Translate this page into:
Prevalence and antibiogram pattern of multidrug-resistant Staphylococcus aureus infections in individuals with sickle cell disease. A retrospective study, hematological and genetic analysis
⁎Corresponding authors. aabdulmanea@ksu.edu.sa (Adel A. Abdulmanea), gkhaled@ksu.edu.sa (Jamal M. Khaled)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The prevalence of Staphylococcus aureus in the circulation (bacteremia) of individuals with sickle cell disease (SCD) may provide a substantial risk for illness and death. Strains of S. aureus exhibit a wide variety of virulence traits, including the ability to generate various toxins and develop treatment resistance. MRSA is characterized by its resistance to a range of beta-lactam antibiotics. This study’s objectives included assessing the frequency of S. aureus infection in SCA patients, analyzing laboratory markers of the CBC test, examining the antibiotic susceptibility pattern, and investigating the mecA gene and heteroresistance, which could potentially link to a higher occurrence of S. aureus infection in SCA patients. The retrospective study was done to gather clinical data on individuals with sickle cell anemia (SCA) who had bloodstream infections. From 2017 to 2021, blood samples and data were gathered by King Saud University Medical City (KSUMC) in Riyadh, Saudi Arabia. The S. aureus strains were re-cultivation, Vitek system and PCR techniques were employed to ascertain the antibiotic resistance patterns. It was confirmed that 2406 patients with SCD had contracted an infection. A total of 138 (5.73 %) patients with SCD were diagnosed with bacteremia. “Of the 138 strains isolated, the MRSA strain revealed as the most prevailing, including 30 isolates (21.7 %), whereas non-MRSA strains constituted 17 isolates (12.3 %).” Males had a higher rate of infection compared to females. The results indicated that all the S. aureus isolates exhibited resistance to ampicillin, while the resistance to penicillin was 98 %. Furthermore, the results indicated that over 65 % of the isolates exhibited resistance to amoxicillin, amoxicillin, cefazolin, imipenem, and oxacillin. The mecA gene was identified in all S. aureus strains obtained from patients with SCD. The current study recognizes the PCR assay as a more dependable technique for assessing antibiotic resistances, in contrast to the Vitek method. A study found strong links between S. aureus infections in individuals with SCD and three different factors: hemoglobin (Hb) level, mean corpuscular volume (RDW), and white blood cell (WBC) count (p < 0.004, 0.005, and 0.02, respectively). The study revealed that 34 % of the S. aureus strains that were examined exhibited heteroresistance to the mecA gene as well as resistance to oxacillin. The results clearly demonstrated that Ampicillin, Penicillin, amoxicillin, cefazolin, imipenem, and oxacillin are unsuitable choices for the treatment of blood infections in individuals with SCD. “The findings of this study confirmed that all S. aureus isolates were sensitive to gentamicin, and more than 80 % of the isolates were sensitive to ciprofloxacin, moxifloxacin, levofloxacin, clindamycin, and vancomycin.” Further investigation is necessary to explore potential factors that increase the susceptibility of individuals with SCD to bacteremia caused by antibiotic-resistant strains of S. aureus.
Keywords
Sickle cell disease
Bloodstream infection
Saudi Arabia
Antibiotics resistance
Staphylococcus aureus
CBC
mecA gene
1 Introduction
Bloodstream infection (BSI) is linked to considerable morbidity and mortality. Staphylococcus aureus is the main reason behind bloodstream infections. S. aureus is a Gram-positive, catalase-positive, mannitol-positive, and coagulase-positive bacterium recognized as a significant human pathogen responsible for various illnesses, such as septicemia, endocarditis, pneumonia, osteomyelitis, toxic shock syndrome, and meningitis. This bacteria manifests as spherical (cocci) cells and exhibits grape-like clusters under microscopic examination due to its division in many planes. The diameter ranges generally from 0.5 to 1.0 μm. It develops on blood agar as smooth, spherical, and slightly elevated colonies, exhibiting the capacity to induce full hemolysis surrounding the colonies (β-hemolytic). It synthesizes staphyloxanthin, a golden pigment that offers antioxidant protection and is believed to enhance its pathogenicity. It thrives best at 37 °C, however it can also proliferate within a temperature range of 15–45 °C. It withstands elevated salt concentrations (up to 7.5 % NaCl), enabling its growth on mannitol salt agar. S. aureus is recognized for generating numerous virulence factors and toxins, such as enterotoxins (which induce food poisoning), toxic shock syndrome toxin (linked to toxic shock syndrome), exfoliative toxins (responsible for staphylococcal scalded skin syndrome), and Panton-Valentine leukocidin (associated with severe skin infections and necrotizing pneumonia). S. aureus frequently harbors plasmids that encode antibiotic resistance and may also be infected by bacteriophages that possess genes for virulence factors such as toxins. S. aureus can produce biofilms that shield the germs from the host immune response and antibiotic therapies. This is especially pertinent in infections associated with medical equipment (e.g., catheters, implants) (Donkor et al., 2010, Appiah et al., 2020). S. aureus is a notable bacterium that has a substantial impact on both healthcare institutions and the general population (Chen et al., 2022).
Methicillin-resistant S. aureus (MRSA) is a variant of S. aureus that exhibits resistance to methicillin, oxacillin, penicillin, and amoxicillin. It is a common source of infections acquired in hospitals, and its occurrence is increasing globally. The resistance of this condition to several treatments is a significant issue for public health, since it limits the available treatment options and creates a major problem for healthcare (Monecke et al., 2012, Aslam et al., 2022, Garoy et al., 2019). MRSA bacteria present a substantial problem in the treatment of infections due to their resistance to many antibiotics, including beta-lactams, aminoglycosides, and macrolides (Duran et al., 2012). The incidence rates of MRSA infections in Saudi Arabia exhibit regional disparities, with figures ranging from 27 % to 42 % (Aljeldah, 2020). The prevalence of MRSA at King Fahad Medical City in Riyadh, Saudi Arabia, in 2011 was 50 % (Alghizzi et al., 2021, Monecke et al., 2012).
Sickle cell anemia (SCA) is a common genetic disease in Saudi Arabia (Alotaibi, 2017). Prior to being diagnosed, sickle cell anemia results in the death of over 90 % of children in sub-Saharan Africa (Modell and Darlison, 2008). Sickle cell disease (SCD) is a collection of inherited disorders distinguished by an anomalous configuration of hemoglobin. People with SCD are more susceptible to severe bacterial infections, which results in frequent hospitalizations and the requirement for antibiotic treatment (Alsultan et al., 2012). Considering the substantial threat that SCA presents to Saudi society, it is crucial to carry out research on these individuals, with a specific focus on bacterial bloodstream infections.
