Translate this page into:
Over-expression of a cyanobacterial gene for 1-deoxy-d-xylulose-5-phosphate synthase in the chloroplast of Chlamydomonas reinhardtii perturbs chlorophyll: carotenoid ratios
⁎Corresponding author at: Applied Biology Section, Applied Sciences Department, Higher College of Technology, University of Technology and Applied Sciences, Al-Khuwair 133, Oman. Umaima.alhoqani@hct.edu.om (Umaima Al Hoqani)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Terpenoids are a diverse class of naturally occurring compounds consisting of more than 50,000 structurally different molecules and are found in all living organisms. Many terpenoid compounds, in particular those isolated from plants, have applications in various commercial sectors including medicine, agriculture and cosmetics. However, these high value terpenoids are produced in relatively small quantities in their natural hosts and their chemical synthesis for large scale production is costly and complicated. Therefore, there is much focus on producing these compounds in novel biological hosts using metabolic engineering technologies. As a photosynthetic system, the unicellular green alga C. reinhardtii is of particular interest as the most well-studied model alga with well-established molecular tools for genetic manipulation. However, the direct manipulation of terpenoid biosynthetic pathways in C. reinhardtii necessitates a thorough understanding of the basic terpenoid metabolism. To gain a better understanding of the methylerythritol phosphate (MEP) pathway that leads to terpenoid biosynthesis in the chloroplast of C. reinhardtii, hence this study has investigated the effect of over-expressing 1-deoxy-d-xylulose-5-phosphate synthase (DXS) on plastidic downstream terpenoids. We produced marker-free chloroplast transformants of C. reinhardtii lines that express an additional cyanobacterial gene for DXS. The analysis of terpenoid content for the transgenic line demonstrates that overexpressing DXS resulted in a two-fold decrease in the chlorophyll levels while carotenoid levels showed variable changes: zeaxanthin and antherxanthin levels increased several-fold, lutein levels dropped to approximately half, but β-carotene and violaxanthin did not show a significant change.
Keywords
Terpenoid
Chlamydomonas reinhardtii
Algal chloroplast
Engineering
DXS
MEP
1 Introduction
Terpenoids are a diverse class of naturally occurring compounds consisting of more than 50,000 structurally different chemicals, which are found in all living organisms (Pateraki et al., 2015). Many terpenoids are of plant origin, and most of them are functionally important in various aspects of cell metabolism, including photosynthesis, membrane permeability and fluidity, respiration, and regulation of growth and development. Another class of plant terpenoids – often referred to as secondary metabolites – is more specialised and limited to certain plant families or genera with functions in defence against plant pathogens, abiotic stress conditions or the attraction of pollinators (Marasco and Schmidt-Dannert, 2008; Vranová et al., 2012). Due to their functional diversity, these secondary metabolic terpenoids are of great industrial interest, largely in areas such as pharmaceuticals, agricultural chemicals, flavours and fragrance additives. For example, the plant derived terpenoid, paclitaxel, is used currently as a potent anticancer drug (Li et al., 2015), while the essential oil from wild mint, menthol, is widely used as a flavour and fragrance additive (Caputi and Aprea, 2011). Additionally, recent studies indicate that some terpenoids also possess properties favourable as fuels, with potential to serve as advanced biofuel precursors (Peralta-Yahya et al., 2011).
Despite their enormous structural diversity, all terpenoids are derived from the five-carbon precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are assembled to produce different classes of terpenoid molecules, such as hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), diterpenes (C20), triterpenes (C30) and tetraterpenes (C40). In photosynthetic eukaryotes, the five carbon precursors IPP and DMAPP are synthesised by two alternative pathways; the methylerythritol phosphate (MEP) and mevalonate (MVA) pathways that are localised in different cellular compartments. The presence of the particular pathway or even both pathways depends on the organism. For example, higher plants and diatoms possess both the MVA and MEP pathways whereas green algae have only the plastidic MEP pathway (Lohr et al., 2012; Matsushima et al., 2012). Many terpenoids with high impact on human health are found in low abundance in nature – normally less than 2–3% of total dry weight (Roberts, 2007). The extraction efficiency of these high value compounds from plant tissue is variable depending on plant source, location and season of harvest, and is normally associated with high extraction costs and low yield recovery (Maury et al., 2005). In addition, the complexity and chirality of the majority of these compounds have generally prevented the development of cost-effective methods for chemical synthesis. Recent genetic engineering efforts therefore have been directed towards finding alternative production platforms for terpenoids other than plants, as genetic manipulations of whole plants often requires in-depth understanding of the metabolic system and regulatory complexity to enable the successful manipulation of the biosynthetic pathway without disturbing other essential processes within the plant (Farhi et al., 2011; Ikram et al., 2015; Saxena et al., 2014). Since all living organisms contain the metabolic precursors needed for terpenoid production, the heterologous expression of terpene synthases in microbial hosts offers sustainable platforms for terpenoid production. The most popular heterologous hosts used for industrially relevant terpenoids are E. coli and S. cerevisiae. These microorganisms in particular are genetically tractable with well-established molecular tools and can be cultured at large scale and high density using existing fermentation infrastructure (Gruchattka and Kayser, 2015; Kirby et al., 2015; Kirby and Keasling, 2009). There has been some success in exploiting these heterologous expression systems for terpenoid supply, where a modified E. coli strain has been used to produce the artemisinin precursor, amorphadiene (Tsuruta et al., 2009). The production of complex terpenoids has been extended to yeast due to the difficulty of expressing in E. coli the plant cytochrome P450 monooxygenases that are often required to modify the carbon skeleton of complex terpenoid metabolites (Putignani et al., 2013).
