Translate this page into:
Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil
⁎Corresponding author. rajerajan@yahoo.com (G. Rajalakshmi) raajeerajan@gmail.com (G. Rajalakshmi)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Lipases are an important hydrolytic enzyme with numerous applications and industrial potential. The present study aimed to produce lipase enzyme from bacterial strains. Eight bacterial strains were isolated from petrol spilled soil by serial dilution technique. Olive oil was used as the substrate in tributyrin agar medium for screening and showed the zone of activity in five of those bacterial strains. Under submerged fermentation conditions, a high level of lipase production was found at 37 °C with pH 6.0 ± 0.5. The presence of 3% sucrose and 5% yeast extract in the medium enhanced enzyme production when compared with other carbon and nitrogen sources. Produced lipases were partially purified by 40–60% (w/v) ammonium sulphate precipitation method followed by dialysis. The molecular weights of the partially purified lipases were estimated to be 32–47 kDa by SDS-PAGE.
Keywords
Lipase
Bacterial strains
Submerged fermentation
Optimization process
SDS-PAGE analysis
1 Introduction
Lipases (triacylglycerol acylhydrolases; EC 3.1.1.3) are class of hydrolytic enzymes which catalyze the hydrolysis of triacylglycerol to glycerol and free fatty acids (Gupta et al., 2004). Lipases are widely found in animals, plants and microorganisms (Sahu and Martin, 2011; Bornscheuer, 2002), especially those originated from bacteria, are more stable than others. Bacterial lipases are commercially more important mainly because of the ease of their cultivation and optimization to obtain higher yield (Hasan et al., 2006). Industrial demand of new sources of lipases with different catalytic characteristics stimulated the isolation and selection of new strains. Lipase producing microorganisms have been found in different sources such as agro industrial waste (Salihu et al., 2012), vegetable oil processing factories (Watanabe et al., 2000), dairy plants (Sorhaug and Stepaniak, 1997) and soil contaminated with oils (Sirisha et al., 2010). Natural oil such as olive oil, coconut oil, vegetable oil, petroleum oil enhance the activity of lipase production.
From the industrial point of view, lipase enzymes are considered very important, due to their greater production potential on a large scale. Recently, research reports proved that submerged fermentation is highly effective in large scale production of industrially important enzymes such as lipases, pectinases, cellulases (Snellman and Colwell, 2004). Submerged fermentation is a promising tool in biotechnology field for the production of microbial enzymes and most appropriate production process for developing countries (Tolan and Foody, 1999). Submerged fermentation has numerous advantages over conventional solid state fermentation, including resembling the natural habitat for several microorganisms, better oxygen circulation, reduced energy and cost requirements, less operational problems, less effect in downstream processing, higher productivity, compactness of fermentation vessel, lower capital and recurring expenditure (Castilho et al., 2000; Singhania et al.,2010).
Lipases have many potential applications in various industries and are selected for each application based on its substrate specificity, position and stereospecificity as well as temperature and pH stability (Bajpai, 1999; Gandhi, 1997). They are mostly used in the detergent, food, pharmaceutical industries (Pandey et al., 1999; Ray, 2012).
In this study we report the production of lipase enzyme from bacterial strains which isolated from petrol spilled soil. We have demonstrated the optimization condition for higher yield, partial purification of lipases and its molecular mass determination.
2 Materials and methods
2.1 Isolation and characterization of bacterial strains
Petrol spilled soil sample was collected from petrol bunk situated in Coimbatore, Tamilnadu, India. Serial dilution technique was used to isolate bacterial strains. Isolated bacterial strains were subjected to Gram’s staining for morphological identification and biochemical tests such as Indole production test, Citrate utilization test, Carbohydrate fermentation test, Triple sugar iron test, Oxidase test, Catalase test, Nitrate reduction test, Hydrogen sulphide test, Methyl red and Voges–Proskauer test were performed according to Cappuccino et al. (1996).
2.2 Screening of lipase producing bacterial strains
Isolated bacterial strains were screened for their lipolytic activity on the basis of Tributyrin Agar plate assay method (TBA). The tributyrin agar was purchased from Himedia. Tributyrin agar media along with 1.0% (v/v) olive oil were prepared and sterilized at 121 °C for 15 min, and then sterilized media were poured into petriplate. Isolated strains were streaked on the tributyrin agar plate and it was incubated at 37 °C for 24 h to observe zone.
