Translate this page into:
The Marshall–Olkin–Weibull-H family: Estimation, simulations, and applications to COVID-19 data
⁎Corresponding author.
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
We define a new extended Weibull-H family and obtain some of its mathematical properties. It is very competitive to the beta-G and Kumaraswamy-G classes, which are highly cited in Google Scholar. The parameters of a specified sub-model are estimated by eight methods and its flexibility is proved in two applications to COVID-19 data.
Keywords
COVID-19 data
Generalized distribution
Maximum likelihood estimation
Weibull distribution
1 Introduction
Adding one or two parameters to parent distributions encourage new concepts for flexible modeling in distribution theory. Among well-established classes of distributions, the exponentiated-G, transmuted-G and Marshall–Olkin-G (MO-G) (Marshall and Olkin, 1997) offer induction of one extra parameter, while the beta-G (Eugene et al., 2002) and Kumaraswamy-G (Cordeiro and de Castro, 2011) classes require two additional shape parameters. Their special cases are explored by Tahir and Nadarajah (2015), among those of other classes.
Composition of distribution generators is emerging as a method to obtain flexible distributions to fit real data in the last five years or so. Some new classes were derived following this method such as the Weibull Marshall–Olkin (Korkmaz et al., 2019), Marshall–Olkin transmuted (Afify et al., 2020), Marshall–Olkin Burr-III (Afify et al., 2021b), among others.
Some other important G families are the exponentiated-G by Gupta et al. (1998), transmuted-G by Shaw and Buckley (2007), gamma-G by Zografos and Balakrishnan (2009), McDonald-G by Alexander et al. (2012), exponentiated-generalized-G by Cordeiro et al. (2013), Burr X-G by Yousof et al. (2017), additive Weibull-G by Hassan and Hemeda (2017), generalized transmuted-G by Nofal et al. (2017), odd Lomax-G by Cordeiro et al. (2019), Kumaraswamy alpha power-G by Mead et al. (2020), modified Kies-G by Al-Babtain et al. (2020), log–logistic tan-G by Zaidi et al. (2021), and generalized linear failure rate-G by Afify et al. (2022). The interested reader can explore more about parameter induction in Tahir and Nadarajah (2015).
Let
be a baseline cumulative distribution function (CDF) with a parameter vector
. Bourguignon et al. (2014) defined the CDF of the Weibull-H (W-H) class with an extra shape parameter
by
The CDF of the MO-G class is defined by
By combining (1) and (2) (and omitting arguments), the CDF of the Marshall–Olkin–Weibull-H (MOW-H) family (with extra parameters
and
) follows as
By differentiating (3), the probability density function (PDF) of the MOW-H family reduces to
Henceforth, denotes a random variable (rv) having density (4).
The hazard rate function (HRF) of X is
By inverting in Eq. (3), we can obtain the quantile function (QF) of X as , where and .
A simple interpretation of the MOW-H family can be given as follows. Consider that the variability of the odds of a rv Z follows a Weibull distribution with unity scale and shape . Let N be a positive integer rv having a geometric distribution with parameter , say (for ). Consider a sequence of N independent copies of Z obtained independently of N. Setting the probability parameters and for and , respectively, the minimum of has PDF (4).