Recently, microalgae have attracted attention as potential heterologous hosts for terpenoid production. As photosynthetic organisms, microalgae have their own advantages including the capabilities to produce valuable metabolites in either open systems or photobioreactors using just CO2 and basic nutrients, where the whole process is driven by sunlight. In addition, there are microalgal species that naturally accumulate very high levels of industrially relevant terpenoids when exposed to adverse environmental conditions. Two prominent examples are Dunaliella salina and Haematococcus pluvialis that accumulate the carotenoids β-carotene and astaxanthin, respectively under stress conditions (Eonseon et al., 2006; Lohr et al., 2012; Ramos et al., 2011). The natural capacity for terpenoid production in these green algae could be improved by metabolic engineering technologies (Lauersen, 2019). However, metabolic engineering of these species is notoriously difficult due to the limited availability of molecular tools. From a biotechnological standpoint, the green microalga Chlamydomonas reinhardtii is the most well studied model species with well-established molecular tools for engineering both the nuclear and chloroplast genomes (Cutolo et al., 2022; Mussgnug, 2015). Several reports of nuclear engineering have highlighted the potential of C. reinhardtii as a low-cost, phototrophic platform. For example, synthesis of the C15 terpenoids patchoulol and (E)-α-bisabolene in the algal cytoplasm was achieved through expression of plant sesquiterpenoid synthases (Lauersen et al., 2016; Wichmann et al., 2018). Conversely, targeting of various diterpenoid synthases into the chloroplast allowed the synthesis of the C20 compounds casbene, taxadiene, and 13R(+) manoyl oxide, with the last compound further modified by also targeting a cytochrome P450 monooxygenase into the organelle (Lauersen et al., 2018).
Given the central role of the algal chloroplast in terpenoid biosynthesis, an alternative engineering strategy is the direct synthesis of heterologous enzymes within the chloroplast rather than their import into the organelle following cytoplasmic synthesis from nuclear-encoded transgenes. Indeed, chloroplast engineering offers several advantages over conventional nuclear engineering including the precise integration of foreign DNA to any loci within the chloroplast genome via homologous recombination, the absence of gene silencing effects, and the possibility to express multiple transgenes as an operon. Chloroplast transformation is well-established for C. reinhardtii and has been exploited for production of a wide range of recombinant products including therapeutic proteins and valuable metabolites (Dyo and Purton, 2018).
With such interest in the use of terpenoids in a variety of industrial and therapeutic applications, this study focused on understanding the chloroplast’s MEP pathway that leads to terpenoid production in C. reinhardtii. We have targeted 1-deoxyxylulose 5-phosphate synthase (DXS) for metabolic engineering as this enzyme catalyses the first biosynthetic step of the pathway and has been reported as a rate limiting step for the production of the C5 precursors in bacteria and plants (Estévez et al., 2001; Gong et al., 2006; Heider et al., 2014). We demonstrate the generation of transgenic C. reinhardtii lines over-expressing DXS by introducing a cyanobacterial version of the dxs gene into a neutral site within the chloroplast genome. The analysis of the transgenic line showed that C. reinhardtii over-expressing DXS had altered levels of chlorophylls and various downstream terpenoids including zeaxanthin, lutein and antheraxanthin.
2 Materials and methods
2.1 Algal strains, media and culture conditions
C. reinhardtii TN72 was used as a recipient line to generate the chloroplast transformant lines named T1.dxs, T2.dxs, T3.Δdxs, T4.Δdxs and Control. TN72 is a photosynthetic mutant of the cell wall deficient strain cw15 (mt+), with the loss of photosynthesis due to disruption of the chloroplast gene psbH with the aadA marker (Wannathong et al., 2016). The selection of transformant lines is therefore based on restoring a functional psbH gene and the ability to grow photosynthetically on acetate-free medium – i.e. high salt minimal (HSM) medium with 2% agar – at a light intensity of 50 µE m−2 s−1. All transformant lines were then maintained mixotrophically on Tris-acetate phosphate (TAP) medium with 2% agar under dim light conditions of approximately 5–10 µE m−2 s−1 at 20 °C (Harris, 2009). Transformant lines were cultured in liquid TAP medium shaking at 120 rpm under continuous illumination of 50–100 µE m−2 s−1 at 25 °C, as a standard growth conditions and for western blot analysis. When required, the growth was monitored by measuring the optical density at 750 nm with a spectrophotometer. For growth tests, the transformant lines were cultured in TAP medium in 1 L Erlenmeyer flasks at 100–200 µE m−2 s−1, 120 rpm shaking and 25 °C in an Algem® photobioreactor (Algenuity, Stewartby, UK) and the cell growth was monitored in situ by direct measurement of optical density at 740 nm.
2.2 Plasmid construction
The dxs coding sequence (locus tag SYNGTS_0514) was amplified from Synechocystis sp. PCC 6803 genomic DNA using primers 5′-CCAGCTCTTCTATGCACATCAGCGAACTGAC-3′ and 5′-GTCAGGCATGCTTATTAAGCGTAATCTGGAACATCGTATGGGTAACTAACTCCAGGAGCGACAAC-3′, and Phusion Polymerase (NEB). The PCR added an HA tag sequence to the 3′ end of the coding sequence and flanked the PCR product with SapI and SphI sites (underlined). The product was cloned between the SapI and SphI sites in the chloroplast expression vector pSRSapI, which has been described previously (Wannathong et al., 2016), to make plasmid pRY134a. Another construct, pRY134aΔdxs, was made by deleting a 141 bp region of dxs between two NcoI sites. This leads to a 47 amino acid in-frame deletion in DXS. All cloning was carried out using chemically competent E. coli DH5α according to standard protocols (Sambrook and Russell, 2001). Restriction enzymes and T4 DNA ligase were purchased from NEB and ThermoFisher.
