Translate this page into:
Screening of rare actinomycetes isolated from natural wetland ecosystem (Fetzara Lake, northeastern Algeria) for hydrolytic enzymes and antimicrobial activities
⁎Corresponding author. tahamenasria@hotmail.com (Taha Menasria)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Actinomycetes from unexplored habitats are considered as a promising source for novel bioactive compounds with a broad range of biological activities. A study was carried out to isolate and identify rare actinomycetes producing antimicrobial from a natural wetland. Water samples from Fetzara Lake (North eastern-Algeria) were collected and subjected to rare actinomycetes isolation using different rich media. Eight selected actinomycetes were screened in vitro for hydrolytic enzymes, antibacterial and anticandidal activities. Based on the 16S rRNA sequencing, the eight actinomycetes isolates were categorized into four different rare genera Actinomadura, Nocardia, Nonomuraea and Micromonospora. Interestingly, significant anticandidal and antibacterial activities against both Gram-positive and Gram-negative bacteria were observed. Furthermore, the actinomycetes isolates were able to produce different hydrolytic enzymes with potential industrial and food processing applications such as amylase, cellulase, protease, and lipase. Overall, the study revealed that the selected aquatic rare actinomycetes recovered from Fetzara Lake presented good candidates to be explored as new sources of bioactive compounds.
Keywords
Wetland
Fetzara Lake
Rare actinomycetes
Hydrolytic enzymes
Antimicrobial activity
1 Introduction
Actinomycetes are present in various ecological habitats and are particularly abundant in soil (Takizawa et al., 1993; Kinkel et al., 2012), as well in fresh water and other marine environments (Lam, 2006; Valli et al., 2012). They are considered highly valuable as they produce various secondary metabolites and other biologically useful compounds such as antibiotics, antitumor agents, immunosuppressive agents, vitamins, nutritional materials, herbicides, pesticides, antiparasitic agents and enzymes (Bérdy, 2012; Mohseni et al., 2013; Abdelmohsen et al., 2015). Actinomycetes have been for decades major storehouse microorganisms for the discovery of natural products (Choi et al., 2015) and Streptomyces, the best-characterized genus of actinomycetes, is considered one of the most important types of industrial bacteria due to its superior capabilities in producing valuable secondary metabolites and bioactive compounds (Pak and Elliot, 2010). They are able to metabolize various compounds of complex structures such as polysaccharides, alcohols, amino acids and aromatic compounds by the production of extracellular enzymes such as amylase, chitinase, cellulase, glucanase, and protease (Antonopoulos et al., 2001).
In recent years, antimicrobial resistance is spreading faster than the introduction of new compounds into clinical practice, causing a public health crisis (Laallam et al., 2015; Ling et al., 2015; Menasria et al., 2015). The research for novel source of a potent bioactive compound for multidisciplinary uses as yet be needed (Rungroch and Nakaew, 2015) and the development of new technologies to find and produce such compounds have again attracted interest in this field (Tilmann et al., 2015).
The rare actinomycetes are considered as a promising source for novel bioactive compounds and hydrolytic enzymes with a broad range of biological activities and pharmacological properties (Arul et al., 2014; Benhadj and Gacemi-Kirane, 2016). Rare actinomycetes are defined as genera in which the isolation frequency by conventional methods is lower than the Streptomyces abundance such as Actinomadura, Actinoplanes, Amycolatopsis, Actinokineospora, Acrocarpospora, Actinosynnema, Catenuloplanes, Cryptosporangium, Dactylosporangium, Kibdelosporangium, Kineosporia, Kutzneria, Microbiospora, Microtetraspora, Nocardia, Nonomuraea, Planomonospora, Planobispora, Pseudonocardia, Saccharomonospora, Saccharopolyspora, Saccharothrix, Streptosporangium, Spirilliplanes, Thermomonospora, Thermobifida and Virgosporangium (Lazzarini et al., 2000; Tiwari and Gupta, 2013).
