Translate this page into:
Pharmacological investigation of ribosome inactivating protein (RIP) – like protein extracted from Annona squamosa L. seeds
⁎Corresponding author at: Department of Biotechnology, School of Agriculture and Biosciences, Karunya Institute of Technology & Sciences, Coimbatore 641 114, India. jannet_r@karunya.edu (J. Jannet Vennila)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
Ribosome inactivating proteins (RIPs), are plant proteins with N Glycosidase activity. Annona squamosa is traditionally used in medicine but RIP presence is less explored. The present study aimed to isolate and structurally analyse RIP- like protein from the seeds of Annona squamosa (ARIP) and investigate its in vitro pharmacological bioactivities.
ARIP was isolated with step wise purification procedures – Cation exchange and Gel filtration column chromatography, Ultrafiltration and SDS PAGE were adopted to purify and separate ARIP. The protein bands were eluted and tested for the presence of ARIP by N Glycosidase activity. The trypsin digested peptides were characterized by LC-MS/ESI-MS and searched for match using MASCOT search engine. Pairwise sequence alignment of peptide sequences were attempted to predict the similarity with known RIP in Magnoliophyta. In vitro antimicrobial, antimutagenic and cytotoxic activity of the the purified fraction of the ARIP was also studied.
Two proteins with band sizes 21 kDa and 28 KDa confirmed to have ARIP like activity. The LC-MS/ESI-MS mass spectra between 9 and 17 min yielded 19 peaks for 21 kDa protein and 18 peaks for 18 peaks for 28 kDa protein. Although database search revealed very low similarity with known RIPs, the closest similarity observed was with Populus trichocarpa which is known to have Type 2 Ricin B chain RIP. On sequence alignment with RIPs, the peptides revealed similarity with RIPs of type II category. Phamacological investigation also suggested strong in vitro antimicrobial, antimutagenic and cytotoxic activity, proving to be a promising candidate as drugs in future.
Our study have observed that ARIP was novel protein with unique sequence but lectin binding properties. They have shown remarkable antimicrobial, antimutagenic and cytotoxic activity. Crystallization of the protein followed by X-ray characterization could help to study the structure of the protein, active sites which would in turn give clear focus on the usage of this RIP in treatment of various diseases.
Keywords
Ribosome-inactivating protein (RIP)
Annona squamosa
Custard apple
LC-MS/ESI-MS
Type II RIP
Antimicrobial
Antimutagenic activity and cytotoxic activity
- RIP
Ribosome inactivating protein
- ARIP
RIP from the seeds of Annona squamosa
- SDS PAGE
Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis
- kDa
Kilo Daltons
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
Abbreviations
1 Introduction
Ribosome inactivating proteins (RIPs) are self defensive proteins, widely present in higher plants (Shu et al., 2009), and have been reported for various bioactive properties. RIPs specifically cleave the N glycosidic bond of the Adenine nucleotide (4324) in the 28S rRNA loop sequence of 40S ribosomes, thus inhbiting the protein synthesis. (Peumans et al., 2001). Three types of RIPs have been reported – Type 1 with single polypeptide chain (Park et al., 2006), Type 2 RIPs with A (RNA N Glycosidase) and B polypeptide chain (galactose specific lectin) (Sharma et al., 2004) and Type 3 RIPs with unknown function (Shu et al., 2009). The applications of RIP have been envisaged in different dimensions(Stirpe and Gilabert-Oriol, 2017). They can be conjugated with targeting ligands and used as immunotoxins to kill the cancer cells (Pizzo and Di Maro, 2016).
Annona squamosa commonly known as Custard apple, is a medicinal plant of Annonaceae family (Kulkarni and Chandrasekar, 2011), which is scientifically reported to have antidiabetic (Shirwaikar et al., 2004), antimicrobial (Mukhlesur Rahman et al., 2005), antioxidant (Kaleem et al., 2006), insecticidal (Jaswanth et al., 2002), cytotoxic (Pardhasaradhi et al., 2005), anti HIV (Wu et al., 1996) and abortifacient activity (Damasceno et al., 2002). The medicinal activities of Annona squamosa seems to resemble with RIP. Till date, there are’nt any studies on RIP in the family of Annonaceae. Henceforth, the present study aimed to isolate and characterize RIP like protein from the seeds of Annona squamosa (ARIP) and investigate its bioactivities.