A number of researchers studying sickle cell disease (SCD) are focused on identifying encapsulated bacterial pathogens in individuals with SCD, including Streptococcus pneumoniae, Haemophilus influenzae, Salmonella spp., Neisseria meningitidis, and Klebsiella spp. The prognosis for individuals born with SCA has been greatly enhanced in industrialized countries with the utilization of combination immunizations that specifically target S. pneumoniae and H. influenzae type B (Williams et al., 2009). Nevertheless, in persons with sickle cell disease (SCD), S. aureus is more prone to causing severe infections in comparison to S. pneumoniae. Although there are worries over antibiotic resistance, it is customary to obtain blood cultures and administer antibiotics to all patients with SCD who exhibit a temperature exceeding 38.5 °C. In recent decades, the widespread use of antibiotics has led to the emergence of drug-resistant forms of S. aureus (Mlynarczyk-Bonikowska et al., 2022, Reddy et al., 2017).
There have been just a few investigations that have documented cases of bloodstream infection caused by S. aureus strains in persons with SCD. Saudi Arabia has not adequately investigated the relationship between S. aureus and MRSA germs in patients with SCD, hematological markers (CBC), and antibiotic resistance. Hence, it is imperative to conduct thorough prospective studies in order to determine the epidemiology of these bacteria in individuals with SCD. These studies should prioritize investigating the prevalence of these bacteria in SCD patients, the factors that contribute to their presence, and the level of antibiotic resistance displayed by the strains that colonize these individuals.
This study is a groundbreaking initiative in Saudi Arabia to identify several bacterial strains in persons with sickle cell disease (SCD) who are experiencing bloodstream infections. Multiple studies have consistently demonstrated an increasing trend in the prevalence of antibiotic resistance in S. aureus infections, specifically MRSA. This highlights the urgent need for continuous and vigilant monitoring. The study aimed to ascertain the prevalence of S. aureus infections, including MRSA infections, and examine the alterations in resistance patterns during a five-year duration in patients with SCD at KSUMC in Riyadh, Saudi Arabia. Furthermore, the study sought to offer suggestions for future interventions in healthcare and therapy to prevent and control these illnesses. This study aims to investigate the frequency of S. aureus infections, namely MRSA, among patients with sickle cell disease (SCD) who have bloodstream infections at King Saud University Medical City (KSUMC) in Riyadh, Saudi Arabia. The study will cover the period from January 2017 to December 2021. Furthermore, they analyzed the CBC, antibiotic susceptibility patterns, the mecA gene, and the occurrence of heteroresistance in S. aureus isolates. The results of this analysis could assist researchers in gaining a deeper understanding of the attributes of these deadly bacteria that endanger persons with SCD.
2 Materials and methods
2.1 Study ethics
The study was conducted in KSUMC's departments of hematology and microbiology. On April 12, 2018, No. 18/0258, the KSUMC institutional review board (IRB) in Riyadh, Saudi Arabia, authorized all protocols and procedures, including the collecting of samples, the isolation of bacteria, and the analysis of data.
2.2 Study design and sample size
We retrospectively identified all BSI that occurring in SCD patients (aged 1–65 years) at KSUMC from January 2017 to December 2021by reviewing the computerized databases of the microbiology and hematology departments. We included and reviewed the medical records of 2406 SCD patients for BSI. The study involved 138 strains were positive of blood culture including 47 strains of S. aureus and MRSA. From blood, we isolated 47 strains of S. aureus and MRSA harboring SCD (HbSS). As a reference, we employed 16 strains of S. aureus that did not harbor SCD. The isolates were kept at −80 ± 5 °C in skim milk medium containing 10 % glycerol until they were required to be re-cultivated for more testing.
2.3 Bacterial isolation and identification
Conventional techniques such as Gram stain, morphological and biochemical tests, VITEK 2 C15, MicroScan configuration auto-mated system (Biomérieux, France), and serology tests were used to identify the 47 strains of S. aureus and MRSA. The identification was then confirmed by the molecular method (PCR assay). A master mix (2x HotStarTaq Plus Master Mix; Qiagen, Hilden, Germany) was used to prepare the PCR reaction. 5 μL of DNA, 12.5 μL of PCR Master Mix, 1.5 μL of each reverse and forward primer (AGAGTTTGATCCTGGCTCAG (16S-F) and AAGGAGGTGATCCAGCCGCA (16S-R)—the usual PCR product for this gene is 1500 pb—and 4.5 μL of nuclease water made up the reaction mixture employed in this work. PCR was used with a thermal cycler to amplify the target 16S rDNA gene (Corbett Research). The initial denaturation at 95 °C for 15 min, 35 cycles (denaturation, 94 °C, 1 min; annealing, 54 °C, 1 min; extension, 72 °C, 1 min), a final extension at 72 °C for 10 min., and holding at 4 °C were the ideal thermal cycling conditions. After the 16S rRNA amplicon was sequenced (Macrogen, Inc., Korea), the identity was verified using BLAST analysis (Pechorsky, Ahmed et al., 2009).
2.4 Antibiotic susceptibility testing
To test several antimicrobial drugs against the S. aureus isolates, we employed MicroScan and the VITEK 2 system (Biomerieux) (Kassim et al., 2016). According to the recommendations of the Clinical Laboratory Standards Institute (CLSI), the gram-positive susceptibility card (AST-GP67Test Kit) was used to identify the resistant, intermediate, and sensitive isolates against 14 different antibiotics, including amoxicillin (AMOX), ampicillin (AMP), cefazolin (CFZ), clindamycin (CLI), erythromycin (ERY), gentamicin (GEN), imipenem (IMP), levofloxacin (LVX), moxifloxacin (MOX), ciprofloxacin (CIP), penicillin (PEN), oxacillin (OXA), tetracycline (TET), and vancomycin (VAN).
2.4.1 Antibiotic susceptibility cluster analysis
The antibiotic susceptibility cluster analysis using dendrogram (Ward linkage method) was done using selected standard antibiotic that included Cloxacillin, Rifampicin, and Tetracycline. It is an effective tool for understanding and visualizing hierarchical clustering. The goal of Ward's approach is to reduce the overall within-cluster variance (Strauss and Von Maltitz, 2017). This analysis was performed using Origin Pro 2018).
2.5 Hemoglobin electrophoresis (Capillary Electrophoresis)
SCD SCD was diagnosed by using Capillary Electrophoresis (CE) (Sebia Capillary’s system). The normal ranges for healthy adults were as follows: HbA1, 96.8 % or more; HbA2, 2.2 % to 3.2 %; HbF, less than 0.5 %; HbC, Absent; and HbS, Absent (Frömmel, 2018). During the study period, 2406 patients were diagnosis with SCD.
2.6 CBC Estimation
A fully automatic blood cell counter hematology analyzer (Bulkman Coulter® Unicel DxH 800) was used to measure hematological values, including mean corpuscular volume (RDW), erythrocyte red blood cell count (RBC), hemoglobin concentration (HGB), platelets (Plt), packed cell volume (PCV), and white blood cell (WBC), in addition to hematocrit (HCT) (Vis and Huisman, 2016). All of the hematological results were compared to the reference and control values, which encompassed sixteen samples.