Because of the exhaustion of the usual terrestrial sources, the discovery of new compounds from marine ecosystems has subsequently increased and relatively few efforts with aquatic rare actinomycetes have been attempted. It has been shown that marine actinomycetes are phylogenetically and physiologically distinct from their terrestrial relatives and were found to represent a rich source for diverse and interest bioactive secondary metabolites and enzymes with potential industrial and clinic applications (Maldonado et al., 2005). Algeria harbors several wetlands and hypersaline lakes, with rare typology and ecology in the world, and of which 50 are classified as being of international importance as Ramsar sites (Menasria et al., 2018). From sub-tropical in the coastal northeast part of the country to semi-arid in the Hauts Plateaux and an arid climate across the Sahara, the Algerian wetlands constitute an important habitat in terms of biodiversity and functional role. However, all aspects related to microbiota (diversity and bioactivity) are poorly investigated and remains unidentified. In response, and for the first time, the aim of this work is to characterize and to study of hydrolytic enzymes, antibacterial and anticandidal activities of rare actinomycetes isolated from Fetzara Lake (northeastern Algeria), a natural representative wetland in the Mediterranean region.
2 Material and methods
2.1 Study area
Located in north-eastern Algeria, the Lake Fetzara (Lat 36°43′and 36°50′N, Long 7°24′ and 7°39′E) is one of the most important coastal wetlands within the Western Mediterranean Basin. In 2003, the lake was included as a wetland with international importance under the Ramsar Convention (Ramsar Convention Official Website, www.ramsar.org). The total site area covers 18600 ha, presenting an important natural reserve for migratory birds and wildfowl species.
2.2 Actinomycetes isolation and their maintenance
The rare actinomycetes were isolated from Fetezara lake, using agar plating method in two different culture media, (i) International Streptomyces Project N°2 (ISP2) supplemented with (2.5 μg/ml of rifampicin, 10 μg/ml of amphotericin B and 75 μg/ml of fluconazole) and (ii) Emmerson agar supplemented with (10 µg/ml of streptomycin, 10 µg/ml amphotericin B and 75 µg/ml of fluconazole). Five water samples were heat treated at 50 °C and dilution series were prepared from samples with aliquots (0.1 ml) being spread plated onto the appropriate agar. The plates were incubated at three different temperatures 10 °C, 28 °C and 37 °C up to 4 weeks and the colonies of actinomycetes were recognized according to their macroscopic and microscopic characteristics. Suspected actinomycetes were subcultured and maintained on ISP2 agar at 4 °C and at -80 °C on the 20% of glycerol as mycelia suspension.
2.3 Phenotypic and physiological characterization
Pure isolates were characterized based on their microscopic, morphological and biochemical characters using standard methods (Shirling and Gottlieb, 1966; Gordon et al., 1974). For the evaluation of growth characteristics, physiological and biochemical characteristics, the actinomycetes isolates were incubated for 15 to 28 days.
The morphological characterization of the isolates was carried out according to the International Streptomyces Project (ISP) using ISP1, ISP2, ISP3, ISP4 and ISP5 media. The observations of growth characteristics were assessed after 3, 7, 14 and 21 days of culture at 30 °C. The ISP6 and ISP7 were used for detection of production of melanoid pigments. Growth was tested at pH 5.0–10.0 (at intervals of 2.0 pH units) and at 4, 25, 30, 37, and 44 °C on nutrient agar. NaCl tolerance was studied on nutrient agar containing NaCl at final concentrations of 0–10% (w/v) (at intervals of 2.5%). The minimal basal medium (ISP9) was used to determine the capacity of the isolates to use different carbon source at 1% of final concentration.
2.4 Molecular identification
2.4.1 Genomic DNA extraction
For DNA extraction, biomasses were obtained by growing the strains in Hickey-Tresner (HT) liquid medium and the genomic DNA was extracted as described by Kieser et al. (2000).
2.4.2 PCR amplification and phylogenetic analysis
The 16S rRNA gene was amplified using universal primers Fd1 and rP2 primers FD1 (5′-AGAGTTTGATCCTGGCTCAG) and rP2 (5′-AAGGAGGTGATCCAGCC) as described by Weisburg et al. (1991) and the purified PCR products were sequenced with the same primers for PCR reaction. The homology search was performed by comparing the sequence with thus present in the public database (NCBI) using the standard Basic Local Alignment Search Tool (BLAST) program as well as with the EzTaxon-server (http://eztaxon-e.ezbiocloud.net/). Phylogenetic analyses were conducted using MEGA software version 6 and the 16S rRNA genes of rare actinomycetes were aligned against neighboring nucleotide sequences using CLUSTALW Larkin et al. (2007). The phylogenetic tree was reconstructed by using the neighbour-joining (NJ) method (Saitou and Nei, 1987) and the topologies were evaluated by bootstrap sampling expressed as a percentage of 1000 replicates (Felsenstein, 1985).