2 Materials and methods
2.1 Isolation and purification of ARIP
200 g of fresh Annona squamosa seeds were collected, authenticated (Botanical Survey of India (BSI), Coimbatore, Number – BSI/SRC/5/23/2012-13/Tech/69) and ground in liquid nitrogen for maximum cell disruption. The proteins were precipitated with 20–80% Ammonium sulphate, dialyzed and fractionated using CM cellulose cation exchange column chromatography (Amersham Biosciences). NaCl gradient varying from 50 mM to 500 mM was used for elution and 150 mM NaCl gradient was optimized to get maximum protein elution. Sephadex G 50 gel filtration column chromatography (Amersham Biosciences) with 100 mM NaCl was used further for protein elution. Each fractions were assayed for N Glycosidase activity and the active fraction was desalted using 5 kDa cutoff ultrafiltration membranes (Sharma et al., 2004; Park et al., 2006). The protein quality check was done at each step of purification. The proteins were separated by SDS PAGE (12% acrylamide discontinuous gels) and detected by staining with Coomassie brilliant blue r250 (Sigma Aldrich, Bangalore, India) (Laemmli, 1970). Protein ladder markers were used as standards for comparison.
2.2 Assay for N Glycosidase activity
Yeast cells were procured from MTCC (MTCC 170), Chandigarh, India and grown in yeast peptone dextrose medium. The yeast ribosomes were isolated as per the protocol specified by Park et al., 2006. The assay for N glycosidase activity/depurination assay was done as per Tumer et al (1997). In brief, ribosomes were incubated with ARIPs followed by rRNA extraction. Half of the extracted rRNA was incubated with 1 M Aniline acetate. 7 M/6% urea polyacrylamide gel electrophoresis was used to separate both the Aniline treated and untreated rRNAs, followed by their detection using Ethidium bromide staining. rRNA incubated in the absence of ARIPs served as a negative control (Park et al., 2006). Ricin was used as positive control.
2.3 Structural analysis
2.3.1 LC-MS/ESI-MS analysis
The protein bands were cut separately from the gels, dissolved, decolourized and subjected to trypsin digestion (Shu et al., 2009). The trypsin digested peptides were identified using LC-MS/ESI-MS (Orbitrap XL, Thermo Scientific, Breman). MASCOT search engine (Matrix Science) was used for peptide match and identification.
2.3.2 Sequence alignment
The peptides obtained during LC MS/ESI MS fragmentation were matched with already known RIPs from the protein database as described by Raj and Vennila (2013). Pairwise alignment was carried out with the Protein Information Resource (PIR) alignment tool. The identity and similarity between the peptides and known RIP was tabulated for comparison.
2.4 Pharmacological investigation
2.4.1 Antimicrobial activity
The bacterial and fungal strains were procured from Microbial Type Culture Collection (MTCC), India and grown in suitable medium for maintainence.
Antimicrobial activity was checked with nosocomial infectious organisms – Gram negative bacteria (Escherichia coli (MTCC 443), Klebsiella pneumonia (MTCC 530) and Pseudomonas aeruginosa (MTCC 799)), Gram positive bacteria (Staphylococcus aureus (MTCC 121) and Bacillus subtilis (MTCC 441)) and fungi (Candida albicans (MTCC 227) and Fusarium oxysporum (MTCC 284)). Agar well diffusion method was adopted (Thomas and Veda, 2007) for evaluation of antimicrobial activity of ARIP. The plates with the media were seeded with overnight grown culture containing 106–107 cells. The purified fractions of ARIP at a concentration of 10, 20 and 30 µg were laid on the wells along with the control. The standard drugs Gentamycin (2 µg) and Cyclohexamide (2 µg) were used as controls for bacteria for fungi respectively. The zone of inhibition (mm) was measured after an overnight incubation.
2.4.2 Antimutagenic activity
The antimutagenic activity was performed by an in vitro Ames assay using Salmonella typhimurium mutant strains (MTCC 1252) as per the protocol specified by Ames et al.(1975). The contents (0.5 ml of ARIP in varying concentration of 0.5 to 2 mg/plate, 0.1 ml of bacterial culture, 0.2 ml of 0.5 mM histidine biotin solution and 0.1 ml of standard mutagen Sodium azide of concentration 1.5 μg/ml) were transferred to 2 ml of molten top agar in a test tube, and mixed thoroughly. This semisolid agar was immediately spread on the layer of minimal glucose agar plates in the petri dish. The dishes were incubated at 37 °C for 48 h and the number of revertants were counted (Issazadeh and Morteza, 2012; Sharififar et al., 2016). A plate containing agar, bacterial strain and the mutagen was considered as positive control. The solvent Dimethyl sulfoxide (DMSO) with agar and bacterial strain was used as a negative control. To detect the indirect mutagenic effect caused by metabolites of the test, liver homogenate of the rat along with the co-factors (Co-factor I- MgCl2 and KCl and Co-factor II Na2HP04) (known as S9 mix) was used. This experiment was done in triplicates in presence and absence of S9 mix. The antimutagenic potential is calculated based on the percent of inhibition as follows:where A and B are the number of revertants in each plate, in the presence of positive control and mutagen or/and sample.