2.7 Detection of mecA gene of S. Aureus
The mecA gene was identified using a polymerase chain reaction using a reverse mecA primer (CTAG-TCCATTCGGTCCA) and a forward mecA primer (CCTAGTAAAGCTCCGGAA), amplifying a 314-bp DNA fragment (Duran et al., 2012). The S. aureus isolates' DNA was extracted using Qiagen's DNeasy Blood & Tissue Kits (Germantown, MD, USA). Using a PCR thermocycler, we amplified DNA under particular reaction conditions (Corbett Research PCR Thermal Cycler). The procedure was as follows: 15 min of initial denaturation at 95 °C; 35 cycles of denaturation at 94 °C for 1 min; 30 s of annealing at 55 °C; 1 min of extension at 72 °C; and 10 min of final extension at 72 °C. Next, we used 1.5–2 % agarose gel electrophoresis to evaluate the amplified genes (Duran et al., 2012). Next, we used a polymerase chain reaction (PCR) that targeted the mecA gene to confirm the isolates resistance to oxacillin/methicillin and heteroresistance in the S. aureus isolates. The phenomenon known as heteroresistance happens when only a small number of cells in a laboratory setting genuinely demonstrate this resistance, despite the fact that all cells in a culture have the capacity to carry the genetic material (mecA) required for resistance.
2.8 Statistical analysis
The data from the study will be initially entered into Microsoft Office Excel, and subsequently transferred to the Statistical Package for the Social Sciences (SPSS) application. Following a number of statistical analyses, we will use frequencies and percentages to summarize the findings. Using independent samples, Chi-square, and basic statistical methods like proportion t-tests and descriptive statistics, we will conduct significance testing. The significance threshold (p-value) will be fixed at less than 0.05.
3 Results
3.1 Incidence of S. Aureus strains among patients with SCD
During the study period, 2406 patients were diagnosed with SCD Fig. 1. Among the 2,406 SCD patients, we identified 138 cases (5.7 %) of bloodstream infections (bacteremia) caused by various types of bacteria. Among them, 30 patients had MRSA, 17 patients had S. aureus, 17 patients had S. epidermidis, 14 patients had S. homins, 7 patients had Enterobacter cloacae complex, 5 patients had S. mitis, 5 patients had Escherichia coli, 4 patients had Salmonella species group D and 2 patients had S. pneumoniae Table 1. Tables 1 and 2 demonstrate that, among SCD patients from 2017 to 2021, the proportion of MRSA and S. aureus was 2 %. Based on the population at high risk of SCA infection, we separated the age groups. Three age groups were separated: those aged 1 to 20, those aged 21 to 40, and those aged 41 to 65. With 61.7 percent of all samples, those over 20 were the most impacted age group. Fig. 2 illustrates the correlation rates of antibiotic resistance among MRSA, S. aureus, male and female. *MRSA = Methicillin-resistant S. aureus, and S. A = S. aureus. ** The value indicates a significant difference when the P-value is less than 0.05.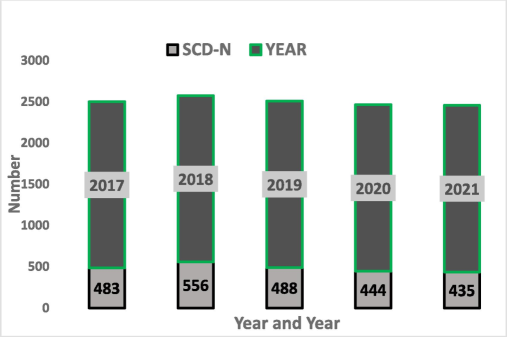
Sickle cell disease (SCD) patients through 2017–2021.
Type of bacteria
Number
Prevalence %
Methicillin-resistant Staphylococcus aureus
30
21.7
Staphylococcus aureus
17
12.3
Streptococcus pneumoniae
2
1.4
Staphylococcus homins
14
10.1
Staphylococcus epidermidis
17
12.3
Staphylococcus capitis
3
2.2
Staphylococcus haemolyticus
3
2.2
Staphylococcus lugaunensis
1
0.7
Fusobacterium nucleatum
1
0.7
Enterobacter cloacae complex
7
5.1
Streptococcus species
1
0.7
Streptococcus mitis
5
3.6
Streptococcus sanguinis
1
0.7
Streptococcus salivarius
1
0.7
Streptococcus parasanquinis
1
0.7
Streptococcus gordonii
1
0.7
Enterococcus faecium
3
2.2
Salmonella species group D
4
2.9
Salmonella species group B
1
0.7
Escherichia coli
5
3.6
Micrococcus species
2
1.4
Corynebacterium jeikeium
2
1.4
Corynebacterium pseudodiphtheriticum
1
0.7
Klebsiella pneumoniae
1
0.7
Neisseria elongata
1
0.7
Clostridium clostridioforme
1
0.7
Acinetobacter baumannii
1
0.7
Actinomyces odontolyticus
2
1.4
Aeromonas hydrophila complex
1
0.7
Bacillus licheniformis
1
0.7
Stenotrophomonas maltophilia
3
2.2
burkholderia cepacia complex
1
0.7
Propionibacterium acens
1
0.7
Providencia rustiqianii
1
0.7
Pseudomonas stutzeri
1
0.7
Year
MRSA and SA
Number
Percent
P-value**
2017
MRSA*
18
38.3
0.001
S.A.
4
8.5
2018
MRSA
4
11.4
0.333
S.A.
7
20
2019
MRSA
1
3.8
0.164
S.A.
4
15.4
2020
MRSA
6
26.1
0.009
S.A.
0
0
2021
MRSA
1
14.3
0.557
S.A.
