA Expression in Virus Infection. In: Tripp, R.A., Tompkins, S.M. (Eds.), Springer International Publishing, Cham, pp. 1–42. https://doi.org/10.1007/82_2017_25.
- In vitro antiviral and antimicrobial activities of Spirulina platensis extract. J. Appl. Pharm. Sci.. 2013;3:52-56.
- [CrossRef] [Google Scholar]
- Biological activities of purified marennine, the blue pigment responsible for the greening of oysters. J. Agric. Food Chem.. 2012;60:3599-3605.
- [CrossRef] [Google Scholar]
- Marennine, promising blue pigments from a widespread Haslea diatom species complex. Mar. Drugs. 2014;12:3161-3189.
- [CrossRef] [Google Scholar]
- A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med. Infect. Dis.. 2020;101646
- [CrossRef] [Google Scholar]
- Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol.. 2006;6:231-243.
- [CrossRef] [Google Scholar]
- Structure-based drug design workflow. Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics. 2018;1–3:273-282.
- [CrossRef] [Google Scholar]
- Hayes, M., 2012. Marine bioactive compounds: Sources, characterization and applications, https://doi.org/10.1007/978-1-4614-1247-2.
- Hayes, M., 2012. Marine bioactive compounds: Sources, characterization and applications, pp. 1–229. https://doi.org/10.1007/978-1-4614-1247-2.
- Huey, R., Morris, G.M., Forli, S., 2012. Using AutoDock 4 and AutoDock Vina with AutoDockTools: A Tutorial. The Scripps Research Institute Molecular 32.
- Synthesis of novel biocomposite powder for simultaneous removal of hazardous ciprofloxacin and methylene blue: central composite design, kinetic and isotherm studies using Brouers-Sotolongo family models. J. Hazard. Mater.. 2020;387:121675
- [Google Scholar]
- Evaluating the potency of Sulawesi propolis compounds as ACE-2 inhibitors through molecular docking for COVID-19 drug discovery preliminary study. J. King Saud Univ. Sci.. 2021;33:101297
- [CrossRef] [Google Scholar]
- In vitro inhibition of influenza A virus infection by marine microalga-derived sulfated polysaccharide p-KG03. Antiviral Res.. 2012;93:253-259.
- [CrossRef] [Google Scholar]
- Extraction and purification of C-phycocyanin from Spirulina platensis (CCC540) Indian J. Plant Physiol.. 2014;19:184-188.
- [CrossRef] [Google Scholar]
- Microalgae in health and disease prevention. Academic Press; 2018.
- Regulation of lipid metabolism in the green microalga Chlorella protothecoides by heterotrophy-photoinduction cultivation regime. Bioresour. Technol.. 2014;192:781-791.
- [CrossRef] [Google Scholar]
- Purification of C-phycocyanin from Spirulina platensis by single-step ion-exchange chromatography. Chromatographia. 2011;73:291-296.
- [CrossRef] [Google Scholar]
- Clinical potential of Spirulina as a source of phycocyanobilin. J. Med. Food. 2007;10:566-570.
- [CrossRef] [Google Scholar]
- Structural insight into the binding of cyanovirin-n with the spike glycoprotein, mpro and plpro of sars-cov-2: Protein–protein interactions, dynamics simulations and free energy calculations. Molecules. 2021;26
- [CrossRef] [Google Scholar]
- Commercial development of microalgal biotechnology: from the test tube to the marketplace. Biomol. Eng. 2003;20:459-466.
- [CrossRef] [Google Scholar]
- Phycobilins as potent food bioactive broad-spectrum inhibitors against proteases of SARS-CoV-2 and other coronaviruses: a preliminary study. Front. Microbiol.. 2021;12:645713
- [CrossRef] [Google Scholar]
- Effect of marennine produced by the blue diatom Haslea ostrearia on behavioral, physiological and biochemical traits of juvenile Mytilus edulis and Crassostrea virginica. Aquaculture. 2017;467:138-148.
- [CrossRef] [Google Scholar]
- Does culture supernatant of Haslea ostrearia containing marennine affect short-term physiological traits in the adult blue mussel Mytilus edulis? Aquac. Rep.. 2019;15:100228
- [CrossRef] [Google Scholar]
- Not only greening: The effects of marennine produced by Haslea ostrearia on physiological traits of three bivalve species. Aquac. Rep.. 2020;18:100546
- [CrossRef] [Google Scholar]
- Effect of the blue pigment produced by the tropical diatom Haslea nusantara on marine organisms from different trophic levels and its bioactivity. Aquac. Rep.. 2020;17:100389
- [CrossRef] [Google Scholar]
- Stabilization of human serum albumin by the binding of phycocyanobilin, a bioactive chromophore of blue-green alga Spirulina: Molecular dynamics and experimental study. PLoS One. 2016;11:1-18.
- [CrossRef] [Google Scholar]
- Oxidative stress, prooxidants, and antioxidants: The interplay. Biomed Res. Int.. 2014;2014
- [CrossRef] [Google Scholar]
- Antiviral compounds obtained from microalgae commonly used as carotenoids sources. J. Appl. Phycol.. 2012;24:731-741.
- [Google Scholar]
- Handbook of surface plasmon resonance (2nd ed.). Cambridge: Royal Society of Chemistry; 2017. https://doi.org/10.1039/9781788010283
- Stable isolation of phycocyanin from Spirulina platensis associated with high-pressure extraction process. Int. J. Mol. Sci.. 2013;14:1778-1787.
- [CrossRef] [Google Scholar]
- Inhibitory effect of microalgae and cyanobacteria extracts on influenza virus replication and neuraminidase activity. PeerJ. 2018;6:e5716.
- [Google Scholar]
- Cyanobacteria: an emerging source for drug discovery. J. Antibiot.. 2011;64:401-412.
- [CrossRef] [Google Scholar]
- Treatment of Influenza A (H1N1) Virus Infections in Mice and Ferrets with Cyanovirin-N. Antiviral Res.. 2008;80:266-271.
- [CrossRef] [Google Scholar]
- A study of the molecular mechanism of binding kinetics and long residence times of human CCR 5 receptor small molecule allosteric ligands. Br. J. Pharmacol.. 2014;171:3364-3375.
- [Google Scholar]
- Adsorption of methylene blue on activated carbon produced from flamboyant pods (Delonix regia): Study of adsorption isotherms and kinetic models. Chem. Eng. J.. 2011;168:722-730.
- [Google Scholar]
- Effectiveness of red alga Asparagopsis taxiformis extracts against Leishmania infantum. Open Life Sci.. 2015;10:490-496.
- [CrossRef] [Google Scholar]
- Marine organisms as a therapeutic source against herpes simplex virus infection. Eur. J. Pharm. Sci.. 2011;44:11-20.
- [CrossRef] [Google Scholar]
- Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol.. 2020;94:1-9.
- [CrossRef] [Google Scholar]
- Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020
- [CrossRef] [Google Scholar]
- Marine compounds and their antiviral activities. Antiviral Res.. 2010;86:231-240.
- [CrossRef] [Google Scholar]
- Antiviral effects of sulfated exopolysaccharide from the marine microalga Gyrodinium impudicum strain KG03. Mar. Biotechnol.. 2004;6:17-25.
- [CrossRef] [Google Scholar]
- Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med.. 2020;46:586-590.
- [CrossRef] [Google Scholar]
- A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273.
- [CrossRef] [Google Scholar]
- Blocking effect of demethylzeylasteral on the interaction between human ACE2 Protein and SARS-CoV-2 RBD protein discovered using SPR technology. Molecules. 2021;26:1-12.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102533.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