Furthermore, the proposed MOW-H family extends the Weibull-G class (Bourguignon et al., 2014) which has 536 citations so far, and then it is more flexible than the Weibull-G class. In fact, the plots in Figs. 1 to 8 reveal that the two extra parameters to the baseline model makes the density and risk functions of the new family much more flexible for the four baseline distributions considered here. Additionally, the proposed family can be a competitive generator to the beta-G (Eugene et al., 2002) and Kumaraswamy-G (Cordeiro and de Castro, 2011) classes, which also require two additional shape parameters. These two classes are among the most cited papers in the distribution theory literature.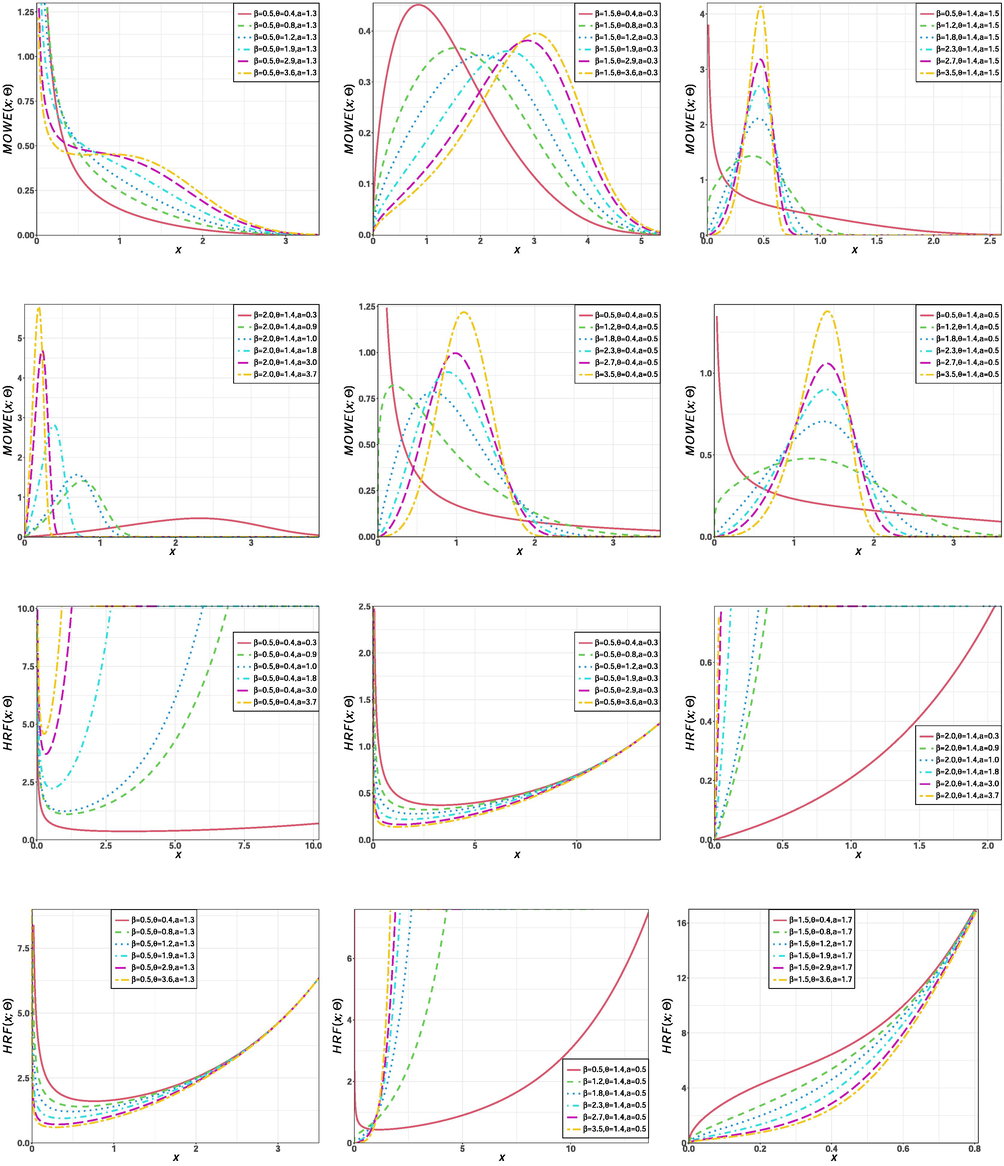
Shapes of density and hazard functions of the MOWE model for different parameter values.
The paper is structured as follows: Section 2 provides four special cases of Eq. (4), and Section 3 addresses some properties of the new family. Section 4 provides the parameter estimation by eight methods. Two applications to COVID-19 data in Section 6 illustrate the utility of the new family. Section 6 ends with some conclusions.
2 Special Models
This section is devoted to introducing some special sub-models of the MOW-H family. The two extra shape parameters of the MOW-H family make the baseline hazard function more flexible to exhibit all important hazard rate shapes, including monotone and non-monotone shapes.
2.1 MOW-exponential (MOWE)
The MOWE density follows from the exponential Exp
distribution, where
. The PDF and CDF of the MOWE distribution are (for
), respectively,
Fig. 1 displays shapes of the PDF and HRF for some parameters. The HRF can assume increasing, decreasing, reversed-J and J shapes.
2.2 MOW-uniform (MOWU)
For the uniform in the interval , where . Then, the PDF of the MOWU model has the form (for )
The Weibull-uniform density when was derived by Phani (1987).