2.5 Extracellular enzyme production
The actinomycetes isolates were screened for amylase, gelatinase, protease, lipase, urease, nitrate reductase and the hemolytic activity according to Larpent and Larpent Gourgaud (1997) using starch, gelatin, casein and tween 80 as substrates.
2.6 Antimicrobial essay
2.6.1 Microorganisms
Fifteen bacterial tests were used for the antibacterial bioassay with seven references strains, S. aureus ATCC25293, S. aureus ATCC43300, Bacillus subtillis ATCC 6633, Micrococcus luteus DSM 1790, Escherichia coli DH5α, E. coli ATCC25422, Pseudomonas aeruginosa ATCC 27853 and eight other clinical isolates. For antifungal activity, five strains of Candida albicans were used. All clinical strains were recovered from hospitalized patients at the Hospital Center of Tebessa (Northeastern Algeria).
2.6.2 Agar diffusion method
Antimicrobial activity of isolated actinomycetes was evaluated using cultures on ISP2, Bennet, and Glucose Yeast Extract Agar (GYEA) by the agar diffusion method (Badji et al., 2007; Kitouni et al., 2005). Actinomycetes were inoculated using spore suspension in three different media (ISP2, Bennet, and GYEA) and incubated for one week at 35 °C. After incubation, plugs of different actinomycete cultures were taken and deposited on the surface of the Luria Bertani soft media (0.7%) (For bacteria) and Sabouraud dextrose agar (For Candida) which had previously been seeded with the indicator strains. The plates were kept at 4 °C for 2 h and then incubated at 37 °C for 24 h. The antimicrobial activities were determined by measuring the diameter of the inhibition zone.
3 Results and discussion
3.1 Isolation and phenotypic characterization of actinomycetes
Based on phenotypic characteristics (macro- and microscopic), eight suspected rare actinomycete strains were isolated from Fetzara Leke. As reported, for successful cultivation of rare actinomycetes groups, major requirement, and appropriate isolation methods are recommended (Bredholdt et al., 2007). In particular, the utilization of enriched selective media supplemented with different antimicrobial agents (antibacterial and antifungal antibiotics) (Shirling and Gottlieb, 1966; Qin et al., 2011).
The morphological and cultural characteristics of the actinomycete isolates were examined using different culture media. Colors of aerial and substrate mycelia were determined using the ISCC-NBS centroid color chart (Kelly and Judd, 1955). The eight actinomycetes isolates showed good growth on ISP1 and ISP2 medium (Fig. 1) and low growth on ISP6 and ISP7 associated with the production of diffusible pigments, after five to 10 days of incubation. The strains were moderately halo tolerates, with a NaCl concentration range for growth of 0– 7%. Growth occurs at 25 to 40 °C (optimum, 37 °C) and pH 6.0–10.0 (optimum, pH 7.0) except for the isolate E3N418 which present a growth at 44 °C (Table 1).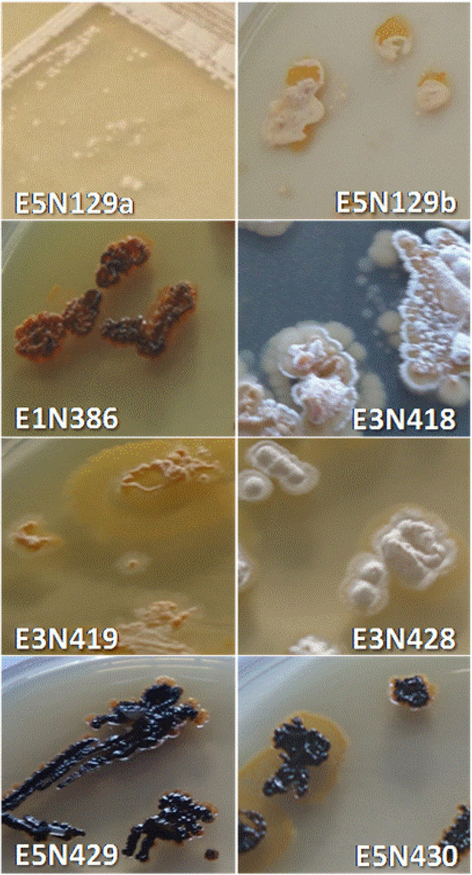
Colony morphology of the eight strains grown on ISP2 at 28 °C for 2 weeks.
