Translate this page into:
Multiple resistance mechanisms in Staphylococcus sp. strain AS6 under arsenite stress and its potential use in amelioration of wastewater
⁎Corresponding author: Department of Microbiology & Molecular Genetics University of the Punjab, New Campus, Lahore 54590, Pakistan. rehman.mmg@pu.edu.pk (Abdul Rehman)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The multiple metal resistant Staphylococcus sp. strain AS6, isolated from wastewater of Pakistan, was able to resist 25 mM arsenite and 150 mM arsenate. SEM analysis showed that no significant change in bacterial morphology under arsenite exposure was observed while EDX and FTIR analyses confirmed the surface adsorption and uptake of arsenite into the bacterial cells exposed to 15 mM arsenite. The GSH/GSSG ratio and NPSHs were increased 45.0 and 76.50% in 15 mM arsenite stress while antioxidant enzymes (SOD, CAT, POX, and APX) showed varied response in arsenic presence. The genome of strain AS6 was sequenced through Illumina NextSeq 550 sequencer and the genes confer resistance to arsenic such as arsR, arsB, and arsC were identified. The bacterial stain AS6 was able to oxidize arsenite 91%/8h and removed 93%/10 h arsenite through its inactivated biomass from the medium. The strain AS6 has great potential, due to its hyper-metal resistance and high arsenite oxidation ability, to be used as a bioresource for green chemistry to eliminate toxic arsenite from the environment.
Keywords
Arsenite
Heavy metals
Staphylococcus sp. strain AS6
Glutathione
Antioxidant enzymes
Bioremediation
1 Introduction
Due to natural geochemical and anthropogenic activities arsenic (As) contamination has become a worldwide problem (Islam et al., 2004). Volcanic activities and weathering of rocks are the main natural sources of arsenic while anthropogenic source is use of arsenic containing compounds such as pesticides, dyes and preservation of wood through which arsenic is increasing in water bodies (Kumari et al., 2018). It is well known carcinogenic for living organisms especially for human beings (Mead, 2005). Prasad et al. (2013) reported that more than 40 million people are being exposed to arsenic above 50 ppb while the limit permitted through the Environmental Protection Agency (EPA) is 10 ppb.
Arsenic toxicity depends on two main factors, one is its chemical form and other is its oxidation states (Rosen, 2002). Inorganic form of arsenic is more toxic than organic one, while arsenite with oxidation state + 3 is 100 times more toxic than arsenate with oxidation state + 5 (Mujawar et al., 2019). Arsenic, which is not only a substantial cause of health risks in Pakistan, has been reported that it effects on human beings in major parts of the world especially northeast India, northwest part of the USA and Bangladesh (Muller et al., 2003). The EPA of the USA places it at the top of list for hazardous substances due to its toxicity (Zhang et al., 2016). The drinking of arsenic containing water for long period of time causes various health related problems in human beings like, change in color of skin or cancer, diabetes, hypertension, chromosomal aberrations, amplification of gene, alternation in eukaryotic cell morphology as well as some disorder which are linked to reproduction system (Smith et al., 2000; Sher and Rehman, 2019).
Microorganisms are found in every kind of environment and have potential to reduce or oxidize arsenic (Oremland and Stolz, 2003; Koechler et al., 2010). The conversion of arsenate (As+5) into arsenite (As+3) is called reduction and is carried out by arsC gene located on chromosomal DNA or plasmid inside the bacteria (Li et al., 2010). On the other hand, oxidation involves the change of As+3 into As+5 which is carried out by aioA and aioB genes present in bacteria (Li et al., 2014). Arsenic toxicity can also be reduced with the process of methylation, in which methyltransferase gene (arsM) uses S-adenosylmethionine (SAM) as a source of methyl group for the addition in arsenic (Huang et al., 2018).
The conventional chemical methods can be employed for arsenic eradication from water, such as membrane filtration, coagulation, ion exchange method, nanoparticles, and some other chemical methods (Ng et al., 2004; Mohanty, 2017). These methods cannot be used further because of non-cost effective and production of secondary toxic compounds (Tariq et al., 2019). The best eco-friendly approach for arsenic detoxification is bioremediation, in which bacteria or other microorganisms used toxic compounds as a source of energy in their metabolism and convert toxic form into less or non-toxic form (Qin et al., 2006; Tariq et al., 2019). Das and Barooah (2018) reported that Staphylococcus sp. AT6 was able to resist As+3 (30 mM) and As+5 (250 mM) along with siderophore production. Similarly, Rathod et al. (2019) isolated a bacterium, Staphylococcus sp. As-3, from a sediment core sample collected from the Budai borehole, Taiwan, that could resist As+3 and As+5 upto 7.5 and 200 mM, respectively. Many researchers have used a variety of microorganisms for the purpose of metals amelioration from the environment (Srivastava et al., 2012; Dey et al., 2016; Das and Barooah, 2018; Tariq et al., 2019; Sher et al., 2020).