2
28.6
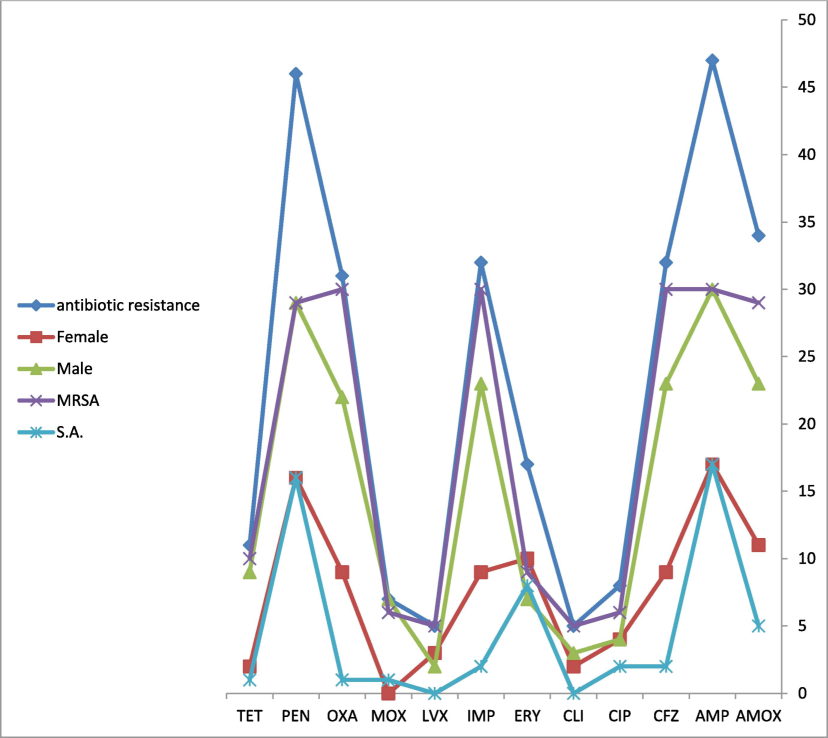
The correlation rates of antibiotic resistance compared with male, female, oxacillin-resistant S. aureus (MRSA), and S. aureus (S. A).
3.2 Haemoglobin electrophoresis (Capillary Electrophoresis
SCA was diagnosed using Capillary Electrophoresis (CE). During the study period, 2406 patients were diagnosis with SCD Fig. 1.
3.3 Identification
Firstly, we extracted the preserved samples from the freezer and allowed them to defrost at ambient temperature. We introduced all the specimens onto agar plates containing chocolate, blood, MacConkey, and mannitol salt. The findings indicated that S. aureus exhibits distinct biochemical characteristics, including a positive gram stain, a particular colony form characterized by mannitol fermentation, and biochemical properties such as positive catalase, oxidase, and coagulase tests. The 16S rRNA analysis confirmed the identification, demonstrating that all organisms tested were S. aureus.
3.4 Antibiotic susceptibility testing
Table 3 displays the correlation between the resistance of S. aureus isolates to antibiotics. The study spans a duration of five years, specifically from 2017 to 2021. The data presents the yearly number and percentage of isolates that show resistance to different antibiotics, together with the p-values that reflect the statistical significance of the observed changes over time. In 2017, the resistance rate of Cefazolin was 56 %, indicating a significant level of resistance. By the year 2021, there will be a notable reduction of 3 %. The p-value is 0.000, which signifies statistical significance. In 2017, the resistance to Penicillin experienced a considerable surge, reaching a rate of 46 % then the projected decrease of 6 % by the year 2021. The p-value is 0.000, which signifies statistical significance. In 2017, the levels of Ampicillin resistance were recorded at 47 %. Nevertheless, as of 2021, the levels of resistance had notably diminished to a mere 6 %. In 2017, the antibiotic Amoxicillin had a resistance rate of 61 %, which reduced to 3 % by 2021. Imipenem the resistance rate in 2017 was 56 %. Nevertheless, by 2021, this rate had markedly declined to a mere 3 %. In 2017, the antibiotic Oxacillin exhibited a resistance rate of 56 %, nevertheless, this rate experienced a substantial decline to 3 % by 2021. In 2017, the antibiotic Erythromycin exhibited a resistance rate of 41 %, nevertheless, this rate will undergo a substantial decline to 6 % by the year 2021. The prevalence of Tetracycline resistance in 2017 was 28 %. Resistance levels exhibited fluctuations, reaching a maximum of 45 % in 2020 and a complete absence of resistance (0 %) in 2021. In 2017, the antibiotic Ciprofloxacin exhibited a resistance rate of 87 %. The rate exhibited fluctuations, reaching a peak of 87 % in 2018 and remaining at a minimum of 0 % in all years except for 2019, when it stood at 13 %. In 2017, the prevalence of resistance to Moxifloxacin was 57 %. The resistance rate exhibited variability over time, reaching 0 % in severe situations. The resistance rate exhibited temporal variability, reaching 0 % in multiple years, and escalating to 29 % in 2019. In 2017, the occurrence of resistance to Levofloxacin was 80 %. The resistance rate was 0 % in the years 2018, 2019, and 2012. However, the p-value of 0.180 did not reach the level of statistical significance.
Antibiotic resistance
Resistant (N, %) *
2017
N, %
2018
N, %
2019
N, %
2020
N, %
2021
N, %
**p-Value
Ampicillin (AMP)
47 (100 %)
22 (47 %)
11 (23 %)
5 (11 %)
6(13 %)
3 (6 %)
0.000
Penicillin (PEN)
46 (98 %)
21 (46 %)
11(24 %)
5 (11 %)
6 (13 %)
3 (6 %)
0.000
Amoxicillin (AMOX)
34 (72 %)
21 (61 %)
4 (12 %)
2 (6 %)
6 (18 %)
1 (3 %)
0.000
Cefazolin (CFZ)
32 (68 %)
18 (56 %)
5 (16 %)
2 (6 %)
6 (19 %)
1(3 %)
0.000
Imipenem (IMP)
32 (68 %)
18 (56 %)
5 (16 %)
2 (6 %)
6 (19 %)
1 (3 %)
0.000
Oxacillin (OXA)
31 (66 %)
18 (59 %)
5 (16 %)
1 (3 %)
6 (19 %)
1 (3 %)
0.000
Erythromycin (ERY)
17 (36 %)
7 (41 %)
4 (23 %)
2 (12 %)
3 (18 %)
1 (6 %)
0.182
Tetracycline (TET)
11 (24 %)
3 (28 %)
2 (18 %)
1 (9 %)
5 (45 %)
0 (0 %)
0.364
Ciprofloxacin (CIP)
8 (17 %)
7 (87 %)
0 (0 %)
0 (0 %)
1 (13 %)
0 (0 %)
0.034
Moxifloxacin (MOX)
7 (15 %)
4 (57 %)
0 (0 %)
2 (29 %)
1(14 %)
0 (0 %)
0.368
Levofloxacin (LVX)
5 (11 %)
4 (80 %)
0 (0 %)
0 (0 %)
1 (20 %)
0 (0 %)
0.180
Clindamycin (CLI)
5 (11 %)
4 80 %)
1 (20 %)
0 (0 %)
0 (0 %)
0 (0 %)
0.180
Vancomycin (VAN)
3 (6 %)
2 (67 %)
0 (0 %)
0 (0 %)
1(33 %)
0 (0 %)
0.180
Gentamicin (GEN)
0 (0 %)
0 (0 %)
0 (0 %)
0 (0 %)
0 (0 %)
0 (0 %)
−
Overall, there is a significant decline in the resistant rate of most antibiotics (ampicillin, penicillin, amoxicillin, cefazolin, imipenem, oxacillin, erythromycin, ciprofloxacin, moxifloxacin, and levofloxacin) with time, as indicated by the p-values of 0.000. This signifies a significant reduction in resistance, indicating the effectiveness of therapies or modifications in clinical processes throughout the investigated period. Nevertheless, the changes in resistance patterns for erythromycin, tetracycline, moxifloxacin, and levofloxacin do not exhibit statistical significance, indicating either uneven trends or less conspicuous adaptations. Regarding the other antibiotics tested in this study (Clindamycin, Gentamicin, and Vancomycin) in Table 3, all bacterial isolates were susceptible to Gentamicin, while 89 % and 94 % of the isolates were susceptible to Clindamycin and Vancomycin, respectively.