2.3 MOW-Lomax (MOWL)
The Lomax distribution has CDF (for ), where is a shape, and is a scale. The PDF of the MOWL model (for ) is
2.4 MOW-Weibull (MOWW)
The CDF of the Weibull is
, where
and
, and the MOWW density (for
) has the form
The MOWW model includes the exponential power (Smith and Bain, 1975) and the Chen (2000) distribution when and , respectively.
3 Properties
The PDF associated with the CDF (2) admits the linear representation (Cordeiro et al., 2014)
is the indicator function of a subset A, and .
By inserting Eq. (1) and its derivative in
and using the expansions for the binomial and exponential function, we obtain (for
)
So, the density of X is a double linear combination of exp-H densities, which can be adopted with most common type of software, MAPLE, Mathematica, Ox and R, among others.
3.1 Moments
Henceforth,
denotes a rv with PDF
. Eq. (11) gives
Expressions for several exp-H moments (under special baselines) are reported in many papers such as Nadarajah and Kotz (2006).
The nth incomplete moment of X, say
, follows as
The mean deviations and Bonferroni and Lorenz curves of X can be determined from (13) with .
3.2 Generating function
The generating function (gf) of X follows from (11) as where is the gf of the exp-H density
4 Estimation in the MOWE model
Let be observations from the MOWE distribution (Section 2.2), and be the order statistics. The CDF and PDF of this model are denoted by and , respectively. Its parameters can be estimated by eight methods described below. For more information about these estimation methods, see Nassar et al. (2018), Ramos et al. (2018), Rodrigues et al. (2018), and Ramos et al. (2019).
The ordinary least-squares estimates (OLSEs) minimize the function
They can also be found by solving the non-linear equations where , and are and
The weighted least-squares estimates (WLSEs) minimize which follow by solving
The maximum likelihood estimates (MLEs) maximize the log-likelihood below for the parameters follows from (6) by classical iterative methods
The maximum product of spacing estimates (MPSEs) are good alternatives to the MLEs. Let be the uniform spacing (for ), where and . The MPSEs maximize the quantity which can be determined from the non-linear equations
The Cramér-von Mises estimates (CVMEs) minimize which also follow by solving
The Anderson–Darling estimates (ADEs) minimize which can be found as solutions of the system
The right-tail Anderson–Darling estimates (RADEs) minimize
They can also be obtained from the non-linear equations
Let be an unbiased estimator of . The PC estimates (PCEs) minimize where .
5 Simulation analysis
The simulation study compares the estimates from the eight methods in Section 4 in terms of the averages of the four quantities: absolute bias ( ), , mean square error (MSE), , and mean relative error (MRE), .
The observations from the MOWE model are simulated from Eq. (8), where U is a uniform random variable in the interval . We generate random samples (for , and 200) from the MOWE model with and .We use R codes (R Core Team, 2020, version 4.0.3) for the simulations and the nlminb function in the stats package (R Core Team, 2020, version 4.0.3).
We estimate its parameters for some parameter combinations and sample sizes, and calculate the MSEs and MREs of the estimates. Four out of twenty-seven simulated outcomes are reported in Tables 1–4, whose numbers in each row have superscripts giving the ranks of the estimates among all methods, and
denotes the partial sum of the ranks.
Est.
Est. Par.
WLSE
OLSE
MLE
MPSE
CVME
ADE
RADE
PCE
30
MSE
MRE
50
MSE
MRE
80
MSE
MRE
120
MSE
MRE
200
MSE
MRE
Est.
Est. Par.
WLSE
OLSE
MLE
MPSE
CVME
ADE
RADE
PCE
30
MSE
MRE
50
MSE
MRE
80
MSE
MRE
120
MSE
MRE
200
MSE
MRE
Est.
Est. Par.
WLSE
OLSE
MLE
MPSE
CVME
ADE
RADE
PCE
30
MSE
MRE
50
MSE
MRE
80
MSE
MRE
120
MSE
MRE
200
MSE
MRE
Est.
Est. Par.
WLSE
OLSE
MLE
MPSE
CVME
ADE
RADE
PCE
30
MSE
MRE
50
MSE
MRE
80
MSE
MRE
120
MSE
MRE
200
MSE
MRE
Table 5 provides the partial and overall ranks of the estimates, thus indicating that the MLEs outperform all other estimates for the MOWE distribution with an overall score of 230.