2.3 C. reinhardtii chloroplast transformation
The chloroplast transformation method used was adapted from the original glass bead method described by Kindle et al., (1991), with some modifications. TN72 was grown in 500 ml TAP to mid-log phase (with a cell density of 1–2 × 106 cells ml−1) under dim light of approximately 5–10 µE m−2 s−1. The cells were harvested by centrifugation at 5000 rpm for 5 min at 16 °C and then re-suspended in fresh TAP to a concentration of 2 × 108 cells ml−1. Aliquots of the cell suspension were pipetted into test tubes containing ∼0.3 g of glass beads (acid washed, 425–600 μm diameter, Sigma). 10 μg of plasmid DNA was added to each tube containing 300 μl of cell suspension and then agitated for 15 s at maximum speed using a Vortex genie-2. Then 3 ml of molten 0.5% (w/v) HSM agar at 42 °C was added to each tube and immediately the mixture was poured onto 2% HSM (w/v) agar plates. The plates were incubated under a light intensity of approximately 50 µE m−2 s−1 at 25 °C for 3–4 weeks, with the selection based on restoration of photosynthesis, i.e. the ability to grow autotrophically on HSM agar plates. The putative transformant colonies were picked and re-streaked three times on HSM agar plates to ensure homoplasmy. To confirm transgene integration and homoplasmic of the chloroplast genome, genomic DNA was extracted using the Chelex 100 method as described by Werner and Mergenhagen (1998) and 2 µl of the DNA extract used in a PCR reaction. PCR analysis was carried out using a combination of three different primers (Table S1) and Phusion Polymerase (NEB) according to the manufacturer’s instructions. PCR products were analysed on 1% agarose gels and the size of PCR products were estimated by reference to a GeneRuler DNA Ladder Mix (Thermo Scientific).
2.4 Western blot analysis
To prepare protein extracts, C. reinhardtii transformant lines were grown to mid-log phase in a shaking incubator and the optical density of cultures at 750 nm was measured using a spectrophotometer. The C. reinhardtii cultures were then harvested by centrifugation at 5800g for 5 min and the pellets re-suspended in 0.8 M Tris.HCl pH 8.3, 0.2 M sorbitol and 1% (v/v) β-mercaptoethanol to equal cell densities. Samples were boiled for 3 min in the presence of 10% SDS (w/v), centrifuged at 21,000g for 2 min and the resulting supernatant was loaded onto a 15% acrylamide SDS-polyacrylamide gel (Laemmli, 1970). Protein separation was carried out in cold electrophoresis buffer (0.25 M Tris, 1.92 M glycine, 1% (w/v) SDS pH 8.3) for 90 min at 150 V. The proteins were transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare) using a Trans-blot semi-dry transfer cell (Bio-Rad) at constant voltage of 19 V for an hour. Following the transfer, the membrane was blocked overnight in 5 % (w/v) skimmed milk powder in TBS-T (20 mM Tris base, pH adjusted to 7.4 with 5 M HCl, 137 mM NaCl, 0.1 % (v/v) Tween) at 4 °C shaking at 50 rpm. The membrane was rinsed briefly with TBS-T, followed by two rounds of 15 min washes in TBS-T while shaking at 50 rpm. Diluted rabbit anti-HA antibody (Sigma-Aldrich product H6908, 1:2000 in TBS-T + 0.5% milk) was used as a primary antibody and the membrane was treated with primary antibody for 1 h at room temperature with shaking at 50 rpm. The membrane was rinsed several times briefly and the membrane was then washed four times in TBS-T for 10 min. The membrane was incubated with secondary antibody ECL anti-rabbit IgG horseradish peroxidase-linked antibody (GE Healthcare product NA934, 1:5000 in TBS-T + 0.5% milk) for 1 h at room temperature with shaking at 50 rpm and the membrane was washed as for the primary antibody. The proteins were detected by the ECL method using a luminescence substrate detection kit according to the manufacturer's instructions (SuperSignal West Pico Chemiluminescent substrate, supplied by Pierce).
2.5 HPLC analysis
Pigments were extracted from 10 mg of freeze-dried biomass by agitation with 25 ml methanol. The separation and chromatographic analysis of pigments were performed on a Merck Hitachi HPLC equipped with a diode-array detector as described by Young and co-workers (Young et al., 1997), using a RP-18 column and a flow rate of 1 ml min−1. The mobile phase consisted of: solvent A, ethyl acetate; solvent B acetonitrile/water (9:1, v/v) and the gradient programme applied was: 0–16 min 0–60%A; 16–30 min 60%A; 30–35 min 100%A. Injection volume was 100 µl. Pigments detection was carried out at 450 nm. Pigments standards were supplied by SIGMA or DHI (Hoersholm, Denmark).