Resistant (N, %)*: number and percentage of antibiotic susceptibility resistance of 47 S. aureus strains (case). p values **: Correlation between antibiotic susceptibility resistance comparing with years. A p-value of less than 5 % indicates statistical significance.
3.4.1 Antibiotic susceptibility cluster analysis
Fig. 3 displays a dendrogram created using the Ward linkage approach, which groups patients together based on their antibiotic susceptibility, age, year, and gender. This hierarchical clustering technique visually organizes patients into groups based on their similarity in the specified parameters. The Ward linkage approach aims to reduce the variance within each cluster, resulting in the formation of more homogeneous groups. The dendrogram illustrates the presence of two primary clusters, suggesting that the patient population may be segregated into two clearly defined categories based on the studied data.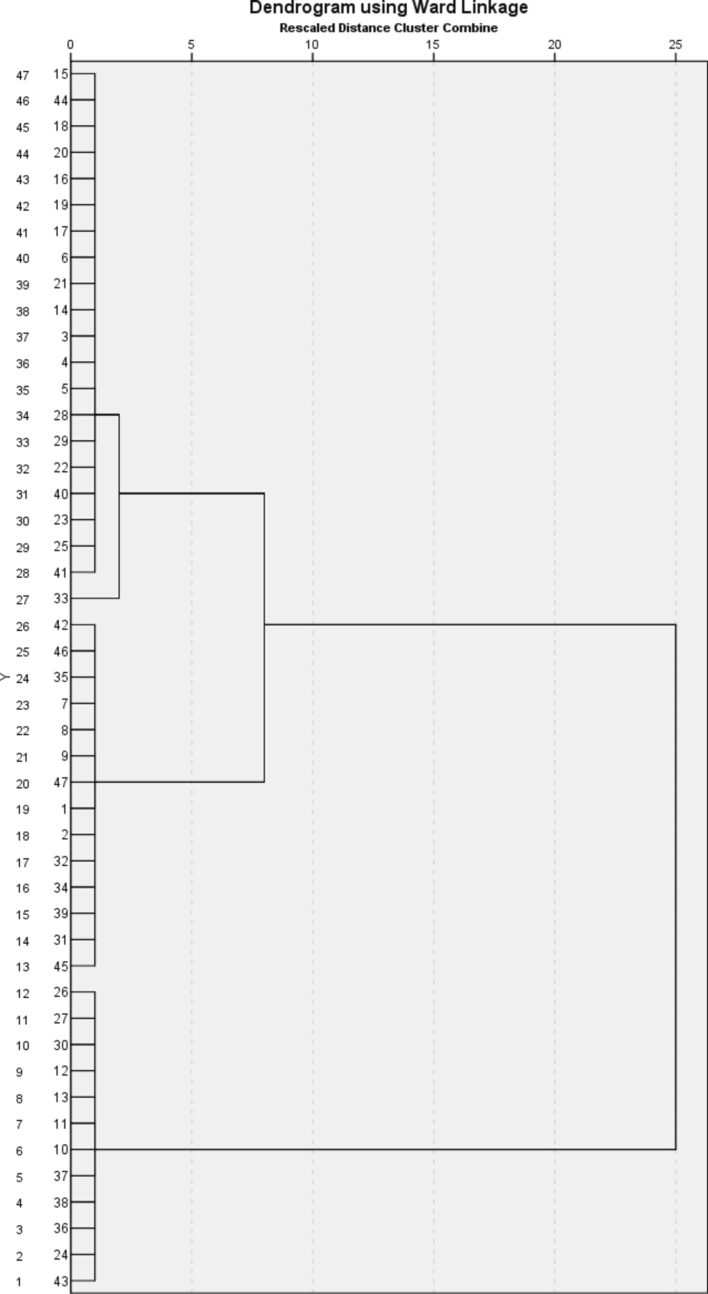
Dendrogram of patient clusters using the Ward linkage method, based on antibiotic susceptibility, age, year, and gender.
Cluster 1 consists of individuals who exhibit comparable antibiotic susceptibility profiles, age ranges, the same year of treatment, and the same gender. This cluster indicates a distinct demographic or clinical segment, such as younger patients or those who had treatment in a given year. Cluster 2 comprises patients who share comparable traits, which are different from those in cluster 1. It could also indicate a different demographic or clinical category, such as elderly patients or those with distinct antibiotic resistance trends. The basis of the difference between the two groups is antibiotic resistance. Healthcare practitioners can develop focused interventions and treatment programs customized to the unique requirements of each cluster by comprehending their distinctive qualities. Clustering can assist in the identification of patterns of antibiotic resistance, hence facilitating the establishment of more efficient antibiotic stewardship programs. Identifying unique patient clusters can aid in optimizing resource distribution, guaranteeing that high-risk populations receive suitable attention and care. The second group stands out due to its inclusion of bacterial strains isolated from age groups younger than eight years, and it also boasts the advantage of not having any resistance to Cloxacillin, Rifampicin, and Tetracycline. In the first group, on the other hand, there were bacterial strains that were found in people older than eight years old. Of these isolates, 38.9 % were not susceptible to Cloxacillin, 5.6 % to Rifampicin, and 30.6 % to Tetracycline.
3.5 CBC Estimation
The CBC was conducted using a fully automated blood cell counter hematology analyzer, specifically the Bulkman Coulter® Unicel DxH 800. We conducted a comparison of all the hematological values with both the control values and the reference values. Table 4 presents a comparison of the average values of all the measured hematological (CBC) parameters between patients with SCD who tested positive for blood cultures with S. aureus and the control groups. There was a statistically significant rise (p < 0.000) in the levels of HGB, RDW, WBCs, and HCT. No statistically significant differences were seen in red blood cell count (RBC), mean corpuscular volume (MCV), and platelet levels between the patient groups and the control group. p-value*: correlation between case and control in CBC parameters. A p-value of less than 5% indicates statistical significance.