2.4.3 Cytotoxic activity
The in vitro cytotoxic activity was assessed using MTT assay (Mosmann, 1983). AGS (Gastric cancer cell lines) was procured from National Centre for cell Science, Pune, India and seeded into 96 well microtitre plates (1.0 × 104 cells /well) along with 100 µl of RPMI-1640 medium followed by 24 h incubation at 37 °C with 5% CO2. Media was replaced with various concentrations of ARIP and incubated the cells for 48 h. After the measurement of viable cell count, 10 µl MTT solution (5 mg/ml in PBS) was added to the well followed by incubation for 4 h. The formazan crystals were dissolved in 100 µl of DMSO, incubated for 5 min in dark and OD was measured at 570 nm in a microplate ELISA reader. A dose–response curve of sample concentration (μg/ml) versus cell viability (%) was plotted. The IC50 values (concentration that is lethal to 50% of the cells) were calculated (Uthaya Kumar et al., 2018). Ricin and 5 Fluorouracil (Chemotherapeutic drug for Gastric cancer) were used as standards for comparison.
2.4.4 Statistical analysis
SPSS software (IBM Corporation) was used for statistical analysis of the data. The differences with P < 0.05 was considered significant.
3 Results and discussion
3.1 Purification of RIP from Annona squamosa seeds (ARIP)
The maximum yield of protein after purification was observed to be 0.23 mg/ml. Among the various fractions eluted from Gel filtration column, three fractions (E20–E22) were found to have rRNA N Glycosidase activity (Fig. 1). These protein fractions were separated by SDS-PAGE and five (5) bands with molecular weight corresponding to 6, 8, 21, 28 and 63 kDa were observed. As the molecular weight of single chain RIPs is reported to range between 30 kDa (Park et al., 2006), we eluted the protein bands corresponding to 21 and 28 kDa, namely ARIP-A and ARIP-B respectively.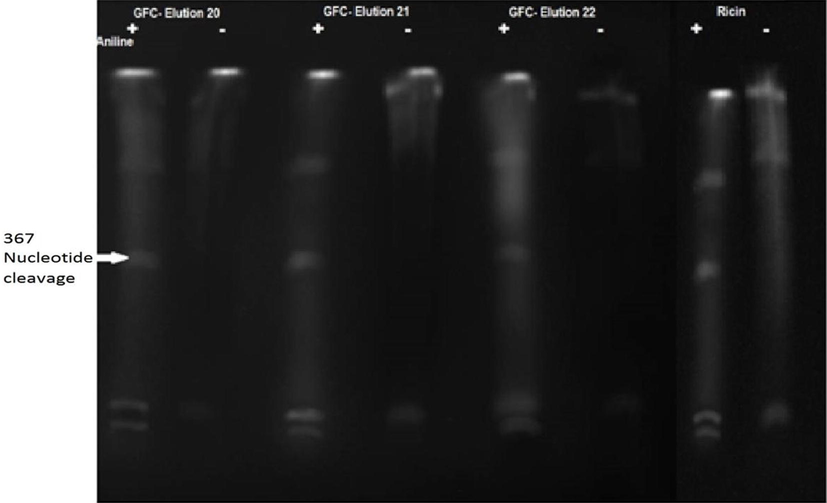
N Glycosidase activity of ARIP. ARIP - RIP from the seeds of Annona squamosa. Yeast ribosmes were isolated and incubated with ARIP fractions (Gel filtration column elution 20, 21 and 22) and positive control Ricin. Ribosomal rRNAs were extracted and treated with Aniline acetate, followed by separation on a 7M/6% Urea polyacrylamide gel, and detection with Ethidium bromide staining. Aniline treated and non treated is denoted as (+) and (−) respectively. The presence of the 367 nucleotide rRNA cleavage product is the characteristic feature of N Glycosylase activity which is is shown by an arrow.