2.3 Enzyme production media
Screened positive bacterial strains were cultivated in lipase producing media for enzyme production. Lipase producing media consist of 3% yeast extract, 3% sucrose, 0.1 g (g/l) CaSO4, 0.5 g/l – KH2PO4, 0.1 g/l – MgSO4.7H20, 1% olive oil and 100 ml distilled water in a 250 ml conical flask as submerged fermentation method. Inoculated flaks were incubated at 37 °C for 24–48 h (Mobarak-Qamsari et al., 2011).
2.4 Optimization of lipase producing media
Production media was supplemented with different carbon and nitrogen source such as sucrose, glucose, lactose, peptone, yeast extract and ammonium sulphate at different concentration (1–5%) to determine the highest yield of lipase enzyme.
Microbial growth was optimized by inoculating bacteria in an autoclaved medium that had pH varying from 5 to 10 by dissolving components of the minimal medium in the buffer of desired pH. Temperature optimization was carried out by growing bacterial strains at temperature 32–40 °C in a shaking incubator. Effect of media components on lipase activity was measured using photoelectric colorimeter at 610 nm. Similar colorimetric measurement method was carried out by Schmidt and Blum (1978).
2.5 Production of lipase by pilot scale fermentor
Among the positive strains one high lipase producing strain were subjected to batch fermentation in 5litre pilot scale fermentor. Fermentations processes were carried out in a Labo controller MDL-BC fermentor. As above optimized protocol, 2.5 L of lipase producing medium was prepared. The medium was sterilized at 121 °C for 20 minutes and then sterilized media was poured into the fermentor containment system under aseptic condition. The reactor was inoculated with 5 ml of bacterial strain. Standard operation conditions were: agitation rate 150–400 rpm, temperature 37 °C, air-flow rate is 1 vvm and fermentation time 48 h, with pH 6 ± 0.5. Samples (20 ml) were taken at regular intervals, cells were removed by centrifugation at 5000 rpm for 30 min, and the culture supernatants were evaluated for enzyme activity. Biomass concentration was measured by turbidimetry at 610 nm in a colorimeter.
2.6 Partial purification of lipase enzyme
All purification steps were performed at room temperature. From the above lipase produced media 20 ml of each bacterial strain medium was taken and the cells were separated by centrifugation at 5000 rpm for 30 min. The supernatant was collected and enzyme was concentrated using addition of 10–100% ammonium sulphate. Fractionated enzyme samples were then subjected to dialysis process for partial purification with the help of dialysis membrane.
2.7 Estimation of protein content by Lowry’s method
Quantitative estimation of the protein content was done by Lowry et al. (1951) method.
2.8 Molecular weight determination by SDS-PAGE
The molecular weight of the lipase partially purified from ammonium sulphate precipitation method and dialysis was checked by Sodium Dodecyl Sulphate Poly Acrylamide Gel Electrophoresis (SDS-PAGE). It was performed as described by Borkar et al. (2009).
3 Results and discussion
Eight strains were isolated on the basis of colony morphology and the appearance on nutrient agar plates by serial dilution technique from petrol spilled soil sample. The oily environment may provide a better environment for isolation on lipase producing microorganism (Mobarak-Qamsari et al., 2011). The isolated 8 bacterial strains were designated as SP1, SP2, SP3, SP4, SP5, SP6, SP7 and SP8. Similar isolation was done by Beller et al. (1996).
By Gram’s staining, it was observed that 5 bacterial strains were gram positive and 3 bacterial strains were gram negative. Saadoun (2002) isolated 4 gram positive bacteria and 2 gram negative bacteria form petrol contaminated oil soil. Results of biochemical tests are noted in Table 1. Through biochemical tests isolated bacterial strains might be the genera of Corynebacterium sp. (SP1), Streptococcus sp. (SP2), Escherichia sp. (SP3), Proteus sp. (SP4), Bacillus sp. (SP5), Staphylococcus sp. (SP6), Pseudomonas sp. (SP7), Klebsiella sp. (SP8). This kind of genera identification of bacterial strains was supported by Cappuccino et al. (1996). Note: MR = Methyl Red; VP = Voges-Proskauer; TSI = Triple Sugar Iron. “+”sign denotes positive and “−” sign denotes negative.