3 Results
3.1 Generation of marker-free transgenic lines of Chlamydomonas reinhardtii expressing a heterologous dxs gene in the chloroplast
Green microalgae such as C. reinhardtii contains only the plastidic MEP pathway for terpenoid biosynthesis (Lohr et al., 2012). A simplified illustration of the pathway is shown in Fig. 1. DXS catalyses the first biosynthetic step of this pathway and has been reported to be a rate limiting step for the production of the terpenoid precursors IPP and DMAPP in bacteria and plants (Estévez et al., 2001; Gong et al., 2006; Heider et al., 2014). To investigate the effect of overexpressing this enzyme on downstream terpenoid metabolites in C. reinhardtii, a plasmid (pRY134a) was constructed containing the dxs gene from the cyanobacterium Synechocystis sp. PCC6803 under the control of the C. reinhardtii psaA exon 1 promoter and 5′ untranslated region (Fig. 2a). The coding sequence of a C-terminal human influenza hemagglutinin (HA) epitope tag was added to the dxs gene to facilitate the detection of the DXS protein. The dxs gene from Synechocystis is more suitable for heterologous expression in the chloroplast than the nuclear-encoded native copy from C. reinhardtii. This is because the GC content and codon bias of the two genomes of C. reinhardtii are markedly different: the nuclear genome has a GC content of 64% and a bias for GC-rich codons whereas the chloroplast genome has a value of 34% and an AT-rich codon bias (Grossman et al., 2003). Specifically, the C. reinhardtii dxs coding sequence is ∼ 65% GC, whereas that of the Synechocystis dxs is ∼50%. The dxs coding sequence was amplified from Synechocystis genomic DNA and cloned into the expression vector pSRSapI to make plasmid pRY134a (Fig. 2a).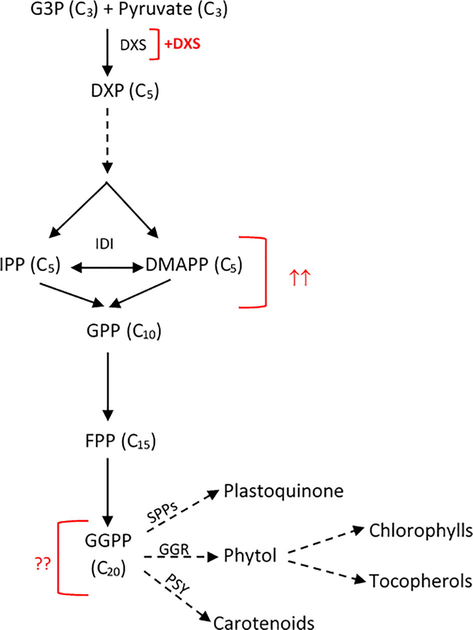
Overview of terpenoid biosynthetic pathway in the chloroplast of C. reinhardtii. The plastidic MEP pathway begins with the condensation of pyruvate and D-glyceraldehyde 3-phosphate (G3P) leading to the formation of isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP). The condensation of DMAPP with one or more IPP units leads to the formation of the following direct precursors; geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), and geranylgeranyl pyrophosphate (GGPP). The introduced additional copy of the gene is indicated by +, whereas the potential flux increase of the corresponding IPP and DMAPP is presented by ↑↑. The subsequent effect on downstream metabolites to be investigated is indicated by??. Dashed arrows indicate a number of steps are involved but are not shown. Abbreviations: DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXP, 1-deoxy-d-xylulose 5-phosphate; IDI, isopentenyl diphosphate isomerase; SPPs solanesyl diphosphate synthase; PSY, phytoene synthase and GGR, geranylgeranyl reductase.
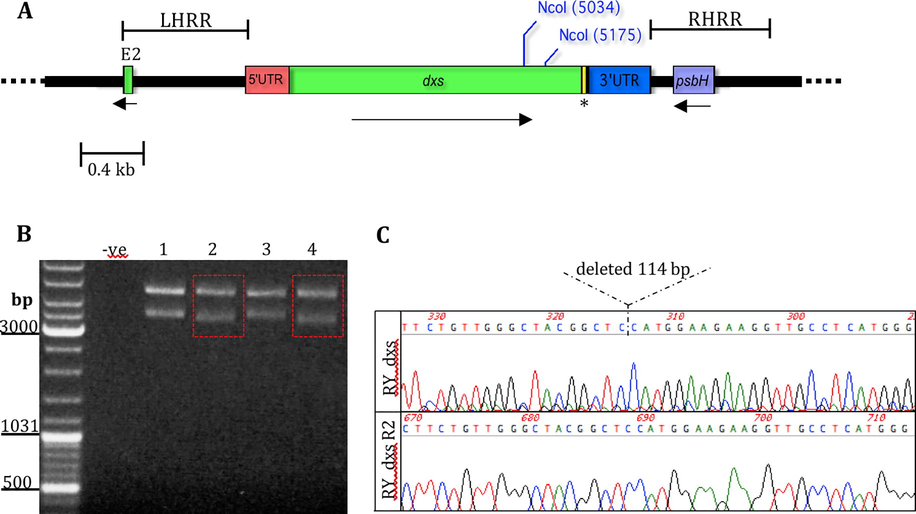
Analysis of partial deletion of dxs in pRY134aΔdxs plasmid. (A) The schematic representation of pRY134a, a cloning vector for introducing a cyanobacterial dxs into the Chlamydomonas chloroplast genome under the control of the psaA exon 1 promoter and the rbcL 3′UTR. The coding sequence of a nine residue haemagglutinin epitope (HA tag) was added to the 3′ end of the dxs gene before the stop codon so that the DXS protein could be detected using anti-HA antibody. The HA tag is indicated by an asterisk. The region of dxs that was deleted in pRY134aΔdxs is between NcoI restriction sites as shown. The flanking regions required for homologous recombination within the chloroplast genome is indicated as LHRR and RHRR, and the intact psbH acts as a selectable marker. The rest of the pRY134a plasmid is depicted as a dashed black line. TrnE2 is abbreviated as E2. (B) Test digestion showing the successful deletion of 141 bp in dxs gene resulting in fragments of the expected sizes, 4599 bp and 3488 bp, as indicated by red box. The lanes 1, 2, 3, and 4 represents different colonies tested for digestion. (C) Sanger sequencing chromatograph confirms the deletion of 141 bp in dxs for clone number 2. The dashed lines indicate the deleted region within the coding sequence of dxs.
High levels of prenyl phosphate precursors (DMAPP/IPP/FPP) have been reported to be toxic in E. coli cells (Martin et al., 2003; Sivy et al., 2011). To investigate the possibility that over-expression of the heterologous dxs in the chloroplast may have similar deleterious effects on C. reinhardtii cells a non-functional version of dxs was created by deleting 141 bp of the coding sequence within plasmid pRY134a. The deletion was achieved by digesting pRY134a with NcoI at two sites to drop out the 141 bp region, and then re-ligating the plasmid. The resulting plasmid, pRY134aΔdxs, was checked by test digestion resulting in the expected fragment sizes, 4599 bp and 3488 bp (Fig. 2b). DNA sequencing confirmed the successful deletion of 141 bp in the gene (Fig. 2c).