Characteristics
Isolates
E5N129a
E5N129b
E1N386
E3N418
E3N419
E5N428
E5N429
E5N430
Temperature
−4 °C
−
−
−
−
−
−
−
−
25 °C
+
+
+
+
±
±
±
±
30 °C
+
+
+
+
+
+
+
+
37 °C
+
±
+
±
±
+
+
+
44 °C
±
±
±
+
−
−
−
±
pH
3
−
−
−
−
−
−
−
−
5
−
−
−
−
−
−
−
−
7
+
+
+
+
+
+
+
+
9
+
+
+
+
+
+
+
+
10
+
+
±
±
+
+
+
+
NaCl%
0
+
+
+
+
+
+
+
+
2.5
±
±
±
±
+
−
+
+
5
±
±
−
±
+
−
+
+
7
−
±
−
±
+
−
±
±
10
−
−
−
±
−
−
−
−
Carbon source
Arabinose
+
−
−
+
−
−
+
+
Fructose
+
−
+
−
−
−
−
+
Galactose
−
−
−
−
−
−
−
+
Glucose
+
+
+
+
+
+
+
−
Melibiose
+
−
−
+
−
−
+
−
Rhamnose
−
−
−
+
−
−
+
−
Ribose
+
+
−
−
+
−
−
−
Saccharose
−
−
−
−
−
−
+
−
Xylose
−
−
−
−
+
+
−
+
Inositol
+
−
−
±
−
−
+
−
Mannitol
+
−
−
+
−
−
+
+
Sorbitol
−
−
−
±
−
−
+
−
Citrate
−
−
−
−
+
−
−
+
Decarboxylation and other
ADH
−
−
−
−
−
−
−
−
LDH
−
−
−
−
−
−
−
−
ODC
−
−
−
−
−
−
−
−
TDA
−
+
−
+
−
+
+
−
IND
−
−
−
−
−
−
−
−
VP
−
−
−
−
−
−
−
−
ONPG
−
+
−
−
−
−
−
−
H2S
−
−
−
−
−
−
−
−
Extracellular enzyme
Amylase
−
−
+
+
+
+
−
−
Gelatinase
+
+
+
−
+
−
+
+
Cellulase
+
+
+
−
−
−
−
−
Lipase
+
+
−
+
+
−
+
+
Protease
−
−
−
+
+
−
+
+
Urease
−
+
−
−
−
−
−
−
Hemolysis
γ
γ
β
γ
γ
α
β
β
As presented in Table 1, different physiological characteristics and carbon substrates utilization were observed. Both of the two isolates EN418 and E3N419 presented potential assimilation profile comparing to other isolates.
3.2 Molecular identification
The obtained sequences of the eight isolates E1N386, E3N418, E3N419, E5N129a, E5N129b, E5N428, E5N429 and E5N430 were subjected to alignment with the homologous closed sequences. The BLAST search of 16S rDNA sequences of the isolated actinomycetes showed highest similarity between (99 and 100%) with four different rare actinomycetes genera as well as Actinomadura, Micromonospora, Nocardia, and Nonomuraea. The constructed phylogenetic tree for partial 16S rRNA (>1400pb) sequences revealed that actinomycetes strains form four distinct phyletic lines within the four described genera species (Fig. 2). In addition, these results are supported and can be easily separated using the combination of physiological properties and the phenotypic characterization.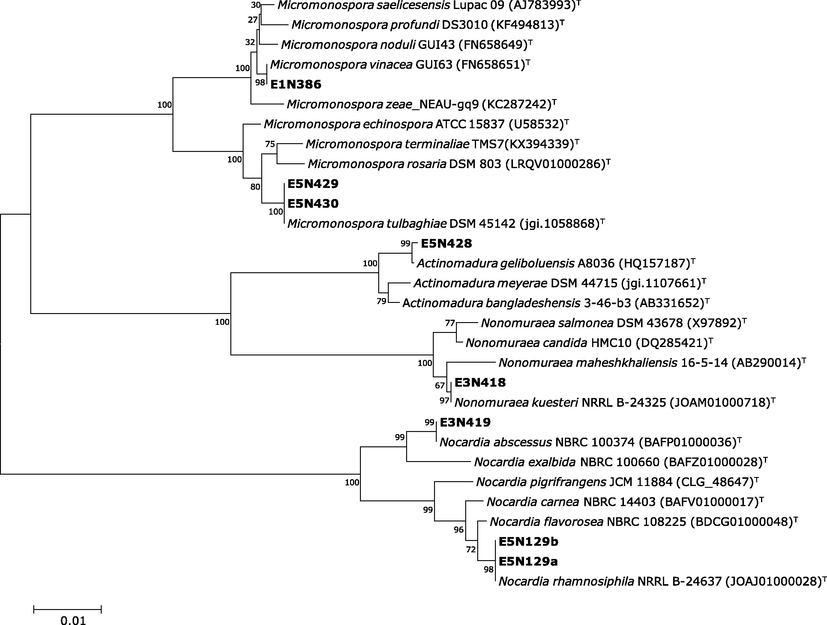
Neighbor-Joining tree based on 16S rRNA sequence. The phylogenetic tree shows the relationships between isolated actinomycetes and related type strains. Percentage bootstrap values based on 1000 resampled data sets are shown at the nodes; only values above 50% are given. The scale bar indicates 0.01 nucleotide substitution per nucleotide position.