WLSE
OLSE
MLE
MPSE
CVME
ADE
RADE
PCE
30
3
5
4
1
6
2
7
8
50
4
5
3
1
6
2
7
8
80
4
5
3
1
6
2
7
8
120
4
5
3
1
6.5
2
6.5
8
200
4
7
3
1
6
2
5
8
30
4
6
1.5
1.5
5
3
7
8
50
4
6
2
1
5
3
7
8
80
4
6
2
1
5
3
7
8
120
4
6
2
1
5
3
7
8
200
4
6
2
1
5
3
7
8
30
4
6
1.5
1.5
5
3
7
8
50
4
6
2
1
5
3
7
8
80
4
5
1
2
6
3
7
8
120
4
6
2
1
5
3
7
8
200
4
6
3
1
5
2
7
8
30
3
6
4
1
7
2
5
8
50
3
6
5
1
7
2
4
8
80
3
5
7
1
6
2
4
8
120
3
4.5
7
1
6
2
4.5
8
200
3
6
7
1
5
2
4
8
30
4
6
2
1
7
3
5
8
50
4
7
1
2
6
3
5
8
80
4
6
2
1
7
3
5
8
120
4
6
2
1
7
3
5
8
200
4
7
1
2
6
3
5
8
30
4
5
2
1
6.5
3
6.5
8
50
4
5
2
1
7
3
6
8
80
4
7
1
2
6
3
5
8
120
4
6
2
1
7
3
5
8
200
4
7
2
1
6
3
5
8
30
3
6
4
1
7
2
5
8
50
3
7
5.5
1
5.5
2
4
8
80
3
5
7
1
6
2
4
8
120
3
5
7
1
6
2
4
8
200
3
6
7
1
5
2
4
8
30
4
5
2
1
7
3
6
8
50
4
6
2
1
7
3
5
8
80
4
6
2
1
7
3
5
8
120
4
5.5
2
1
7
3
5.5
8
200
4
7
2
1
6
3
5
8
30
4
5
2
1
7
3
6
8
50
4
7
2
1
6
3
5
8
80
4
5
1
2
7
3
6
8
120
4
6
2
1
7
3
5
8
200
4
6
2
1
7
3
5
8
30
4.5
6
1
2
7
3
4.5
8
50
4.5
7
1
2
6
3
4.5
8
80
4.5
7
1
2
6
3
4.5
8
120
5
6
1
2
7
3
4
8
200
5
7
1
2
6
3
4
8
30
5
7
1
2
6
3
4
8
50
4.5
7
1
2
6
3
4.5
8
80
4
7
1
2
6
3
5
8
120
5
7
1
2
6
3
4
8
200
5
6
1
2
7
3
4
8
30
5
7
1
2
6
3
4
8
50
5
7
1
2
6
3
4
8
80
4.5
7
1
2
6
3
4.5
8
120
4
7
1
2
6
3
5
8
200
4
7.5
1
2
6
3
5
7.5
30
4
8
1
2
5
3
6
7
50
4
7
1
2
5
3
6
8
80
4
6
1
2
5
3
7
8
120
4
5
1
2
6
3
7
8
200
4
5
1
2
6
3
8
7
30
4
7.5
1
2
5
3
6
7.5
50
4
5
1
2
7
3
6
8
80
4
6
1
2
5
3
7
8
120
4
7
1
2
5
3
6
8
200
4
6
1
2
5
3
7
8
30
4
6
1
2
5
3
7
8
50
4
6
1
2
5
3
7
8
80
4
5
1
2
6
3
7
8
120
4
6
2
1
5
3
7
8
200
4
6
2
1
5
3
7
8
30
4
6
1
2
5
3
7
8
50
4
6
1
2
5
3
8
7
80
4
6
1
2
5
3
7
8
120
4
6
1
2
5
3
7
8
200
4
7
1
2
5
3
8
6
30
4
8
1
2
5
3
6
7
50
4
6.5
1
2
5
3
8
6.5
80
4
5
1
2
6
3
8
7
120
4
6
1
2
5
3
8
7
200
4
6
1
2
5
3
8
7
30
4
5
2
1
6
3
8
7
50
4
6
2
1
5
3
8
7
80
4
5
2
1
6
3
7
8
120
4
6
2
1
5
3
8
7
200
4
6
2
1
5
3
7
8
30
6
8
1
2
7
3
5
4
50
6
7
1
2
8
4
5
3
80
6
8
1
2
7
4
5
3
120
6
7
1
2
8
4
5
3
200
6
7
1
2
8
4
5
3
30
6
7
1
2.5
8
2.5
5
4
50
5
7
1
2
8
4
6
3
80
6
7
1
2
8
4
5
3
120
6
8
1
2
7
4
5
3
200
5
8
1
2
7
4
6
3
30
6
8
1
3
7
2
4.5
4.5
50
6
7.5
1
2
7.5
3
4
5
80
6
8
1
2
7
3
5
4
120
6
8
1
2
7
4
5
3
200
5
8
1
2
7
4
6
3
30
6
8
1
5
4
2
7
3
50
4
8
1
5
6
2
7
3
80
5
7
1
4
6
2
8
3
120
5
7
1
2
6
3
8
4
200
5
7
1
2
6
3
8
4
30
7
8
1
5
4
2
6
3
50
4
8
1
5
6
2
7
3
80
5
7
1
3
6
4
8
2
120
5
7
1
4
6
3
8
2