Parameters
Observation
SP1
SP2
SP3
SP4
SP5
SP6
SP7
SP8
Motility test
−
+
+
−
−
+
+
+
Indole production test
−
+
+
+
+
−
−
+
MR test
−
+
+
+
−
−
−
+
VP test
−
+
+
−
−
−
+
+
Citrate utilization test
+
+
−
+
−
+
+
−
Carbohydrate fermentation test
+
−
−
+
+
−
−
+
TSI test
−
−
−
−
−
+
+
−
Oxidase test
+
−
+
−
−
−
+
+
Urease test
−
+
−
−
+
−
−
Catalase test
+
−
+
−
+
−
+
+
Nitrate reduction test
−
+
−
−
−
−
−
+
Hydrogen sulphide test
−
+
+
+
+
+
−
+
Based on the clear zone production on tributyrin agar plate, 5 bacterial strains were identified as lipase producers (Fig. 1). Among the 8 bacterial strains, SP1, SP5, SP7 showed high intensity of clear zone, SP3 and SP8 strains showed moderate intensity of clear zone and other 3 (SP2,SP4,SP6) strains did not show any zone around the colonies. Olive oil used as the substrate for screening of lipase producing bacterial strains. Among natural oils, olive oil has been referred as one of the best inductors and substrate for lipase production (Bornscheuer, 2002).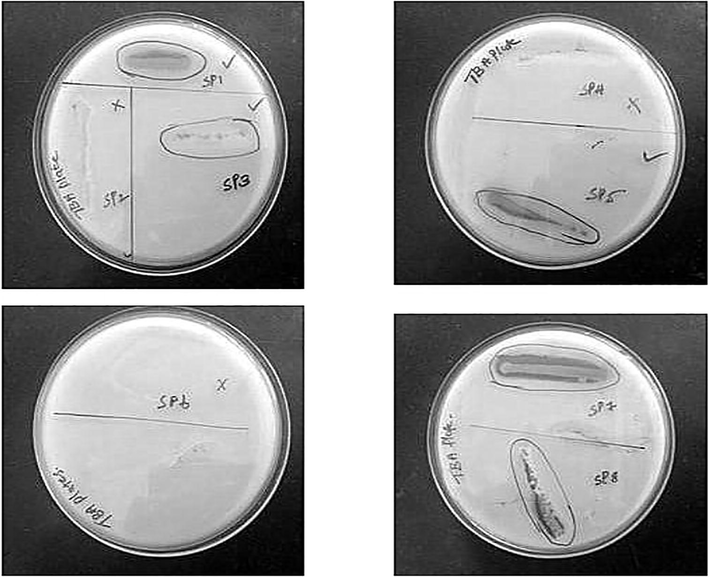
Screening of lipase activity in Tributyrin Agar plates. SP1, SP3, SP5, SP7, SP8 showed clear zone and other SP2, SP4, SP6 strains did not show any zone around the colonies. SP1 – Corynebacterium sp., SP2 – Streptococcus sp., SP3 – Escherichia sp., SP4 – Proteus sp., SP5 – Bacillus sp., SP6 – Staphylococcus sp., SP7 – Pseudomonas sp. and SP8 – Klebsiella sp. SP denotes sample from petrol.
Media components were optimized by single factor analysis. 3% Sucrose showed higher lipase production as a carbon source. Sucrose stimulate the gene expression which produce high lipase content (Mazhar et al., 2017) It has been reported that maximum lipase production was obtained when sucrose used as carbon source (Veerabagu et al., 2013). 5% Yeast extract produced higher biomass in lipase producing media as a nitrogen source. Increasing concentration of nitrogen source produced higher biomass, but reduces the lipase activity (data not shown).
Bacterial strains showed high yield of lipase production at pH 6 ± 0.5. It has been reported that maximum lipase production was achieved at pH at 6–7 (Larbidaouadi et al., 2015). Optimum temperature plays a vital role for the production of enzyme in shake flask method. Higher biomass concentration of lipase was observed at temperature 37 °C. Researchers reported that the slight increase in temperature up to 38 °C enhance the lipase production (Zhang and Hebin, 2005; Yuan et al., 2016; Gaur et al., 2008).