The three isolates E1N386, E5N430 and E5N429 belonged to the genus Micromonospora, of which the E1N386 is closely related (99.86%) to the newly Micromonosporasa vinacea GUI63(T) isolated from Pisum sativum nodules (Carro et al., 2016) and both isolates E5N430 and E5N429 shared 100% of similarity with Micromonospora tulbaghiae DSM45142 (Fig. 1). The isolates E5N129a, E5N129b and E3N419 are belonged to the genus Nocardia and closely related to the Nocardia abscessus NBR1003774 and Nocardia rhamnosiphila 202GMO, respectively. The only isolate E3N418 belonged to the genus Nonumeraea was identified as Nonomuraea kuesteri GW 14-1925 with 100% of similarity. The isolate E5N428 belonged to the genus Actinomadura and closely related to Actinomadura geliboluensis A8036.
3.3 Extracellular enzyme production
The metabolic characterization revealed other actinomycetes potency to produce different extracellular hydrolytic enzymes (Table 1). Furthermore, seven isolates produced at least three different enzymes and the majority produced the lipase in the first place followed by caseinase, gelatinase, cellulose, and amylase. These enzymes represent the largest groups of industrial enzymes (Kirk et al., 2002), which are extensively exploited commercially, in food, pharmaceutical, and detergent industry. The actinomycetes have a great capacity for biodegradation of different and complex substrate present in their natural habitats (McCarthy and Stanley, 1992; Tuomela et al., 2000) indicating the variety for their complex metabolites and genomic organization (Bentley et al., 2002) (in particular Streptomyces genus) (Narayana et al., 2007).
The scarcity of reports on industrially relevant enzymatic activities from the identified rare actinomycetes indicated their potential for the production of various hydrolytic enzymes with a promising prospect for industrial application.
3.4 Antimicrobial activity
The antimicrobial activity of the rare actinomycetes isolates is presented in Tables 2 and 3. The primary screening on plates showed that the actinomycetes strains exhibited a significant and variable antibacterial activity against both Gram-negative and Gram-positive bacteria (more frequent) and against at least one indicator organism except for the isolate E5N429 from which no activity was recorded. Such differences in susceptibility are in concordance with other studies by the fact that the Gram-negative strains were highly resistant to many antibiotics (Lucet and Birgand, 2011). In addition, different antibacterial activities were obtained using the three different media (ISP2, Bennet, and GYEA). These results were confirmed by Vijayakumar et al. (2012) reported the influence the culture conditions and the medium composition on the production of antimicrobial molecules. Furthermore, many actinomycetes presented a relatively different spectrum and antimicrobial activities due to different bioactive substances secreted rather than a single inhibitory compound (Benhadj et al., 2018; Mitra et al., 2008).