200
5
7
1
2
6
4
8
3
30
6
8
1
4
5
2
7
3
50
5
7
1
3
6
2
8
4
80
5
7
1
3
6
2
8
4
120
5
7
1
2
6
4
8
3
200
5
7
1
2
6
3
8
4
30
5
8
1
6
4
2
7
3
50
5
8
1
3
6
2
7
4
80
4
7
1
2
6
3
8
5
120
5
7
1
2
6
4
8
3
200
5
7
1
2
6
4
8
3
30
5
8
1
4
6
2
7
3
50
6
8
1
4
5
2
7
3
80
5
7
1
4
6
2
8
3
120
5
7
1
2
6
3
8
4
200
5
7
1
2
6
4
8
3
30
6
8
1.5
3
5
1.5
7
4
50
6
7
1
4
5
2
8
3
80
5
7
1
2.5
6
2.5
8
4
120
5
7
1
2
6
3.5
8
3.5
200
5
7
2
1
6
4
8
3
Ranks
599.5
881
230
260
808
389
836
856.5
Overall Rank
4
8
1
2
5
3
6
7
6 Modeling biological data
The applicability of a sub-model of the new family is proved empirically in modeling two COVID-19 data sets.
The first set refers to 36 COVID-19 mortality rates in Canada: 1.5157, 1.5806, 1.9048, 2.1901, 2.4141, 2.4946, 2.5261, 2.6029, 2.7704, 2.7957, 2.8349, 2.8636, 2.9078, 3.0914, 3.1091, 3.1091, 3.1444, 3.1348, 3.2110, 3.2135, 3.2218, 3.2823, 3.3592, 3.3769, 3.3825, 3.5146, 3.6346, 3.6426, 3.8594, 4.0480, 4.1685, 4.2202, 4.2781, 4.9274, 4.9378, 6.8686. The second set refers to 53 COVID-19 survival times of patients in critical conditions in China in the first two months of 2020. The times measured from the admission to the hospital until death are: 0.054, 0.064, 0.087, 0.087, 0.235, 0.352, 0.364, 0.421, 0.437, 0.458, 0.479, 0.548, 0.568, 0.704, 0.787, 0.796, 0.816, 0.865, 0.976, 0.976, 0.978, 1.756, 1.978, 2.089, 2.643, 2.869, 3.079, 3.348, 3.543, 3.646, 3.867, 3.890, 4.092, 4.093, 4.190, 4.237, 5.028, 5.083, 6.174, 6.743, 7.058, 7.274, 8.273, 9.324, 10.827, 11.282, 13.324, 14.278, 15.287, 16.978, 17.209, 19.092, 20.083. The two data sets were analyzed by Liu et al. (2021).
We compare the fits of the MOWE model and some other extensions of the exponential (E): the beta-E (BE) (Jones, 2004), Marshall–Olkin-generalized E (MOGE) (Ristic and Kundu, 2015), Marshall–Olkin-Nadarajah–Haghighi (MONH) (Lemonte et al., 2016), transmuted generalized-E (TGE) (Khan et al., 2017), modified-E (ME) (Rasekhi et al., 2017), Marshall–Olkin-alpha power E (MOAPE) (Nassar et al., 2019) and Topp–Leone-odd log–logistic E (TLOLLE) (Afify et al., 2021a) distributions.
We adopt the information criterion (IC) measures: Akaike-IC (AIC), consistent Akaike-IC (CAIC), Hannan–Quinn IC (HQIC), Bayesian-IC (BIC), Cramér–Von Mises ( ), Anderson–Darling ( ), and Kolmogorov–Smirnov (K–S) (and K–S p-value).