Variables
Gender
Patients
Number
Patients
(Mean ± S.D)
Control
Number
Control
(Mean ± S.D)
t-test
(p-Value)*
Normal
range
Hgb
Male
30
7.7 ± 1.8
11
9.9 ± 2.3
0.002
13.2–16.6 g/dL
Female
17
9.2 ± 1.4
5
9.9 ± 1.5
11.6–15 g/dL
Hct
Male
30
22.8 ± 5.3
11
29.6 ± 7.1
0.004
38.3 %-48.6 %
Female
17
27.3 ± 4.2
5
29.6 ± 7.1
35.5 %-44.9 %
WBC
Male
30
17797 ± 9511
11
13736 ± 8685
0.079
3,400 to 9,600 cells/mcL
Female
17
15906 ± 9065
5
9740 ± 4666
RBC
Male
30
3.6 ± 4.7
11
3.5 ± 4.7
0.649
4.35–5.65 million cells/mcL
Female
17
5.0 ± 6.7
5
3.5 ± 6.7
MCV
Male
30
85.0 ± 9.5
11
85.0 ± 7.3
0.789
80–94 fl.
Female
17
82.0 ± 10.8
5
83.0 ± 6.6
80–94 fl.
RDW
Male
30
21 ± 5.3
11
15.4 ± 3.2
0.005
11.4 % to 13.5 %
Female
17
18 ± 2.0
5
16.9 ± 7.5
11.4 % to 13.5 %
Platelet
Male
30
342 ± 188
11
346 ± 180
0.422
135–317 billion/L
Female
17
264 ± 109
5
376 ± 271
157–371 billion/L
3.6 Determination of mecA antibiotic resistance gene of S. Aureus strains
The results of the PCR assay indicated that the oxacillin resistance gene (mecA) was present in all S. aureus isolates of SCD patients, with the expected amplicon size (314 bp). The findings indicated notable disparities (p < 0.05) between the case and control cohorts concerning mecA Table 5. * Correlation between S. aureus mecA antibiotic resistance gene in case and control groups. A p-value of less than 5% indicates statistical significance.
Gene
Group
Positive N, Positive%
Negative N, Negative%
p Values *
mecA
Case
47 (100)
0 (0)
0.00
Control
9 (56.3)
7 (43.7)
3.6.1 Comparison of the mecA gene-based PCR and vitek method
We examined the disparities between the oxidase resistance identified by the Vitek approach and the detection of the mecA gene using PCR. The Vitek method identified that 66 % of the samples showed resistance to oxacillin, however the mecA gene PCR test detected the presence of the mecA gene in all samples, indicating a 100 % detection rate. The mecA gene polymerase chain reaction (PCR) can be employed as a valuable tool for verifying these findings. Presence of the mecA gene in S. aureus isolates and their resistance to oxacillin exhibit significant disparities compared to isolates that are not resistant to oxacillin (Table 6). This study demonstrates that 34 % of the isolates examined exhibited varying degrees of resistance to oxacillin as determined by both phenotypic testing and the presence of the mecA gene as determined by genotypic testing. (OR1): The odds ratio of Oxacillin resistance with a positive mecA gene. (OR2): The odds ratio of Oxacillin resistance with a positive mecA gene between the case and control groups.
group
Gene
Result
OXA
S
R
Total
OR1
OR2
Case
mecA
Positive
16
31
47
0.464
1.305556
mecA
Negative
0
0
0
Control
mecA
Positive
5
5
10
0.355
mecA
Negative
7
0
7
4 Discussion
Bacterial infections significantly contribute to the illness and death of individuals with sickle cell anemia (Williams et al., 2009). S. aureus is the main cause of bloodstream infections in most developed nations (Yu et al., 2012). Scientists acknowledge S. aureus as a notable pathogen that causes bloodstream infections, which have a high likelihood of causing illness and death due to an infectious agent (Cheung et al., 2021). We recognize the increased vulnerability of persons with sickle cell disease (SCD) to infections. The occurrence of S. aureus has escalated in Saudi Arabia. The present study determined the prevalence of S. aureus and identified the clinical and laboratory indicators that were most strongly associated with a higher occurrence of S. aureus in the blood samples of sickle cell disease (SCD) patients who visited King Saud University Medical City (KSUMC). We performed susceptibility testing using a total of 14 antibiotics. We employed polymerase chain reaction (PCR) to identify the existence of MecA antibiotic resistance. During the 5-year study period, we observed a prevalence of 5.7 % of bacteremia among patients with sickle cell disease at a hospital in Riyadh, Saudi Arabia. Nevertheless, our findings diverged slightly from the reported results in King Abdulaziz Hos-pital (KAH), Al Ahsa, Saudi Arabia, Johns Hopkins Aramco Healthcare (JHAH) (Saudi Arabia), Ibadan University College Hospital (Nigeria), and Cameroon. These studies indicated that the incidence of blood-stream infections (BSI) in patients with sickle cell disease (SCD) was below 10 %, 8.3 %, 13.8 %, and 9.7 %, respectively (Brown et al., 2017, Alsaif et al., 2021, Al-Tawfiq et al., 2021, Alima Yanda et al., 2017).
The predominant bacteria responsible for bacteremia in this study were Gram-Positive Microorganisms, including MRSA (21.7 %), S. aureus (12.3 %), S. epidermidis (12.3 %), and S. homins (10.1 %). In contrast to a study conducted in Ghana (Appiah et al., 2020), the predominant bacterial species in SCD patients were S. aureus (33.3 %), MRSA (3.3 %), and coagulase-negative staphylococci (7.5 %). The variability of these results can be attributed to variations in the research populations and geographical locations. The current study demonstrated a prevalence of S. aureus bloodstream infection in 2 % of patients with sickle cell disease (SCD). On the other hand, a prior study conducted by Indira Gandhi, Shinde, and their colleagues in 2015 found that 11.1 % of patients with sickle cell disease had an infection caused by S. aureus (Shinde et al., 2015), Differences in geographical areas and urban centers, along with the high occurrence of sickle cell disease, may explain the discrepancy in outcomes. In 2012, Saudi Arabia saw a prevalence rate of 39.5 % for MRSA infection, which is typically linked to infections of wounds, skin, and soft tissues (El Amin and Faidah, 2012). The study found that 34 % of SCD patients had MRSA and S. aureus infections. The present investigation documented a prevalence of 1.4 % for S. pneumoniae, which deviates slightly from the findings of previous studies (Makani et al., 2015, Shinde et al., 2015). The difference in these results may be due to variations in the study populations, geographical areas, and vaccination. The study found that 34 % of SCD patients had MRSA and S. aureus infections. The present study documented a prevalence of 1.4 % for S. pneumoniae, which deviates slightly from the findings of previous studies (Makani et al., 2015, Shinde et al., 2015).