Tested bacteria
Actinomyctes isolates
E5N129a
E5N129b
E1N386
E3N418
E3N419
E5N428
E5N429
E5N430
I
B
G
I
B
G
I
B
G
I
B
G
I
B
G
I
B
G
I
B
G
I
B
G
E. coli ATCC25422
−
−
−
−
−
−
−
−
−
++a
−
−
−
−
−
−
−
−
−
−
−
−
−
−
E. coli DH5α
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
P. aeruginosa ATCC27853
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
S. aureus ATCC25293
−
−
−
−
−
−
−
−
−
−
−
−
−
++
−
−
−
−
−
−
−
++
−
−
S. aureus ATCC43300
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
M. luteus DSM1970
−
−
−
−
+++
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
B. subtilis ATCC6633
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
Staphylococcus sp.IC13
+++
+++
−
−
−
+++
+++
−
−
+++
−
−
−
−
+++
−
−
−
−
−
−
−
−
++
Citrobacter koseri IC8
−
−
−
−
−
−
−
++
++
−
−
++
−
−
−
++
−
−
−
−
−
−
−
−
Enterobacter sakazakii IC11
++
−
−
−
−
++
++
−
−
++
−
−
−
−
−
−
−
−
−
−
−
−
−
++
Klebsiella sp.IC10
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
+
−
−
−
−
−
−
−
Morganella sp.IC1
−
−
−
−
−
−
−
−
−
−
−
−
−
++
−
−
−
−
−
−
−
++
−
−
Porteus mirabilis IC2
−
−
−
−
−
−
−
−
−
−
−
−
−
++
−
−
−
−
−
−
−
+
−
−
Serratia sp.IC4
−
−
−
−
−
−
−
−
−
−
−
−
−
++
−
−
−
−
−
−
−
+
−
−
Serratia sp.IC7
−
−
−
−
+++
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
−
Isolates
Anticandidal activity (mm)
Candida albicans ICF 18
Candida albicans ICF 19
Candida albicans ICF 22
Candida albicans ICF 23
Candida albicans ICF 24
E5N129a
NA
33.5 ± 1.5
(D10)
31.5 ± 1.5
(D10)
25 ± 1
(D10)
15 ± 2
(D10)
E5N129b
NA
NA
NA
NA
NA
E1N386
21 ± 4
(D3)
25.5 ± 4.5
(D3)
24.5 ± 2.5
(D10)
23.5 ± 0.5
(D10)
10.5 ± 1.5
(D7)
E3N418
NA
31 ± 3
(D10)
22 ± 3
(D10)
23 ± 2
(D10)
12 ± 1
(D10)
E3N419
NA
NA
NA
NA
NA
E5N428
NA
9 ± 0
(D7)
NA
10 ± 0
(D7)
9 ± 0
(D7)
E5N429
NA
NA
NA
NA
NA
E5N430
NA
NA
NA
NA
NA
Antifungal activity of rare actinomycetes has been highlighted during the tests carried out on the clinical isolates. Among the tested actinomycetes, the isolate E5N129a (Nocardia) and the two Micromonospora strains (E1N386 and E3N418) were significantly effective against the tested pathogenic yeasts (10.5 ± 1.5 to 33.5 ± 1.5 mm) (Table 3). In contrast, weak or no activity was recorded using the Nonomuraea isolate (E5N428), Nocardia sp. E5N428 and the two Micromonospora strains (E5N429, E5N430). The anticandidal activity of a rare actinomycete has been reported recently (Tanvir et al., 2016). Further, the two genera Micromonospora and Nocardia were previously reported for the production of antimicrobial compounds which showed broad-spectrum against both bacterial and fungal pathogens (Bredholdt et al., 2007; Kavitha et al., 2010).
Actinomycetes from several unexplored environments have been studied intensively in last few decades and more than 50 rare actinomycete taxa are reported to be producing more than 2000 bioactive compounds (Mitra et al., 2008; Subramani and Aalbersberg, 2013). Several studies reported the production of various antimicrobial compounds by Actinomadura, Nocardia and Nonumeraea strains (Jalali et al., 2016; Kodani et al., 2016). Similar studies indicated the antagonistic activity of actinomycetes isolates such as Micromonospora, against human pathogens (Lee et al., 2012; Talukdar et al., 2012). However, no previous reports on antibacterial activity were found for Micromonosporasa vinacea, Nocardia abscessus, Nocardia rhamnosiphila, Nonomuraea kuesteri or Actinomadura geliboluensis strains.
Overall, the actinomycetes are one of the most attractive sources of new enzymes and bioactive metabolites. Recently, rare actinomycetes have been shown to be an important source of novel secondary metabolites and useful antibiotics. In spite of the limited number of isolates tested, this work constitutes a primary investigation on rare actinomycetes isolated from underexploited habitat (Fetzara Lake) which is a very specific ecosystem with regard to the occurrence of novel micro-flora that hold promising sources of extracellular enzymes and antibacterial compounds.
Conflict of interest
The authors declare that they have no conflict of interest
Authors’ contributions
MB and DGK conceived and designed the study. MB, KG, and ZA conducted the experiment and laboratory work. MB and TM analysed data and drafted the manuscript. All authors read and approved the manuscript.
References
- Elicitation of secondary metabolism in actinomycetes. Biotech. Adv.. 2015;33(6):798-811.