3.2 Structural analysis
3.2.1 LC-MS/ESI-MS
The LC-MS/ESI-MS mass spectra between 9 and 17 min yielded 19 peaks for ARIP-A and 18 peaks for ARIP-B (Figs. 2 and 3). The MS/MS fragmentation spectra were searched for matching with MASCOT search engine (Matrix Science) and the peptides were identified as shown in Supplementary files 1 and 2. It has been observed that the peptides of ARIP A and ARIP B purified from the seeds of Annona squamosa did not show remarkable match with known RIPs so far reported. The matching was in the range of 1–15% and 1–11% for ARIP-A and ARIP-B (Tables 1 and 2). In the total protein plant description, one uncharacterized protein from Populus trichocarpa showed maximum matches with ARIP-A (% coverage – 2 + 3 + 3 + 5 = 13%) (Table 1) and ARIP-B (% coverage – 2 + 1 + 1 + 3 + 3 = 10%) (Table 2). This plant is reported to have RIP with single ricin B-chain domain, which suggested the possibility of ARIPs could be of Type 2 or single ricin B-chain domain. Interstingly, 13 out of 19 tryptic peptides from ARIP-A and 15 out of 18 tryptic peptides from ARIP-B matched with uncharacterized proteins from various plants reported to have RIP activity. The genome sequence of Annona squamosa genome with the annotated information is not available until now. This could be ascertained as the reason for their low similarity.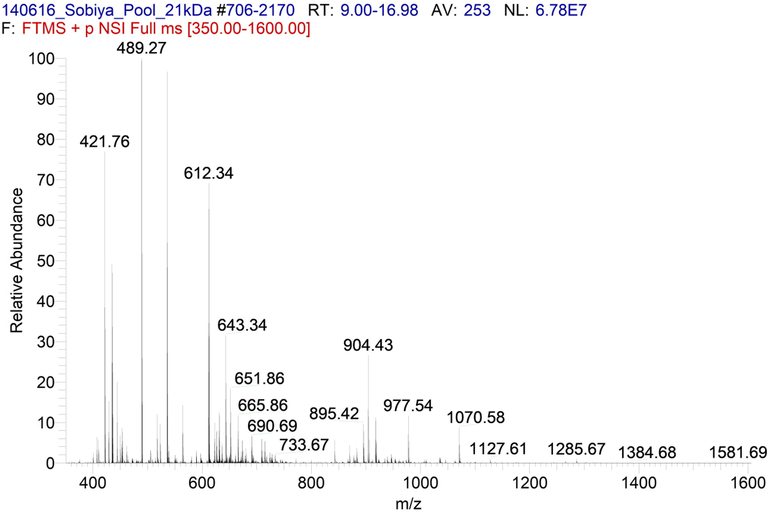
ESI MS/MS mass spectra of ARIP-A. 19 peaks were observed within the Retention Time of 9 to 17 min.
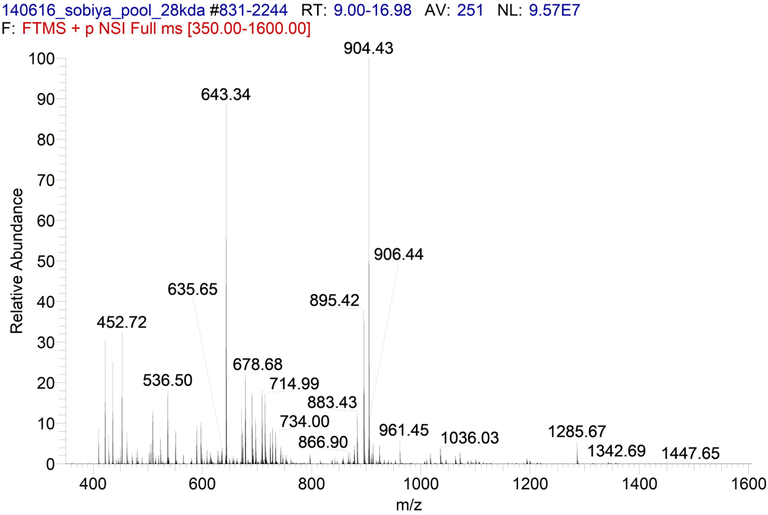
ESI MS/MS mass spectra of ARIP-B. 18 peaks were observed within the Retention Time of 9 to 17 min.
S. No
Plant description
Match Coverage (%)
Total Coverage (%)
RIP activity reported
RIP gene identified
Type of RIP
References
1
Populus trichocarpa
2 + 3 + 3 + 5
13
Yes
Yes
Type 2 (B domain only)
Peumans and Vandamme (2010)
2
Brachypodiumdistachyon
1
1
Yes
Yes
Type 3
Peumans and Vandamme (2010)
3
Morusnatabilis
2
2
No
No
–
–
4
Solanum lycopersicum
4
4
Yes
No
–
Barbieri et al. (2006)
5
Arabidopsis thaliana
2 + 2
4
Yes
No
–
Pena et al. (2008)
6
Arabidopsis lyrata sub sp.