The Vitek test revealed that many strains of S. aureus exhibited resistance to cefazolin, penicillin, ampicillin, amoxicillin, imipenem, oxacillin/methicillin, erythromycin, tetracycline, ciprofloxacin, and moxifloxacin. Nevertheless, they exhibited a higher level of sensitivity towards the remaining medications. Our results are consistent with other studies conducted in Kuwait, which also found a significant frequency of antibiotic resistance (Udo et al., 2009). The antibiotic susceptibility pattern identified in our investigation aligns with the findings reported in earlier studies (Appiah et al., 2020). The current study classifies S. aureus isolates among SCD patients into two categories based on antibiotic susceptibility tests, age, and year. The isolates were obtained from the initial cohort, including solely of male patients, between 2017 and 2018. The second group consisted of both male and female people, with a greater proportion of men.
In this study found that the S. aureus strains exhibited susceptibility to both gentamicin and vancomycin. These options are the most optimal for therapy and for investigating their synergistic effects. The results we obtained differed slightly from the findings reported in Nigeria, where 71.7 % of S. aureus strains were found to be susceptible to gentamicin (Olufunmiso et al., 2017). The study found that individuals with Sickle Cell Disease (SCD) who tested positive for S. aureus in their blood culture had lower levels of hemoglobin (7.7 g/dL), higher values of red blood cell distribution width (RDW), and slightly higher white blood cell (WBC) counts compared to individuals in the control group. These differences were statistically significant (p < 0.002, p < 0.004, and p < 0.005). This conclusion is supported by research conducted in Tanzania (Makani et al., 2015). The mecA gene is responsible for the production of a protein called PBP2a. The bacteria become resistant to methicillin and other medications in the β-lactam group. Bacteria containing the mecA gene are able to survive in the presence of β-lactam antibiotics because PBP2a has a low affinity for them. Treating infections caused by bacteria carrying the mecA gene can be challenging due to the resistance conferred by PBP2a to other β-lactam medicines (Grazul et al., 2023). It is hereditary. This genetic segment contains the methicillin resistance (mecA) gene, facilitating its trans-mission to other strains of S. aureus (McClure et al., 2006).
Approximately 100 % of the mecA-positive isolates in patients with sickle cell disease (SCD) were found to have the mecA gene, which is in contrast to a study conducted in Ghana by (Dayie et al., 2021). This study revealed that 20 % of children with SCD tested positive for the mecA gene and were diagnosed with MRSA infections. The variation in these findings could be attributed to disparities in the study cohorts, sample types, and geographic regions. The current study recognizes the PCR assay as a method for detecting the mecA gene. The current study recognizes the PCR assay for detecting the mecA gene as a more dependable technique for assessing oxacillin resistance, in contrast to the Vitek and MicroScan methods. Both Vitek and MicroScan exhibited fewer significant mistakes than anticipated. Our findings indicate that both the MicroScan and Vitek systems had poor performance in detecting oxacillin resistance in S. aureus. The findings were consistent with the results obtained by Skuluick and his colleagues (Skulnick et al., 1992). Dy makes a substantial contribution to the current body of literature. Nevertheless, there Nevertheless, it is crucial to note that we did not conduct tests on certain antibiotic-resistant genes that hold significance in other research on the SCA condition. One challenge we encounter is the limited sample size for our investigation.
Heteroresistance in S. aureus refers to the phenomenon where subpopulations within a bacterial culture exhibit varying levels of resistance to antibiotics, even though they are genetically identical or similar. This can complicate treatment, as standard susceptibility testing might not detect these resistant subpopulations, leading to treatment failures. S. aureus strains resistant to oxacillin are often referred to as methicillin-resistant S. aureus (MRSA) (Heidarian et al., 2024, Ryffel et al., 1994). Oxacillin resistance is primarily mediated by the presence of the mecA gene, which encodes for an altered penicillin-binding protein (PBP2a) that has a low affinity for β-lactam antibiotics, including methicillin and oxacillin. In heteroresistant populations, a small number of cells within a predominantly susceptible population can survive antibiotic treatment due to their higher resistance levels. Heteroresistance can be due to genetic mutations, gene amplification, or the presence of plasmids that confer resistance. Standard susceptibility tests (e.g., disk diffusion or broth microdilution) may fail to detect heteroresistant subpopulations, as the majority of the population might appear susceptible. Infections caused by heteroresistant S. aureus may lead to treatment failures if the resistant subpopulation is not detected and appropriately managed. Infections caused by heteroresistant S. aureus may lead to treatment failures if the resistant subpopulation is not detected and appropriately managed (Schimidt and Póvoa, 2023, Finan et al., 2002). This work reported that 34 % of the S. aureus strains isolated from sickle cell disease patients at KSUMC, Riyadh, Saudi Arabia were heteroresistance.
5 Conclusions
Conclusively, to our current understanding, this study is the first in Saudi Arabia to investigate the occurrence of S. aureus among patients with SCA. Our research revealed that 2 % of patients with Sickle Cell Disease (SCD) were discovered to have the presence of S. aureus and MRSA. Furthermore, the incidence of infection was higher in males compared to females. The study has provided insight into the seldom presence of Streptococcus pneumoniae among individuals with sickle cell disease in Saudi Arabia. This finding contributes to the ongoing acknowledgment of the significance of delivering pneumococcal vaccines to children with SCD in order to combat these germs. Patients with S. aureus bloodstream infection in the present study exhibit decreased levels of hemoglobin (Hgb) and hematocrit (Hct), as well as increased red cell distribution width (RDW) and white blood cell (WBC) counts. On the other hand, the PCR assay is widely recognized as a more dependable technique for detecting oxacillin resistance through the mecA gene, in comparison to the Vitek approach. To improve the findings of this study, we can analyze the sequencing of the mecA gene, which is responsible for resistance, to evaluate genetic variation and molecular typing for clonality. These factors play a crucial role in determining and affecting the resistance profile of isolates worldwide. Furthermore, a more in-depth analysis is necessary to comprehend the correlation between hematological parameters and S. aureus infections in persons with SCD in Saudi Arabia. Furthermore, further research is required to investigate potential underlying mechanisms that contribute to the increased vulnerability of individuals with sickle cell disease (SCD) to bacteremia. Heteroresistance in S. aureus involving oxacillin resistance and mecA gene identification is a significant clinical problem. Efficient identification and control of heteroresistant bacteria are crucial for guaranteeing favorable treatment results. PCR continues to be a good technique for identifying the mecA gene, but additional approaches are frequently required to comprehensively comprehend and manage heteroresistance in clinical environments.
6 Future perspective
In the future, we recommend conducting a comprehensive study to compare the chemical and biochemical properties of the microbe isolated from patients with SCD and comparing them with those of other common diseases.
CRediT authorship contribution statement
Adel A. Abdulmanea: Writing – review & editing, Validation, Methodology, Investigation, Formal analysis, Data curation. Naiyf S. Alharbi: Writing – review & editing, Validation, Methodology. Ali M. Somily: Methodology, Formal analysis. Osamah T. Khojah: Investigation. Mohamed A. Farrag: Validation, Formal analysis. Ahmed S. Alobaidi: Data curation. Jamal M. Khaled: Methodology, Investigation.