- [CrossRef] [Google Scholar]
- The Use of Extracellular Enzymes from Streptomyces albus ATCC 3005 for the Bleaching of Eucalyptus Kraft Pulp. Appl. Microbiol. Biotechnol.. 2001;57(1–2):92-97.
- [CrossRef] [Google Scholar]
- Characterization of Antibiotic Producing Rare Actinomycete Nonomuraea sp. JAJ18 Derived from an Indian Coastal Solar Saltern. Sci. World J.. 2014;2014
- [CrossRef] [Google Scholar]
- Isolation and partial characterization of antimicrobial compounds from a New Strain Nonomuraea sp. NM94”. J. Ind. Microbiol. Biotechnol.. 2007;34(6):403-412.
- [CrossRef] [Google Scholar]
- Benhadj, M., Gacemi-kirane, D., 2016. Les Actinomycètes: source de biomolécules d'intérêt. Éditions universitaires européennes, p 60.
- Diversity and antimicrobial activities of Streptomyces isolates from Fetzara Lake, northeastern Algeria. Ann. Biol. Clin.. 2018;76(1):81-95.
- [CrossRef] [Google Scholar]
- Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141-147.
- [CrossRef] [Google Scholar]
- Thoughts and facts about antibiotics: Where We Are Now and Where We Are Heading”. J. Antibiot.. 2012;65(8):385-395.
- [CrossRef] [Google Scholar]
- Rare actinomycete bacteria from the shallow water sediments of the Trondheim fjord, Norway: isolation, diversity and biological activity. Environ. Microbiol.. 2007;9:2756-2764.
- [CrossRef] [Google Scholar]
- Micromonospora ureilytica sp. nov., Micromonospora noduli sp. nov. and Micromonospora vinacea sp. nov., isolated from Pisum sativum nodules. Int. J. Syst. Evol. Microbiol.. 2016;66:3509-3514.
- [CrossRef] [Google Scholar]
- Genome mining of rare actinomycetes and cryptic pathway awakening. Proc. Biochem.. 2015;50(8):1184-1193.
- [CrossRef] [Google Scholar]
- Confidence Limits on Phylogenies: An Approach Using the Bootstrap”. Evolution. 1985;39(4):783-791.
- [CrossRef] [Google Scholar]
- Nocardia Coeliaca, Nocardia Autotrophica, and the Nocardin Strain. Int J Syst Evol Microbiol. 1974;24(1):54-63.
- [CrossRef] [Google Scholar]
- Antagonistic activity of Nocardia brasiliensis PTCC 1422 against isolated Enterobacteriaceae from urinary tract infections. Probiotics Antimicrob Prot.. 2016;8(1):41-45.
- [CrossRef] [Google Scholar]
- Isolation, characterization and biological evaluation of bioactive metabolites from Nocardia levis MK-VL_113. Microbiol Res. 2010;165:199-210.
- [Google Scholar]
- The ISCC-NBS color names dictionary and the universal color language. NBS Circular. 1955;553
- [Google Scholar]
- Pratical streptomyces genetics. Colney: Jean Innes Foundations; 2000.
- Streptomyces competition and co-evolution in relation to plant disease suppression. Res Microbiol.. 2012;163(8):490-499.
- [CrossRef] [Google Scholar]
- Industrial enzyme applications. Curr. Opin. Biotechnol.. 2002;13:345-351.
- [CrossRef] [Google Scholar]
- Isolation of actinomycetes producing bioactive substances from water, soil and tree bark samples of the North–east of Algeria. J. Med. Mycol.. 2005;15(1):45-51.
- [CrossRef] [Google Scholar]
- Isolation and structure determination of a new lantibiotic cinnamycin B from Actinomadura atramentaria based on genome mining. J. Ind. Microbiol. Biotechnol.. 2016;43:1159-1165.
- [CrossRef] [Google Scholar]
- Modeling the synergistic antibacterial effects of honey characteristics of different botanical origins from the Sahara Desert of Algeria. Front. Microbiol.. 2015;6:1239.
- [Google Scholar]
- Discovery of Novel Metabolites from Marine Actinomycetes. Curr. Opin. Microbiol.. 2006;9(3):245-251.
- [CrossRef] [Google Scholar]
- Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23(21):2947-2948.
- [CrossRef] [Google Scholar]
- Mémento Technique de Microbiologie (3eme edition). Lavoisier Tec & Doc.; 1997.