The pRY134a and pRY134aΔdxs plasmids were used to transform C. reinhardtii TN72 (a non-photosynthetic psbH mutant) by agitating the cells with glass beads in the presence of the DNA. The expression cassette in both plasmids is flanked by homologous sequence that contains an intact copy of psbH, so homologous recombination into the chloroplast genome results in restoration of functional psbH and integration of the transgene, and allows the selection of restored photosynthetic transformants on minimal medium in the light (see Fig. S1 for transformation strategy to generate marker-less transgenic lines). Putative transformant colonies appeared after four weeks of incubation. Independent transformant lines named T1.dxs and T2.dxs were obtained using pRY134a, while the colonies named T3.Δdxs and T4.Δdxs were obtained using pRY134aΔdxs. These lines were re-streaked for a further three rounds of phototrophic selection on minimal medium to ensure the homoplasmic state of the chloroplast genome since C. reinhardtii has approximately 80 copies per cell. The integration of the transgene and homoplasmy of the genome in all transgenic lines were then examined by PCR using a specific set of three primers, as illustrated in Fig. 3. Successful transgene integration in all transgenic lines was confirmed by the presence of a 1134 bp band, while the homoplasmic state was indicated by the absence of the 880 bp band that arises from the untransformed copies of the parental plastome. A control strain (C) was included from previous studies and this strain was generated by transforming the TN72 strain with the pSRSapI plasmid but lacking a transgene (Young and Purton, 2014).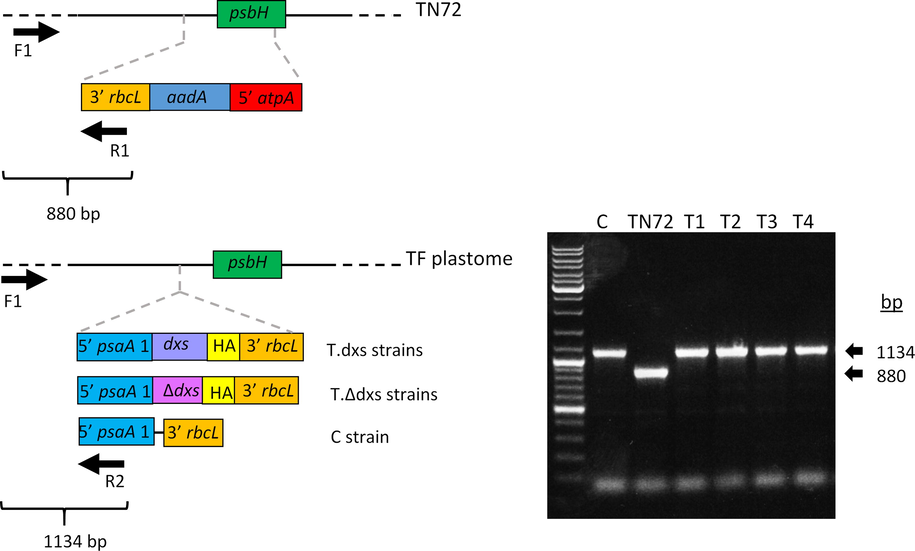
Three primer PCR to confirm transgene integration and homoplasmy of transformants. Left panel: schematic representation of PCR based method to confirm the integration of GOI and homoplasmy of transformants. PCR was performed using three primers in one reaction (F1, R1 and R2). The first primer pair (F1 and R1) amplifies part of the recipient line TN72 plastome, resulting in an 880 bp band, whereas the second primer pair (F1 and R2) amplifies part of the transformant plastome, resulting in a 1134 bp band. The successful integration of the gene of interest is indicated by the presence of the 1134 bp band and the homoplasmy is indicated by the absence of the 880 bp. The C. reinhardtii lines are TN72 recipient strain, T.dxs strains containing the dxs gene, T.Δdxs strains containing the mutant dxs gene and an empty-vector transformant as a control (C). TF is an abbreviation of transformant. Right panel: PCR confirming homoplasmic integration of the transgene into the chloroplast genome.
3.2 Accumulation of the transgenic DXS protein in C. reinhardtii
To demonstrate the accumulation of the transgenic DXS protein, crude cell extracts were prepared from C. reinhardtii lines and analysed by western blot using antibodies against the HA-tag. The C. reinhardtii lines used were T1.dxs and T2.dxs for intact DXS protein, T3.Δdxs and T4.Δdxs for mutant DXS protein and C as a negative control. The DXS protein with an HA tag has an expected size of 70.4 kDa, and as shown in Fig. 4, there is a detectable level of the tagged DXS protein in the extracts of both T.dxs lines. For the T.Δdxs lines, the accumulation of the smaller mutant DXS protein was not detected at 65 kDa (Fig. 4). The deletion does not cause a frameshift in the coding sequence (Fig. S2), but might result in conformational changes of the protein structure that make it more vulnerable to degradation.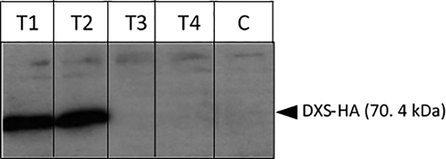
Expression of the cyanobacterial dxs gene in the C. reinhardtii lines demonstrated by western blot analysis of equalised lysate from T.dxs strains containing an intact cyanobacterial dxs gene, T.Δdxs strains containing the mutant dxs gene and C strain as a negative control (i.e. TN72 transformed with pSRSapI, an empty vector). Analysis with anti-HA antibody shows expression of dxs by a strong band running at an apparent mass of 70.4 kDa in cell lines T1.dxs (T1) and T2.dxs (T2) but not in T3.Δdxs (T3) and T4.Δdxs (T4), or in the negative control line (C). The immunoblot was visualised by ECL with one-minute exposure. Protein size was determined using the PageRulerTM Prestained Protein ladder (Thermo Scientific).