2
2
Yes
No
–
–
7
Canavalia ensiformis
4
4
Yes
No
–
Carlini and Guimarães (1991)
8
Amborella trichopoda
15
15
No
No
–
–
9
Mimulus guttatus
2
2
No
No
–
–
10
Triticum urartu
5
5
Yes
No
–
–
11
Seteria italica
1
1
Yes
Yes
–
Uniprot
12
Fagopyrum esculentum
3
3
No
No
–
–
13
Lotus japonicus
5 + 5
10
No
No
–
–
14
Volvox carteri
1
1
No
No
Peumans and Vandamme (2010)
S. No
Plant description
Match Coverage (%)
Total Coverage (%)
RIP activity reported
RIP gene identified
Type of RIP
References
1
Populus trichocarpa
2 + 1 + 1 + 3 + 3
10
Yes
Yes
Type 2 (B domain only)
Peumans and Vandamme (2010)
2
Astragalus membranaceus
10
10
No
No
–
–
3
Polygala tenuifolia
1
1
No
No
–
–
4
Vitis vinifera
0 + 2
2
Yes
No
–
Peumans and Vandamme (2010)
5
Triticum aestivum
2
2
Yes
Yes
Type 1
Raj and Vennila (2013)
6
Glycine max
2 + 1
3
Yes
No
–
Peumans and Vandamme (2010)
7
Brachypodiumdistachyon
1
1
Yes
Yes
Type 3
Peumans and Vandamme (2010)
8
Hordeum vulgare
3
3
Yes
Yes
Type 1
Raj and Vennila (2013)
9
Ricinus communis
2
2
Yes
Yes
Type 2
–
10
Micromonaspusilla
1
1
Yes
No
–
Peumans and Vandamme (2010)
11
Musa acuminata subsp.
11
11
No
No
–
–
12
Arabidopsis lyrata subsp.
3
3
No
No
–
Peumans and Vandamme (2010)
3.2.2 Sequence alignment
As Annona squamosa is a flowering plant belonging to Magnoliophyta division, the FASTA sequences of RIP proteins from Magnoliophyta families, were collected from the protein database (PDB) as described by Raj and Vennila (2013), matched with LC-MS/ESI-MS data of ARIP-A and ARIP-B (Supplementary files 3 and 4). Nine (9) peptides from ARIP-A showed 100% similarity with a part of amino acid sequence present in RIPs reported from Fabaceae (Abrus precatorius), Viscaceae (Viscum album), Cucurbitaceae (Momordica balsamina, Momordica charantia and Cucurbita moschata), Nyctaginaceae (Bougainvilliea spectabilis) and Phytolaccaceae (Phytolacca americana, Phytolacca dioica and Phytolacca accinosa). However, in ARIP-B, only 7 out of 18 peptides showed 100% similarity with a part of amino acid sequence in RIPs reported in Cucurbitaceae (Momordica balsamina, Momordica charantia and Cucurbita moschata), Phytolaccaceae (Phytolacca americana, Phytolacca dioica and Phytolacca accinosa), Viscaceae (Viscum album), Fabaceae (Abrus precatorius) and Euphorbiaceae (Ricinus communis). The maximum number of peptides identity of ARIP-A was noticed with RIPs reported in Fabaceae and Viscaceae. While ARIP-B showed maximum identity with Phytolaccaceae, Cucurbitaceae and Viscaceae. Most of the RIPs found in these families are reported as Type 2. Hence, we presumed that the ARIPs are likely belong to Type 2. RIP. The presence of carbohydrates during purification steps also substantiates this fact that Type 2B-chain lectin domain have bound to carbohydrates. Although the genome sequence of the Annona squamosa is unavailable for comparison, the peptides of the ARIPs still showed little similarity with part of the amino acid sequence in already known RIPs. This emphasizes that in spite of their difference, all RIPs in Magnoliophyta illustrated a definite motif/pattern in their structure.
All these assumptions led to the conclusion that the ARIP-A and ARIP-B could be the two peptide chains A and B of ricin like Type II ARIP which would be have been denatured and separated as two bands during SDS PAGE.