Shake flask method were used for the production of lipase enzyme from 5 lipase producing bacterial strains. After 48 h of incubation, biomasses from all five bacterial flasks were collected for further studies. From above 5 strains, SP5 strain showed high biomass production in lipase producing media. Potential bacterial strain SP5 Bacillus sp. was cultivated in the laboratory batch fermentor in constant pH at 6 ± 0.5. The extracellular lipase secretion was started about 12–16 h and then reached maximum lipase activity at 46–48 h in pilot scale fermentor. The increase of the lipase production was mainly through the increase in cell mass. There is overall increase in the production of extracellular lipase as compared to shake flask method. Under the experiment conditions, the growth (O.D.610) of organisms started (0.4 at 610 nm) at 2 h and reached maximum (3.4) at 48 h.
The cell-free supernatant was used as a crude enzyme for precipitation and dialysis process. The protein was precipitated using ammonium sulphate fractions from 20 to 80% (w/v). 40–60% (w/v) of ammonium sulphate fractions showed higher lipase precipitation activity than the rest of the fractions. The activity was not observed in and above 80% (w/v) saturation. Precipitated enzyme was purified by dialysis method with the help of dialysis membrane.
The total protein content was determined by Lowry’s method. Protein content of lipases was shown in Table 2. Maximum protein content was observed in FS followed by SP1, SP7, SP3, SP8 and SP5. Separated proteins of supernatants from different bacterial strains were separated by SDS-PAGE and are depicted in Fig. 2. Form SDS-PAGE analysis, molecular weight of the lipase has different range starting from 32 to 47 kDa. It suggested that each bacterial strain has different molecular weight of lipase enzyme and also different amount of protein content. This is due to the genetic diversity of bacterial species. There are also different molecular weight of lipase has been reported from Pseudomonas aeruginosa ATCC 27,853 (Izrael‐Zivkovic et al., 2009), Bacillus sp. MPTK 912 (Mukesh Kumar et al., 2012), Pseudomonas sp. ADT3 (Dey et al., 2014) possessing of 30 kDa, 66 kDa and 13.9 kDa, respectively. Note: SP denotes sample from petrol and FS denotes fermentor sample.
Bacterial strains
Estimated protein content (mg/ml)
SP1
1.52
SP3
1.37
SP5
1.10
SP7
1.46
SP8
1.20
FS
1.83

Molecular weight determinations by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). M – Middle level marker (32–64 kDa), SP1 – Corynebacterium sp. (32 kDa), SP3 – Escherichia sp. (44 kDa), SP5 – Bacillus sp. (44 kDa), SP7 – Pseudomonas sp. (37 kDa), SP8 – Klebsiella sp. (43 kDa), and FS – Bacillus sp. (47 kDa). Samples SP1, SP3, SP5, SP7 and SP8 were produced by shake flask method and FS sample was produced by pilot scale fermentor. SP denotes sample from petrol and FS denotes fermentor sample, kDa denotes Kilo Daltons.
4 Conclusion
From the present study it can be concluded that petrol spilled soil has formed an industrially useful source of bacteria, that can be used for potential industrial importance. The lipase enzyme production was optimized with different physiological conditions and high yield produced using pilot scale fermentor. Sucrose and yeast extract could be a better source for maximum lipase production. Partial purification of the enzyme was done using 40–60% ammonium sulphate precipitation method. The results obtained after molecular weight determination by SDS–PAGE indicate that the produced lipases have different molecular weight this might be the production by different bacterial strains.
Conflicts of interest
The authors of the present work report that there is no conflict of interest in this work.
References
- Application of enzymes in the pulp and paper industry. Biotechnol. Progr.. 1999;15(2):147-157.
- [Google Scholar]
- Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. Appl. Environ. Microb.. 1996;62(4):1188-1196.
- [Google Scholar]
- Purification and characterization of extracellular lipase from a new strain: Pseudomonas aeruginosa SRT 9. Braz. J. Microbiol.. 2009;40(2):358-366.