The MLEs of the parameters from the fitted models, their standard errors (SEs), and the previous measures are given in Tables 6 and 7 for both data sets. The numbers in these tables indicate that the MOWE distribution gives a superior fit over the other models tested. The PDF, CDF, survival function (SF) and probability–probability (PP) plot for the MOWE model are reported in Fig. 2 for both data sets.Fig. 3 provides the total time on test (TTT) plots for both data sets and it also illustrates that the HRF of the first data is increasing because it has a concave shape. The HRF of the second data is decreasing because the TTT plot has a convex shape. Hence, the MOWE distribution can capture all data sets with monotone HRF properly.
Model
Par.
Estimates
(SEs)
AIC
CAIC
BIC
HQIC
K–S
K–S p-value
MOWE
5.50558
(0.67807)
100.042
100.792
104.793
101.700
0.06008
0.34880
0.09773
0.88177
0.00279
(0.00094)
0.09308
(0.01173)
MOAPE
633804.1
(8388.73)
101.659
102.409
106.410
103.317
0.08217
0.45835
0.10992
0.77716
1.88309
(0.32586)
30.70694
(33.68783)
TLOLLE
0.20969
(0.03824)
101.103
101.853
105.854
102.761
0.07514
0.43090
0.10240
0.84465
2.84413
(0.96418)
2.02808
(1.55940)
MONH
0.78838
(0.22230)
101.433
102.183
106.184
103.091
0.07280
0.40724
0.10341
0.83613
4.30678
(4.48412)
1539.80599
(2478.19647)
BE
15.14949
(10.67618)
101.991
102.741
106.742
103.649
0.09263
0.53929
0.10492
0.82298
2.21092
(1.95802)
0.78561
(0.38565)
TGE
26.86514
(14.61735)
101.799
102.549
106.549
103.457
0.09225
0.54625
0.11143
0.76270
1.31236
(0.17246)
−0.65410
(0.34168)
MOGE
31.58462
(24.81951)
101.064
101.814
105.815
102.722
0.08096
0.45622
0.10652
0.80864
1.78986
(0.36869)
9.09406
(13.42972)
ME
1.83639
(1.47673)
103.578
104.869
109.912
105.789
0.08547
0.49574
0.10581
0.81504
5.41691
(9.80746)
17.07664
(17.24968)
1.04794
(0.56020)
E
0.30473
(0.05078)
159.560
159.677
161.143
160.112
0.09950
0.57412
0.40970
0.00001
Model
Par.
Estimates
(SEs)
AIC
CAIC
BIC
HQIC
K–S
K–S p-value
MOWE
0.81250
(0.15573)
270.385
270.875
276.296
272.658
0.06537
0.41252
0.11296
0.50828
0.33504
(0.22823)
0.07572
(0.02532)
MOAPE
0.99990
(1.84449)
273.312
273.802
279.223
275.585
0.08339
0.49486
0.13065
0.32606
0.12275
(0.04762)
0.35277
(0.37871)
TLOLLE
0.22384
(0.11090)
270.538
271.027
276.448
272.811
0.06146
0.39944
0.11682
0.46466
0.47642
(0.15364)
2.21427
(1.02144)
MONH
0.40812
(0.21923)
273.830
274.319
279.741
276.103
0.08558
0.51428
0.12036
0.42635
2.54060
(7.71478)