- Rare genera of actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek.. 2000;78(3–4):399-405.
- [CrossRef] [Google Scholar]
- Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol.. 2012;28:2125-2137.
- [CrossRef] [Google Scholar]
- A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517(7535):455-459.
- [CrossRef] [Google Scholar]
- Les Bacilles À Gram-Négatif Multi-Résistants: Où va-T-On? J. Anti-Infectieux. 2011;13(2):122-132.
- [Google Scholar]
- Salinispora Arenicola gen. nov., sp. nov. and Salinispora Tropica Sp. Nov., Obligate Marine Actinomycetes Belonging to the Family Micromonosporaceae”. Int J Syst Evol Microbiol.. 2005;55(5):1759-1766.
- [CrossRef] [Google Scholar]
- Actinomycetes as Agents of Biodegradation in the Environment—a Review. Gene. 1992;115(1):189-192.
- [CrossRef] [Google Scholar]
- Diversity and Bioprospecting of Extremely Halophilic Archaea isolated from Algerian Arid and Semi-Arid Wetland Ecosystems for Halophilic-Active Hydrolytic Enzymes. Microbiol. Res. 2018;207:289-298.
- [Google Scholar]
- External bacterial flora and antimicrobial susceptibility patterns of Staphylococcus spp and Pseudomonas spp. isolated from two house hold cockroaches, Blattella germanica and Blatta orientalis. Biomed. Environ. Sci. 2015;28:316-320.
- [Google Scholar]
- Distribution of actinomycetes, their antagonistic behaviour and the physico chemical characteristics of the world's largest tidal mangrove forest. Appl. Microbiol. Biotechnol.. 2008;80:685-695.
- [CrossRef] [Google Scholar]
- Screening of antibacterial producing Actinomycetes from sediments of the Caspian Sea. Int. J. Mol. Cell Med.. 2013;2(2):64-71.
- [Google Scholar]
- Biological activity of phenylpropionic acid isolated from a terrestrial streptomycetes”. Pol. J. Microbiol.. 2007;56(3):191-197.
- [Google Scholar]
- Regulation of a novel gene cluster involved in secondary metabolite production in Streptomyces coelicolor. J. Bacteriol.. 2010;192(19):4973-4982.
- [CrossRef] [Google Scholar]
- Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol.. 2011;89(3):457-473.
- [CrossRef] [Google Scholar]
- The genus Nonomuraea: a review of a rare actinomycete taxon for novel metabolites. J. Basic Microbiol.. 2015;55(5):554-565.
- [CrossRef] [Google Scholar]
- The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.. 1987;4(4):406-425.
- [CrossRef] [Google Scholar]
- Method for Characterization of Streptomyces Species. Int. J. Syst. 1966:16-313.
- [CrossRef] [Google Scholar]
- Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery. Appl Microbiol Biot.. 2013;97:9291-9321.
- [CrossRef] [Google Scholar]
- Isolation and Diversity of Actinomycetes in the Chesapeake Bay. Appl Environ Microbiol.. 1993;59(4):997-1002.
- [Google Scholar]
- Bioprospecting Micromonospora from Kaziranga National Park of India and their anti-infective potential. World J. Microbial. Biotechnol.. 2012;28:2703-2712.
- [CrossRef] [Google Scholar]
- Rare actinomycetes Nocardia caishijiensis and Pseudonocardia carboxydivorans as endophytes, their bioactivity and metabolites evaluation. Microbiol. Res.. 2016;185:22-35.
- [Google Scholar]
- Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol.. 2015;33(1):15-26.
- [CrossRef] [Google Scholar]
- Diversity and isolation of rare actinomycetes: an overview. Crit. Rev Microbiol.. 2013;39(3):256-294.
- [CrossRef] [Google Scholar]
- Biodegradation of lignin in a compost environment: a review”. Biores Technol.. 2000;72(2):169-183.
- [CrossRef] [Google Scholar]
- Antimicrobial potential of actinomycetes species isolated from marine environment. Asian Pac J. Trop. Biomed.. 2012;2(6):469-473.
- [CrossRef] [Google Scholar]
- Antimicrobial potentiality of a halophilic strain of Streptomyces sp. VPTSA18 isolated from the saltpan environment of Vedaranyam. India. Ann. Microbiol. 2012;62:1039-1047.
- [CrossRef] [Google Scholar]
- 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol.. 1991;173(2):697-703.
- [CrossRef] [Google Scholar]