- [Google Scholar]
- Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev.. 2002;26(1):73-81.
- [Google Scholar]
- Microbiology: A Laboratory Manual (No. QR 63. C36 1996). 1996.
- Economic analysis of lipase production by Penicillium restrictum in solid-state and submerged fermentations. Biochem. Eng. J.. 2000;4(3):239-247.
- [Google Scholar]
- Production, partial purification and characterization of an extracellular psychrotrophic lipase from Pseudomonas sp. ADT3. J. Bioremed. Biodeg.. 2014;5(242):2.
- [Google Scholar]
- Purification and characterization of lipase from solvent tolerant Pseudomonas aeruginosa PseA. Process Biochem.. 2008;43(10):1040-1046.
- [Google Scholar]
- Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol.. 2004;64(6):763-781.
- [Google Scholar]
- Industrial applications of microbial lipases. Enzyme Microb. Tech.. 2006;39(2):235-251.
- [Google Scholar]
- Enzymatic characterization of 30 kDa lipase from Pseudomonas aeruginosa ATCC 27853. J. Basic Microbiol.. 2009;49(5):452-462.
- [Google Scholar]
- Screening selection identification production and optimization of bacterial lipase isolated from industrial rejection of gas station. Int. J. Biotechnol. Allied Fields. 2015;3(9):146-153.
- [Google Scholar]
- Optimized production of lipase from Bacillus subtilis PCSIRNL-39. Afr. J. Biotechnol.. 2017;16(19):1106-1115.
- [Google Scholar]
- Isolation and identification of a novel, lipase-producing bacterium, Pseudomnas aeruginosa KM110. Ran. J. Micro.. 2011;3(2):92-98.
- [Google Scholar]
- Production, optimization and purification of lipase from Bacillus sp. MPTK 912 isolated from oil mill effluent. Adv. Appl. Sci. Res.. 2012;3(2):930-938.
- [Google Scholar]
- The realm of microbial lipases in biotechnology. Biotechnol. Appl. Bioc.. 1999;29(2):119-131.
- [Google Scholar]
- Isolation and characterization of bacteria from crude petroleum oil contaminated soil and their potential to degrade diesel fuel. J. Basic Microbiol.. 2002;42(6):420-428.
- [Google Scholar]
- Optimization of growth conditions for the production of extracellular lipase by bacterial strains from dairy industry effluents. Biotechnol. Bioinf. Bioeng.. 2011;1(3):305-311.
- [Google Scholar]
- Lipase production: an insight in the utilization of renewable agricultural residues. Resour. Conserv. Recycl.. 2012;58:36-44.
- [Google Scholar]
- The biochemical constituents of the venom of the harvester ant, Pogonomyrmex badius. Comp. Biochem. Phys. C. 1978;61(1):239-247.
- [Google Scholar]
- Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb. Tech.. 2010;46(7):541-549.
- [Google Scholar]
- Isolation and optimization of lipase producing bacteria from oil contaminated soils. Adv. Bio. Res.. 2010;4(5):249-252.
- [Google Scholar]
- Acinetobacter lipases: molecular biology, biochemical properties and biotechnological potential. J. Ind. Microbiol. Biot. 2004;31(9):391-400.
- [Google Scholar]
- Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends food sci. Tech.. 1997;8(2):35-41.
- [Google Scholar]
- Cellulase from submerged fermentation. Rec. Progr. Bioconv. Lignocellulosics 1999:41-67.
- [Google Scholar]
- Screening selection identification production and optimization of bacterial lipase from oil spilled soil. Asian J. Pharm. Clin. Res.. 2013;6(3):62-66.
- [Google Scholar]
- Continuous production of biodiesel fuel from vegetable oil using immobilized Candida antarctica lipase. J. Am. Oil Chem. Soc.. 2000;77(4):355-360.
- [Google Scholar]
- Screening and characterization of thermostable lipase from marine Streptomyces Sp. Strain w007. Biotech. Appl. Biochem.. 2016;63(1):16-22.
- [Google Scholar]
- Characterization of thermostable lipase from thermophilic Geobacillus Sp.TW1. J. Pep.. 2005;42(1):153-159.
- [Google Scholar]