3.3 Overexpression of dxs does not affect C. reinhardtii growth
As high levels of prenyl phosphate precursors (DMAPP/IPP/FPP) were reported to be toxic in transgenic E. coli cells (Martin et al., 2003; Sivy et al., 2011), the growth of the transgenic lines was investigated using an Algem® photobioreactor system. As illustrated in Fig. 5, the growth of lines T1.dxs and T2.dxs was not inhibited compared to both the empty-vector control and T.Δdxs lines. Indeed, the growth rates of all four we very similar, and superior to that of the control; possibly because of an unrelated mutation that has arisen in our stock of the control strain. The results demonstrate that the presence of a cyanobacterial DXS protein in addition to the native one does not have a deleterious effect on growth.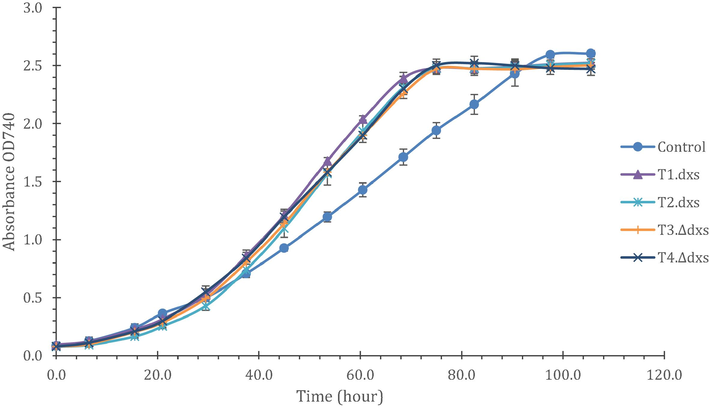
Growth curves of C. reinhardtii cell lines with and without the dxs gene from Synechocystis 6803. All cultures were grown in TAP medium using the Algem® system and the optical density was measured at 740 nm. The growth conditions were 25 °C, 200 µE light, and continuous shaking at 120 rpm. Error bars represents ± standard deviation of triplicate cultures.
3.4 Analysis of terpenoid profile of the genetically engineered C. reinhardtii
In order to determine the impact of expressing an additional dxs gene on terpenoid content, the quantity of plastidic terpenoids –i.e. carotenoids– such as violaxanthin, lutein and β-carotene was measured in the T1.dxs and compared with the levels found in the control line. Carotenoids were chosen because they encompass a large group of compounds derived from geranylgeranyl diphosphate precursor and represent one of the major product classes derived from the MEP pathway. The analysis of the T1.dxs line showed an overall increase in the levels of plastidic carotenoids such as β-carotene, zeaxanthin, antheraxanthin and violaxanthin (Fig. 6 and Fig. S3 for terpenoid profile). However, since the carotenoid levels are normalized to the chlorophyll content in the samples, then it is possible that the observed percentages changes are affected by changes in total chlorophyll in the transgenic line. This is possible since chlorophylls are tetrapyrrole molecules possessing a phytol ‘tail’ composed of a 20-carbon diterpene alcohol derived from GGPP. We therefore compared chlorophyll content in the control and transgenic lines, and indeed identified a significant change in chlorophyll levels when expressed per gram of algal dry weight. As shown in Fig. 6, there is a significant reduction by two folds in the levels of both chlorophyll a and b in the transgenic line – to 54% and 47% of control levels, respectively. Such a reduction in the chlorophyll levels is unexpected since the overexpression of dxs is presumed to increase the precursor pool of GGPP and this precursor pool is shared by carotenoid, phytol and plastoquinone pathways. In addition, several studies in plants reported that the overexpression of dxs resulted in an enhanced accumulation of both carotenoids and total chlorophyll content (Estévez et al., 2001; Munoz-Bertomeu et al., 2006). Nevertheless, this observation allows us to normalize the carotenoid changes to dry weight by dividing the transgenic values by approximately two-fold, and reveals that β-carotene and violaxanthin do not show a significant change in the DXS transformant, whereas zeaxanthin and antherxanthin levels have increased several-fold and lutein levels have dropped to approximately half.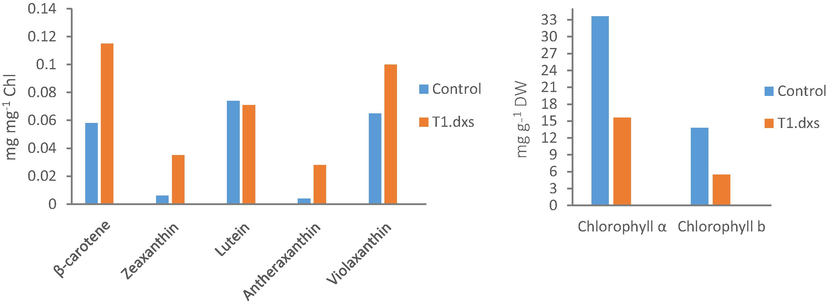
Terpenoid analysis of the C. reinhardtii lines. The effect of expressing an additional dxs gene on carotenoid level (left panel) and chlorophyll level (right panel). Carotenoid and chlorophyll levels were determined by HPLC analysis. Abbreviations are Chl: chlorophyll and DW: dry weight.
4 Discussion
Because of the biotechnological application of terpenoids in animal feed, human nutrition, pharmaceuticals and biofuels, this study aimed at gaining a better understanding of the MEP pathway that leads to terpenoid production in the green microalga C. reinhardtii. A number of studies have proven that the DXS enzyme catalyses one of the rate limiting steps in the MEP pathway in a variety of systems. For example, the overexpression of dxs led to increased accumulation of essential oil in lavender, β-carotene accumulation in the oil palm Elaseis guineensis and carotenoid accumulation in Arabidopsis thaliana (Estévez et al., 2001; Munoz-Bertomeu et al., 2006). Similarly in Corynebacterium glutamicum, where the overexpression of dxs improved the lycopene accumulation (Heider et al., 2014). To extend these studies further in the algal chloroplast and investigate the fundamental aspects of flux through the MEP pathway in C. reinhardtii, a reverse genetic approach was used to supplement the DXS activity within the chloroplast through chloroplast expression of a cyanobacterial gene encoding DXS, and then study the corresponding changes in the downstream terpenoid profile.