Funding
Researchers Supporting Project Number (RSPD2024R679), King Saud University, Riyadh, Saudi Arabia.
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project number (RSPD2024R679), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alghizzi, M. J., Alansari, M. & Shami, A. 2021. The Prevalence of Staphylococcus aureus and Methicillin Resistant Staphylococcus aureus in Processed Food Samples in Riyadh, Saudi Arabia. Journal of Pure & Applied Microbiology, 15.
- Burden and spectrum of bacterial infections among sickle cell disease children living in Cameroon. BMC Infect. Dis.. 2017;17:1-7.
- [Google Scholar]
- Prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) in Saudi Arabia: A Systematic Review. Journal of Pure & Applied. Microbiology. 2020;14
- [Google Scholar]
- Sickle cell disease in Saudi Arabia: A challenge or not. Journal of Epidemiology and Global Health. 2017;7:99.
- [Google Scholar]
- Prevalence of serious bacterial infections in children with sickle cell disease at King Abdulaziz Hospital, Al Ahsa. Mediterranean Journal of Hematology and Infectious Diseases. 2021;13
- [Google Scholar]
- Sickle cell disease subphenotypes in patients from Southwestern Province of Saudi Arabia. J. Pediatr. Hematol. Oncol.. 2012;34:79-84.
- [Google Scholar]
- al-Tawfiq, J. A., Rabaan, A. A. & Aledreesi, M. H. 2021. Frequency of bacteremia in patients with sickle cell disease: a longitudinal study. Annals of Hematology, 100, 1411-1416.
- Staphylococcus aureus nasal colonization among children with sickle cell disease at the Children’s Hospital, Accra: Prevalence, risk factors, and antibiotic resistance. Pathogens. 2020;9:329.
- [Google Scholar]
- Antimicrobial susceptibility pattern of Methicillin-resistant Staphylococcus aureus in a tertiary care Hospital. Journal of University Medical & Dental College. 2022;13:468-472.
- [Google Scholar]
- Prevalence and etiology of bacteremia in febrile children with sickle cell disease at a Nigeria tertiary hospital. In: Mediterranean Journal of Hematology and Infectious Diseases. 2017. p. :9.
- [Google Scholar]
- Exploring the Role of Staphylococcus aureus in Inflammatory Diseases. Toxins. 2022;14:464.
- [Google Scholar]
- Nasopharyngeal carriage of methicillin-resistant Staphylococcus aureus (MRSA) among sickle cell disease (SCD) children in the pneumococcal conjugate vaccine era. Infectious Disease Reports. 2021;13:191-204.
- [Google Scholar]
- Invasive disease and paediatric carriage of Streptococcus pneumoniae in Ghana. Scand. J. Infect. Dis.. 2010;42:254-259.
- [Google Scholar]
- Antibiotic resistance genes & susceptibility patterns in staphylococci. Indian J. Med. Res.. 2012;135:389-396.
- [Google Scholar]
- Methicillin-resistant Staphylococcus aureus in the western region of Saudi Arabia: prevalence and antibiotic susceptibility pattern. Ann. Saudi Med.. 2012;32:513-516.
- [Google Scholar]
- Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob. Agents Chemother.. 2002;46:24-30.
- [Google Scholar]
- Newborn screening for sickle cell disease and other hemoglobinopathies: a short review on classical laboratory Methods—Isoelectric focusing, HPLC, and capillary electrophoresis. International Journal of Neonatal Screening. 2018;4:39.
- [Google Scholar]
- Garoy, E. Y., Gebreab, Y. B., Achila, O. O., Tekeste, D. G., Kesete, R., Ghirmay, R., Kiflay, R. & Tesfu, T. 2019. Methicillin-resistant Staphylococcus aureus (MRSA): prevalence and antimicrobial sensitivity pattern among patients—a multicenter study in Asmara, Eritrea. Canadian Journal of Infectious Diseases and Medical Microbiology, 2019.
- Analysis of the presence of the virulence and regulation genes from Staphylococcus aureus (S. aureus) in coagulase negative staphylococci and the influence of the staphylococcal cross-talk on their functions. Int. J. Environ. Res. Public Health. 2023;20:5155.
- [Google Scholar]
- High prevalence of heteroresistance in Staphylococcus aureus is caused by a multitude of mutations in core genes. PLoS Biology. 2024;22
- [Google Scholar]
- Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: a cross-sectional study. Ann. Clin. Microbiol. Antimicrob.. 2016;15:1-7.
- [Google Scholar]
- Bacteraemia in sickle cell anaemia is associated with low haemoglobin: a report of 890 admissions to a tertiary hospital in Tanzania. British Journal of Haematology. 2015;171:273-276.
- [Google Scholar]
- Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant staphylococci. J. Clin. Microbiol.. 2006;44:1141-1144.
- [Google Scholar]
- Molecular mechanisms of drug resistance in Staphylococcus aureus. Int. J. Mol. Sci.. 2022;23:8088.
- [Google Scholar]
- Global epidemiology of haemoglobin disorders and derived service indicators. Bull. World Health Organ.. 2008;86:480-487.
- [Google Scholar]
- Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol.. 2012;12:1-9.
- [Google Scholar]
- Multidrug and vancomycin resistance among clinical isolates of Staphylococcus aureus from different teaching hospitals in Nigeria. Afr. Health Sci.. 2017;17:797-807.
- [Google Scholar]
- Identification of pathogenic bacteria in blood cultures: comparison between conventional and PCR methods. J. Microbiol. Methods. 2009;78:325-330.
- [Google Scholar]
- An update on clinical burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infectious Diseases: Research and Treatment. 2017;10
- [Google Scholar]
- Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother.. 1994;38:724-728.
- [Google Scholar]
- Schimidt, D. B. & Póvoa, H. C. C. 2023. Exposure to oxacillin subinhibitory concentrations and in vitro induction of resistance expression in heteroresistant and non-heteroresistant oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA). bioRxiv, 2023.01. 24.521449.
- Evaluation of commercial and standard methodology for determination of oxacillin susceptibility in Staphylococcus aureus. J. Clin. Microbiol.. 1992;30:1985-1988.
- [Google Scholar]
- The prevalence of antimicrobial resistance and carriage of virulence genes in Staphylococcus aureus isolated from food handlers in Kuwait City restaurants. BMC. Res. Notes. 2009;2:1-6.
- [Google Scholar]
- Verification and quality control of routine hematology analyzers. Int. J. Lab. Hematol.. 2016;38:100-109.
- [Google Scholar]
- Bacteraemia in Kenyan children with sickle-cell anaemia: a retrospective cohort and case–control study. Lancet. 2009;374:1364-1370.
- [Google Scholar]
- Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn. Microbiol. Infect. Dis.. 2012;74:363-368.
- [Google Scholar]