2.47644
(5.15816)
BE
0.69168
(0.12646)
272.840
273.330
278.751
275.113
0.07697
0.50460
0.13050
0.32739
1.00261
(2.99548)
0.16232
(0.51865)
TGE
0.73423
(0.13934)
272.628
273.118
278.539
274.901
0.075175
0.48307
0.12597
0.36956
0.15049
(0.04462)
0.21354
(0.46487)
MOGE
0.80648
(0.19856)
272.353
272.842
278.263
274.626
0.07403
0.46144
0.11861
0.44508
0.13806
(0.04808)
0.60440
(0.44413)
ME
3.84904467
(6.14941)
274.365
275.198
282.246
277.396
0.07294
0.45987
0.12148
0.41461
823.54185
(383.2700)
0.43063
(0.12777)
0.03553
(0.08355)
E
0.20892
(0.02869)
273.977
274.055
275.947
274.734
0.07751
0.50704
0.21143
0.01751
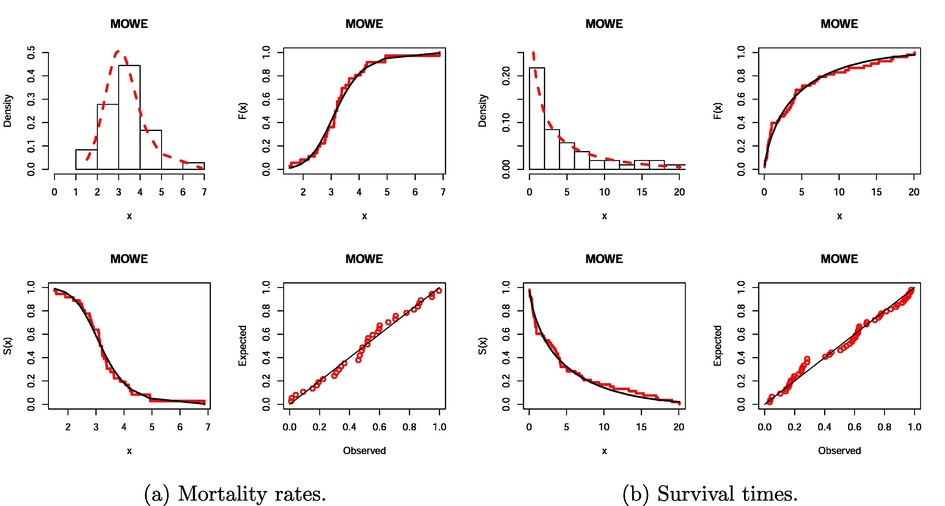
Fitted functions for the MOWE model for the two data sets.
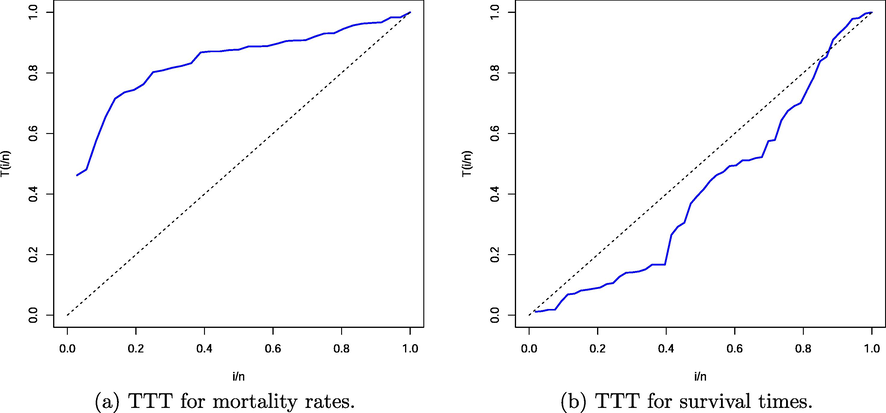
TTT plots for the two analyzed data sets.
7 Concluding remarks
We constructed a new competitive family of distributions to the well-established beta-G and Kumaraswamy-G classes. Some of its mathematical properties were determined. We addressed eight estimation methods for a special model called the MOW-exponential (MOWE) distribution. The simulation results showed that the maximum likelihood approach is the best estimation method for the MOWE parameters. We proved the utility of this distribution to analyze COVID-19 data from Canada and China.
The topics of this article can be extended in several ways. For example, a discrete version of the new family can be established and its properties can be explored. Bivariate extensions of the new family can also be investigated.
Data Availability
This work is mainly a methodological development and has been applied on secondary data, but, if required, data will be provided.
Fund
This study was funded by Taif University Researchers Supporting Project number (TURSP-2020/279), Taif University, Taif, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Topp–Leone odd log-logistic exponential distribution: Its improved estimators and applications. Anais da Academia Brasileira de Ciências. 2021;93:e20190586
- [Google Scholar]
- The Marshall-Olkin odd Burr III-G family: theory, estimation, and engineering applications. IEEE Access. 2021;9:4376-4387.
- [Google Scholar]
- The extended failure rate family: properties and applications in the engineering and insurance fields. Pakis. J. Stat.. 2022;38):165-196.
- [Google Scholar]
- The Marshall-Olkin transmuted-G family of distributions. Stochast. Qual. Control. 2020;35:79-96.