Transgenic lines were generated, in which the dxs gene from Synechocystis PCC 6803 was integrated successfully into the chloroplast genome under the control of the promoter/5′UTR from psaA exon 1, and the 3′UTR from rbcL. Another set of transgenic lines carrying non-functional dxs was also created to examine the possibility that over-accumulation of DXS in C. reinhardtii has a toxic effect on the cells, as the high levels of prenyl phosphate precursors (DMAPP/IPP/FPP) was reported to be toxic in E. coli cells (Sivy et al., 2011). Nonetheless, the growth analysis indicated that there was no toxic effect as demonstrated by normal growth rate and healthy appearance in all transgenic lines.
In principle, the over-accumulation of DXS provides the ‘push’ to increase the precursor pools for downstream metabolites, as opposed to the ‘pull’ of introducing a downstream enzyme that competes with other pathways for the same precursor pool. Successful examples of the latter are seen in several reports of nuclear engineering in C. reinhardtii whereby over-expression of carotenoid biosynthesis genes resulted in elevated levels of carotenoid products such as β-carotene, lutein of violaxanthin (Cordero et al., 2011; Rathod et al., 2020; Tokunaga et al., 2021). Conversely, as example of undesirable collateral effects was seen following the over-expression of phytoene synthase (the first committed step in a carotenoid biosynthesis pathway) in tomato fruit. The overexpression severely affected the transgenic lines resulting in dwarfism and reduce chlorophyll content due to the alteration in the synthesis of both abscisic acid and phytol from the precursor GGPP shared with carotenoids (Fray et al., 1995).
The overexpression of DXS should increase the precursor pools of IPP and DMAPP for the downstream terpenoid metabolites, where three main pathways are most likely to be affected. These pathways share the same precursor GGPP and include phytol, carotenoids and plastoquinone. Ent-kaurene is an additional biosynthetic pathway that is found in plants and is involved in the synthesis of a group of plant hormones called gibberellins. However, hormone studies have been neglected in microalgae and whilst there is a possibility of involvement of some these terpenoid hormones in algae these have not been identified yet (Lohr et al., 2012). Although there are three pathways that diverge from the GGPP precursor, and so all three pathways could be assessed upon the upregulation of the DXS enzyme activity in the MEP pathway, the main focus was to analyse the quantitative changes in the carotenoid levels of various classes due to their industrial applications, and also to analyse the quantitative changes in phytol-derived compounds, chlorophyll a and b to gain a better insight as a result of changing DXS levels. Although several studies reported that DXS overexpression resulted in increased levels of carotenoids and chlorophylls, the general finding of this study for two divergent terpenoid pathways is that by increasing the levels of DXS, the carotenoid levels were perturbed with some being elevated and other reduced or unchanged, whereas the levels of chlorophylls decreased by a factor of ∼2. The reason for such variation in the abundance of plastidic terpenoid is unclear, although it is possible that the phytol pathway could be affected by feedback inhibition as a result of the increase in precursor pool GGPP. Alternatively, it could be due to an indirect effect on the other moiety of chlorophyll since it consists of two components; i) the terpenoid phytol, and ii) chorophyllide, which is not a terpenoid but a tetrapyrrole formed from the precursor molecule 5-aminolevulinate (Estévez et al., 2001).
5 Conclusions
The strategy used here for engineering terpenoid biosynthesis relies on enhancing the flux to increase the precursor pools of IPP and DMAPP by introducing an additional transgenic copy of the gene for DXS, which is involved in catalysing the first step in the plastidic MEP pathway. DXS overexpression influenced the level of several plastidic downstream terpenoids as well as chlorophyll suggesting that DXS catalyses one of the rate limiting steps of the MEP biosynthetic pathway in the C. reinhardtii. The study also provided a supportive evidence for the effect of over-expression on downstream terpenoid metabolites, where the over-expression of DXS disturbs chlorophyll: carotenoid ratios. This work opens up the possibility of engineering carotenogenic pathways for improved carotenoid production in the C. reinhardtii.
Acknowledgements
We thank the Ministry of Higher Education, Research and Innovation, Sultanate of Oman for covering the publication costs through grant no. TRC/BFP/HCT/01/2019. Umaima Al Hoqani was funded by a doctoral scholarship from the Ministry of Labor previously known as Ministry of Manpower, Sultanate of Oman. The research was supported by grant BB/R016534/1 from the UK’s Biotechnology and Biological Sciences Research Council (BBSRC). Umaima Al Hoqani is grateful to Syed Najmul Hejaz Azmi for his continuous support and guidance to get this work published.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Use of terpenoids as natural flavouring compounds in food industry. Recent Patents Food, Nutr. Agric.. 2011;3(1):9-16.
- [CrossRef] [Google Scholar]
- Enhancement of carotenoids biosynthesis in Chlamydomonas reinhardtii by nuclear transformation using a phytoene synthase gene isolated from Chlorella zofingiensis. Appl. Microbiol. Biotechnol.. 2011;91(2):341-351.
- [CrossRef] [Google Scholar]
- Harnessing the algal chloroplast for heterologous protein production. Microorganisms. 2022;10(4):743.
- [Google Scholar]
- The algal chloroplast as a synthetic biology platform for production of therapeutic proteins. Microbiology (United Kingdom). 2018;164(2):113-121.
- [CrossRef] [Google Scholar]
- Secondary carotenoid accumulation in Haematococcus (Chlorophyceae): Biosynthesis, regulation, and biotechnology. J. Microbiol. Biotechnol.. 2006;16(6):821-831.
- [Google Scholar]
- 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem.. 2001;276(25):22901-22909.
- [CrossRef] [Google Scholar]
- Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol.. 2011;29(12):1072-1074.
- [CrossRef] [Google Scholar]
- Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J.. 1995;8(5):693-701.