3.3 Investigation of pharmacological activities
Based on the above assumptions, the ARIP–A and ARIP-B were pooled together and single ARIP protein and studied for their in vitro pharmacological activities
3.3.1 Antimicrobial activity
The antimicrobial activity against bacterial and fungal strains was found to increase with increase in the concentration of the ARIP (Fig. 4). The ARIP was more effective against bacteria when compared to fungal strains. The antibacterial and growth inhibitory activity of ARIP could be attributed to the presence of lectin (Al-Mamun et al., 2016). Similar antimicrobial activities was also noticed with RIPs reported in Mirabilis expansa (ME1 and ME2) against Pseudomonas aeruginosa (Vivanco et al., 1999). Ricin also have been reported to show antibacterial effect against S. aureus P. aeruginosa, E. coli and E. aerogenes (Al-Mamun et al., 2016). Tobacco ribosomal inactivating protein (TRIP) have also shown strong inhibitory activity against many bacteria (Sharma et al., 2004; Vivanco et al., 1999). Considering the antifungal activities of RIP, Park et al.(2002) have reported that ricin was less toxic to C. albicans ribosomes than saporin. Another study on the RIP protein BE27 from sugar beet by Citores et al (2016) have observed antifungal activity against Penicillium digitatum.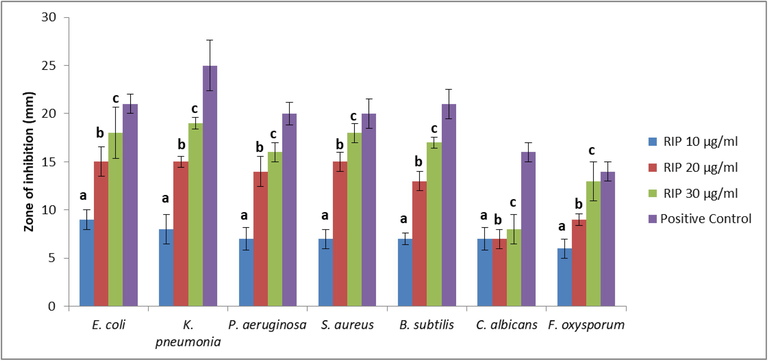
Antimicrobial activity of ARIP. ARIP - RIP from the seeds of Annona squamosa. All the experiments were done in triplicates and P < 0.05 was considered statistically significant. Positive controls (C) – Gentamycin and Cyclohexamide were used for bacteria and fungi respectively. a denotes comparision of control (C) with ARIP 10 µg/ml; b denotes comparision of control (C) with ARIP 20 µg/ml; c denotes comparision of control (C) with ARIP 30 µg/ml.
3.3.2 Antimutagenic activity
Antimutagenic activity helps in cancer prevention. Antimutagenic agents present in plants inhibit the pathogenic effects of mutagenic and carcinogenic substances (Makhafola et al., 2016). With increase in the concentration from 0.5 to 2.0 mg/plate, ARIP exhibited strong antimutagenic activity and inhibition range of of 38.43 to 69.64%. This was found to significantly increase when S9 mix was added (52.71–72.78%) (Table 3). The antimutagenic activity of ARIP was found to be stronger than Viscum album Agglutinin (Hong and Lyu, 2012) but weaker than Ricin (Abbas et al., 2018) and Sechuimin (Wu et al., 1998). All these findings, suggest that ARIP can serve as antimutagenic agents in mutagenesis associated diseases. Each value represents the Mean ± Standard deviation in triplicates. CFU – Colony forming unit, S9+ – with S9 mix (a liver homogenate of the rat along with the co-factors to detect the indirect mutagenic effect), S9− – without S9 mix. Based on the Inhbition %, the following consideration was adopted: No antimutagenic effect – Inhibition <25%. Moderate antimutagenic effect – Inhibition ranging from 25%−40%. Strong antimutagenic effect – Inhibition greater than 40%.
S. No
Concentration (mg/plate)
Salmonella typhi + S9−
Salmonella typhi + S9+
Revertants (CFU/Plate)
Inhibition (%)
Revertants (CFU/Plate)
Inhibition (%)
1
0.5
324.51 ± 12.51
38.43 ± 9.21
394 ± 18.23
52.71 ± 9.45
2
1.0
289.19 ± 21.34
52.91 ± 5.33
304 ± 22.34
61.53 ± 6.66
3
2.0
178.35 ± 13.64
61.81 ± 8.45
125 ± 17.59
69.89 ± 6.48
4
4.0
151.28 ± 14.43
69.64 ± 9.47
104 ± 21.54
72.87 ± 8.36
3.3.3 Cytotoxic activity
The percentage viability of the AGS Gastric cancer cells treated with different concentrations of ARIP was studied and the IC50 value was observed as 43.02 µg/ml (Fig. 5). The cytotoxic effectiveness was found to be in the order Ricin (IC50 value: 25.57 µg/ml) > ARIP (IC50 value: 43.02 µg/ml) >5 Fluorouracil (IC50 value: 55.34 µg/ml). RIPs have been reported for their potential cytotoxic effect on cancer cells – Riproximin (Ximenia americana) (Voss et al., 2006), Curcin (Jatropha curcas) (Zhang et al., 2017) and Mamorin (Hypsizigus marmoreus) (Pan et al., 2013).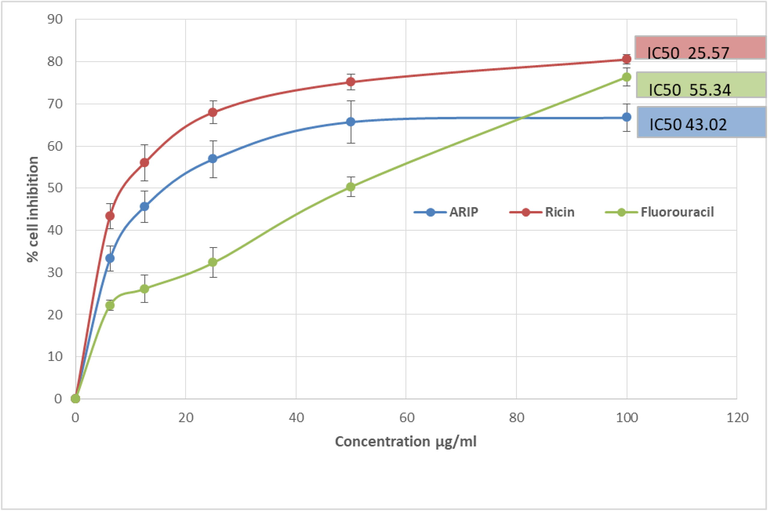
Cell growth inhibitory activity of ARIP against AGS Gastric cancer cell lines. ARIP - RIP from the seeds of Annona squamosal. Ricin and 5 Fluorouracil were used as controls for comparison.