- [Google Scholar]
- A new modified Kies family: properties, estimation under complete and type-II censored samples, and engineering applications. Mathematics. 2020;8:1345.
- [Google Scholar]
- Generalized beta-generated distributions. Comput. Stat. Data Anal.. 2012;56:1880-1897.
- [Google Scholar]
- A new two-parameter lifetime distribution with bathtub shape or increasing failure rate function. Stat. Prob. Lett.. 2000;49:155-161.
- [Google Scholar]
- The odd Lomax generator of distributions: properties, estimation and applications. J. Comput. Appl. Math.. 2019;347:222-237.
- [Google Scholar]
- A new family of generalized distributions. J. Stat. Comput. Simul.. 2011;81:883-898.
- [Google Scholar]
- The Marshall-Olkin family of distributions: mathematical properties and new models. J. Stat. Theory Practice. 2014;8:343-366.
- [Google Scholar]
- Beta-normal distribution and its applications. Commun. Stat. Theory Methods. 2002;31:497-512.
- [Google Scholar]
- Modeling failure time data by Lehman alternatives. Commun. Stat. Theory Methods. 1998;27:887-904.
- [Google Scholar]
- A new fmily of additive Weibull-generated distributions. Int. J. Math. Appl.. 2017;4:151-164.
- [Google Scholar]
- Families of distributions arising from distributions of order statistics. Test. 2004;13:1-43.
- [Google Scholar]
- Transmuted generalized exponential distribution: a generalization of the exponential distribution with applications to survival data. Commun. Stat. Simul. Comput.. 2017;6:4377-4398.
- [Google Scholar]
- The Weibull Marshall-Olkin family: regression model and application to censored data. Commun. Stat. Theory Methods. 2019;8:4171-4194.
- [Google Scholar]
- A new useful three-parameter extension of the exponential distribution. Statistics. 2016;50:312-337.
- [Google Scholar]
- Modeling the survival times of the COVID-19 patients with a new statistical model: a case study from China. PloS One. 2021;16:e0254999
- [Google Scholar]
- A new method for adding a parameter to a family of distributions with application to the exponential and Weibull families. Biometrika. 1997;84:641-652.
- [Google Scholar]
- The modified Kumaraswamy Weibull Distribution: properties and Applications in Reliability and Engineering Sciences. Pakistan J. Stat. Oper. Res.. 2020;16:433-446.
- [Google Scholar]
- A new extension of Weibull distribution: properties and different methods of estimation. J. Comput. Appl. Math.. 2018;336:439-457.
- [Google Scholar]
- The Marshall-Olkin alpha power family of distributions with applications. J. Comput. Appl. Math.. 2019;351:41-53.
- [Google Scholar]
- The generalized transmuted-G family of distributions. Commun. Stat. Theory Methods. 2017;46:4119-4136.
- [Google Scholar]
- Reliability-centered maintenance: analyzing failure in harvest sugarcane machine using some generalizations of the Weibull distribution. In: Modelling and Simulation in Engineering, 2018. 2018. p. :1241856.
- [Google Scholar]
- Modeling traumatic brain injury lifetime data: improved estimators for the generalized gamma distribution under small samples. PLoS one. 2019;14:e0221332
- [Google Scholar]
- The modified exponential distribution with applications. Pakistan J. Stat.. 2017;33:383-398.
- [Google Scholar]
- Poisson-exponential distribution: different methods of estimation. Journal of Applied Statistics. 2018;45:128-144.
- [Google Scholar]
- Shaw, W.T., Buckley, I.R., 2007. The alchemy of probability distributions: beyond Gram-Charlier expansions, and a skew-kurtotic-normal distribution from a rank transmutation map, UCL discovery repository. http://discovery.ucl.ac.uk/id/eprint/643923.
- An exponential power life testing distribution. Commun. Stat.– Theory Methods. 1975;4:469-481.
- [Google Scholar]
- Parameter induction in continuous univariate distributions: well-established G families. Anais da Academia Brasileira de Ciências. 2015;87:539-568.
- [Google Scholar]
- The Burr X Generator of Distributions for Lifetime Data. J. Stat. Theory Appl.. 2017;16:288-305.
- [Google Scholar]
- A new generalized family of distributions: properties and applications. Aims Math.. 2021;6:456-476.
- [Google Scholar]
- On families of beta-and generalized gamma-generated distributions and associated inference. Stat. Methodol.. 2009;6:344-362.
- [Google Scholar]