- [CrossRef] [Google Scholar]
- Molecular cloning and expression profile analysis of Ginkgo biloba DXS gene encoding 1-deoxy-D-xylulose 5-phosphate synthase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Planta Med.. 2006;72(4):329-335.
- [CrossRef] [Google Scholar]
- Chlamydomonas reinhardtii at the crossroads of genomics. Am. Soc. Microbiol.. 2003;2(6):1137-1150.
- [CrossRef] [Google Scholar]
- In vivo validation of in silico predicted metabolic engineering strategies in yeast: Disruption of α-ketoglutarate dehydrogenase and expression of ATP-citrate lyase for terpenoid production. PLoS ONE. 2015;10(12):1-25.
- [CrossRef] [Google Scholar]
- Harris, E.H., 2009. The Chlamydomonas Sourcebook: Introduction to Chlamydomonas and Its Laboratory Use (Issue v. 1). Elsevier Science. https://books.google.co.uk/books?id=xTjJGV5GWY0C.
- Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front. Bioeng. Biotechnol.. 2014;2(August):28.
- [CrossRef] [Google Scholar]
- Stable heterologous expression of biologically active terpenoids in green plant cells. Front. Plant Sci.. 2015;6(March):129.
- [CrossRef] [Google Scholar]
- Engineering the chloroplast genome: techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. PNAS. 1991;88(March):1721-1725.
- [CrossRef] [Google Scholar]
- Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant. Biol.. 2009;60:335-355.
- [CrossRef] [Google Scholar]
- Enhancing Terpene yield from sugars via novel routes to 1-deoxy-D-xylulose 5-phosphate. Appl. Environ. Microbiol.. 2015;81(1):130-138.
- [CrossRef] [Google Scholar]
- Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680-685.
- [Google Scholar]
- Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta. 2019;249(1):155-180.
- [CrossRef] [Google Scholar]
- Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng.. 2016;38:331-343.
- [CrossRef] [Google Scholar]
- Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng.. 2018;49(July):116-127.
- [CrossRef] [Google Scholar]
- Current and emerging options for taxol production. In: Schrader J., Bohlmann J., eds. Biotechnology of Isoprenoids. Springer International Publishing; 2015. p. :405-425.
- [CrossRef] [Google Scholar]
- Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci.. 2012;185–186:9-22.
- [CrossRef] [Google Scholar]
- Exploring and accessing plant natural product biosynthesis in engineered microbial hosts. In: Kayser O., Quax W.J., eds. Medicinal Plant Biotechnology: From Basic Research to Industrial Applications. WILEY-VCH Verlag; 2008. p. :287-317.
- [CrossRef] [Google Scholar]
- Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotech. 2003;21(7):796-802.
- [CrossRef] [Google Scholar]
- The single cellular green microalga Botryococcus braunii, race B possesses three distinct 1-deoxy-d-xylulose 5-phosphate synthases. Plant Sci.. 2012;185–186:309-320.
- [CrossRef] [Google Scholar]
- Microbial isoprenoid production: an example of green chemistry through metabolic engineering. Adv. Biochem. Eng. Biotechnol.. 2005;100
- [Google Scholar]
- Munoz-Bertomeu, J., Arrillaga, I., Ros, R., & Segura, J. (2006). Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils. Plant P, 142(November), 890–900. https://doi.org/10.1104/pp.106.086355.
- Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol.. 2015;99(13):5407-5418.
- [CrossRef] [Google Scholar]
- Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv. Biochem. Eng. Biotechnol.. 2015;148:107-139.
- [CrossRef] [Google Scholar]
- Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun.. 2011;2:483.
- [CrossRef] [Google Scholar]
- Engineered Escherichia coli as new source of flavonoids and terpenoids. Food Res. Int.. 2013;54(1):1084-1095.
- [CrossRef] [Google Scholar]
- The unicellular green alga Dunaliella salina Teod. as a model for abiotic stress tolerance: genetic advances and future perspectives. Algae. 2011;26(1):3-20.
- [CrossRef] [Google Scholar]
- Metabolic engineering of Chlamydomonas reinhardtii for enhanced β-carotene and lutein production. Appl. Biochem. Biotechnol.. 2020;190(4):1457-1469.
- [CrossRef] [Google Scholar]
- Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol.. 2007;3(7):387-395.
- [Google Scholar]
- Molecular cloning: A laboratory manual (Third). Cold Spring Harbor Laboratory press; 2001.
- Metabolic engineering of chloroplasts for artemisinic acid biosynthesis and impact on plant growth. J. Biosci.. 2014;39(1):33-41.
- [CrossRef] [Google Scholar]
- Evidence of isoprenoid precursor toxicity in Bacillus subtilis. Biosci. Biotechnol. Biochem.. 2011;75(12):2376-2383.
- [CrossRef] [Google Scholar]
- Enhanced lutein production in Chlamydomonas reinhardtii by overexpression of the lycopene epsilon cyclase gene. Appl. Biochem. Biotechnol.. 2021;193(6):1967-1978.
- [CrossRef] [Google Scholar]
- High-level production of amorpha-4, 11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE. 2009;4(2):e4489.
- [Google Scholar]
- Structure and dynamics of the isoprenoid pathway network. Mol. Plant. 2012;5(2):318-333.
- [CrossRef] [Google Scholar]
- New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol.. 2016;100(12):5467-5477.
- [Google Scholar]
- Mating Type Determination of Chlamydomonas reinhardtii by PCR. Plant Molecular Biology Reporter. 1998;16(4):295-299.
- [CrossRef] [Google Scholar]
- Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng.. 2018;45:211-222.
- [Google Scholar]
- Methods for caroternoid analysis. In: Pessarakli M., ed. Handbook of Photosynthesis. Marcel Dekker; 1997. p. :597-622.
- [Google Scholar]
- Cytosine deaminase as a negative selectable marker for the microalgal chloroplast: A strategy for the isolation of nuclear mutations that affect chloroplast gene expression. Plant J.. 2014;80(5):915-925.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102141.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