The current study clearly demonstrates the cytotoxic effect of ARIP on cancer cells. These findings suggest that ARIP could be potentially considered as new and novel anticancer compounds for possible drug development against cancer.
4 Conclusion
Our study have observed that ARIP was novel protein with unique sequence and but proposed to have lectin binding properties. They have shown remarkable antimicrobial, antimutagenic and cytotoxic activity. Crystallization of the protein followed by X-ray characterization could help to study the structure of the protein, active sites which would in turn give clear focus on the usage of this RIP in treatment of cancer and other diseases. In vivo studies and clinical findings could serve these RIPs as potential candidates in targeting drugs and treatment of diseases.
Acknowledgement
The authors wish to acknowledge the financial assistance as Senior Research Fellow by Indian Council of Medical Research(ICMR) (Project number- 45/12/11/BMS-BIF dated 09.11.2011), New Delhi, India. They also thank the technical support received from the Department of Biotechnology, Karunya Institute of Technology & Sciences, Coimbatore, India
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Mutagenicity, cytotoxic and antioxidant activities of Ricinus communis different parts. Chem. Cent. J.. 2018;12(3):1-3.
- [CrossRef] [Google Scholar]
- Characterization and evaluation of antibacterial and antiproliferative activities of crude protein extracts isolated from the seed of Ricinus communis in Bangladesh. BMC Complement Altern. Med.. 2016;16
- [CrossRef] [Google Scholar]
- Methods for detecting carcinogens and mutagens with the salmonella/mammalian-microsome mutagenicity test. Mutat. Res.. 1975;31(6):347-363.
- [Google Scholar]
- Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta. 2006;1760(5):783-792.
- [CrossRef] [Google Scholar]
- Plant and microbial toxic proteins as hemilectins: Emphasis on canatoxin. Toxicon. 1991;29:791-806.
- [Google Scholar]
- Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum: Antifungal activity of the N -glycosylase BE27. Mol. Plant Pathol. 2016;17(2):261-271.
- [Google Scholar]
- Effects of Annona squamosa extract on early pregnancy in rats. Phytomedicine. 2002;9:667-672.
- [Google Scholar]
- The Antimutagenic Effect of Mistletoe Lectin (Viscum album L. var. coloratum agglutinin): antimutagenicity of mistletoe lectin. Phytother. Res.. 2012;26:787-790.
- [Google Scholar]
- Antimutagenic activity of olive leaf aqueous extract by Ames test. Adv. Stud. Biol.. 2012;4(9):397-405.
- [Google Scholar]
- Evaluation of mosquitocidal activity of Annona squamosa leaves against filarial vector mosquito, Culex quinquefasciatus say. Ind. J. Exp. Biol.. 2002;40:363-365.
- [Google Scholar]
- Antidiabetic and antioxidant activity of Annona squamosa extract in streptozotocin-induced diabetic rats, Singapore Med. J.. 2006;47:670-675.
- [Google Scholar]
- Evaluation of antimicrobial activity of Annona squamosa. L. J. Pharm. Biomed. Res.. 2011;1:12-17.
- [Google Scholar]
- Genoprotection and cytotoxicity of Cassia surattensis seed extract on vero cell evaluated by comet and cytotoxicity assays. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci.. 2018;88:313-320.
- [Google Scholar]
- Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-685.
- [Google Scholar]
- The correlation between antimutagenic activity and total phenolic content of extracts of 31 plant species with high antioxidant activity. BMC Complement Altern. Med.. 2016;16
- [CrossRef] [Google Scholar]
- Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55-63.
- [Google Scholar]
- Differential inhibitory potencies and mechanisms of the type I ribosome inactivating protein marmorin on estrogen receptor (ER)-positive and ER-negative breast cancer cells. Biochim. Biophys. Acta. 2013;1833(5):987-996.
- [Google Scholar]
- Differential cytotoxic effects ofAnnona squamosa seed extracts on human tumour cell lines: Role of reactive oxygen species and glutathione. J. Biosci.. 2005;30:237-244.
- [Google Scholar]
- Isolation and Purification of Ribosome-Inactivating Proteins, Methods Mol. Biol.. 2006;318:335-347.
- [CrossRef] [Google Scholar]
- Enzymatic specificity of three ribosome-inactivating proteins against fungal ribosomes, and correlation with antifungal activity. Planta. 2002;216(2):227-234.
- [Google Scholar]
- Novel role for pectin methylesterase in Arabidopsis: a new function showing ribosome-inactivating protein (RIP) activity. Biochim. Biophys. Acta. 2008;1780:773-783.
- [Google Scholar]
- Ribosome‐inactivating proteins from plants: more than RNA N‐glycosidases? FASEB J.. 2001;15:1493-1506.
- [Google Scholar]
- Evolution of plant ribosomal inactivating proteins. In: Michael Lord J., Hartley M.R., eds. Toxic Plant Proteins Plant Cell Monographs. New York: Springer Heidelberg Dordrecht London; 2010. p. :1-26.
- [Google Scholar]
- A new age for biomedical applications of Ribosome Inactivating Proteins (RIPs): from bioconjugate to nanoconstructs. J. Biomed. Sci.. 2016;23(1)
- [CrossRef] [Google Scholar]
- Antimicrobial and cytotoxic constituents from the seeds of Annona squamosa. Fitoterapia. 2005;76:484-489.
- [Google Scholar]
- Progress in Ribosomal inactivating protein (RIP) studies: Recent review of potential applications. Int. J. Pharm. Biol. Sci.. 2013;3:88-100.
- [Google Scholar]
- Sharififar, F., Dehghan-Noudeh, G., Moshafi, M., Ohadi, M., Zaman-Basir, M., Yazdanpanah, E., 2016. Antimutagenic activity of major fractions of Zataria multiflora Boiss by Ames method. Asian J. Pharm. 9, 195–199.
- Isolation and characterization of an rip (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol.. 2004;134(1):171-181.
- [Google Scholar]
- Antidiabetic activity of aqueous leaf extract of Annona squamosa in streptozotocin–nicotinamide type 2 diabetic rats. J. Ethnopharmacol.. 2004;91(1):171-175.
- [Google Scholar]
- Purification and characterization of a novel ribosome-inactivating protein from seeds of Trichosanthes kirilowii Maxim. Protein Exp. Purif.. 2009;67:120-125.
- [CrossRef] [Google Scholar]
- Ribosome-inactivating proteins: an overview. In: Gopalakrishnakone P., Carlini C., Ligabue-Braun R., eds. Plant Toxins. Springer, Dordrecht: Toxinology; 2017. p. :153-182.
- [Google Scholar]
- Screening of ten Indian medicinal plants for their antibacterial activity against shigella species and Escherichia coli. Afr. J. Infect. Dis.. 2007;1:36-41.
- [Google Scholar]
- C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci.. 1997;94:3866-3871.
- [Google Scholar]
- Characterization of two novel type I Ribosomal inactivating proteins from the storage roots of the Andean Crop Mirabilis Expansa. Plant Physiol.. 1999;119:1447-1456.
- [CrossRef] [Google Scholar]
- Identification and characterization of riproximin, a new type II ribosome‐inactivating protein with antineoplastic activity from Ximenia americana. FASEB J.. 2006;20:1194-1196.
- [Google Scholar]
- Sechiumin, a ribosome-inactivating protein from the edible gourd, Sechium edule Swartz- Purification, charaterization, molecular cloning and expression. Eur. J. Biochem.. 1998;255(2):400-408.
- [Google Scholar]
- Identification of ent-16 beta, 17-dihydroxykauran-19-oic acid as an anti-HIV principle and isolation of the new diterpenoids annosquamocins A and B from Annona squmosa. J. Nat. Prod.. 1996;59:635-637.
- [CrossRef] [Google Scholar]
- Curcin C, a novel type I ribosome-inactivating protein from the post-germinating cotyledons of Jatropha curcas. Amino Acids. 2017;49(9):1619-1631.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.08.002.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