The present work objectives were to isolate heavy metals resistant bacteria from the industrial waste, characterize and metal-microbe interaction through SEM, EDX, and FTIR analyeses. Moreover, antioxidant enzymes activities, glutathione, non-protein thiols concentration, and arsenic bioremediation potential of the strain were also ascertained. This investigation would present an efficient strategy for the arsenite oxidation, and provides a novel microbial resource for arsenic eradication.
2 Materials and methods
2.1 Samples collection and arsenite resistant bacterial isolation
The wastewater samples were collected from the industrial area of District Sheikhupura (Fig. S1), Punjab, Pakistan. Physicochemical characteristics of wastewater samples including pH, temperature, color, turbidity, total dissolved solid (TDS), electrical conductivity, and arsenic concentration were determined. For bacterial isolation, sample of 100 µl was spread on LB-agar plates already supplemented with arsenite and incubated at 37 °C for overnight. Afterwards, bacteria were purified by streaking and re-streaking on LB-agar plates.
Minimum inhibitory concentration (MIC) against arsenic was determined by growing the bacteria in different flasks containing MS-broth according to procedure described in Naureen and Rehman (2016). Flasks containing As+3 from 5 to 50 mM and As+5 from 5 to 250 mM separately were placed in a shaking incubator at 37 °C with 100 rpm for 48 h. After incubation, one ml was drawn from each flask and optical density (OD), as a function of cell growth, was measured at OD600 nm with the help of spectrophotometer.
2.2 Morphological, biochemical and molecular characterization of bacterial isolate
The isolate AS6 showed maximum resistance against arsenite i.e. 25 mM was selected for further research work. Various morphological and biochemical characteristics of the bacterial isolate were monitored (Table 1) according to procedures described in Cappucino and Sherman (2001). Molecular characterization was performed according to Sher and Rehman (2019). Briefly, DNA was extracted by using the MasterPure™ complete DNA and RNA purification kit (Lucigen, WI, USA). Illumina sequencing libraries were prepared using the Nextera XT sample preparation kit (Illumina, CA, USA), and sequencing was performed by an Illumina NextSeq 550 sequencer. The obtained sequences were submitted to GenBank for the assignment of accession number.
Genes and its position
Product
Putative function
Closest related sequence
% Query coverage
% Ident
arsC2_1 79013–79408
Arsenate mycothiol transferase ArsC2
Play role in arsenic reduction
WP_019469651.1
99%
100%
arsB_1 79426–80013
Arsenical pump membrane protein
Arsenite efflux transporter
WP_064264181.1
91%
100%
arsB_2 80117–80716
Arsenical pump membrane protein
Regulate the genes responsible for arsenic reduction
KKI63246.1
92%
100%
arsC2_1 13183–13578
Arsenate mycothiol transferase ArsC2
Play role in the reduction of arsenic
WP_002509695.1
99%
100%
arsB_3 13596–14885
Arsenical pump membrane protein
Arsenite efflux transporter
WP_013730039.1
96%
100%
arsA 16863–18590
Arsenical pump- driving ATPase
Transport arsenite, arsenate and antimony from out of cell
WP_115041147.1
99%
100%
arsD 18571–18918
Arsenical resistance operon trans-acting repressor ArsD
Arsenic efflux transporter metallochaperone
WP_019467779.1
90%
100%
arsR 19447–19767
Arsenical resistance operon repressor
Regulate arsenic resistance operon
WP_013730044.1
99%
100%
2.3 Optimum growth conditions and growth curves
The bacterium optimum growth conditions i.e. pH and temperature were determined according to procedure described in Elahi and Rehman (2019). Bacterial growth curves were prepared according to Elahi and Rehman (2019).
2.4 Heavy metals resistance
Multiple metal resistance of strain AS6 was ascertained against cadmium chloride, cobalt chloride, potassium dichromate, lead nitrate, mercuric chloride, nickel chloride and sodium selenite according to procedure described in Elahi and Rehman (2019).
2.5 Scanning electron microscope (SEM) and energy dispersive X-ray (EDX) and fourier transform infrared spectroscopy (FTIR) analysis
The strain AS6 was cultured in LB-broth containing arsenite (15 mM) and without arsenite under its optimum growth conditions for 24 h. For SEM analysis, cell's suspension was put on aluminum stub. The fixation of cells was done with glutaraldehyde (2.5%) in PBS with pH 7 and was placed at room temperature for 30 min. The cells were washed with PBS and then dehydrated with different concentrations of acetone i.e. 30, 50, 70, 80, 90, and 100% in a step-by-step manner with a regular interval of 10 min. The treatments were covered with gold film by a sputter coater (Denton, Desk V HP) and assessed through scanning electron microscope (Nova NanoSEM 450) equipped with Oxford energy dispersive X-ray (EDX) microanalysis system (Khan et al., 2016).
The FTIR samples were prepared according to procedure described in Mujawar et al. (2019). First of all, strain AS6 was grown under arsenite stress (15 mM) and without arsenite at its optimum conditions for 24 h. The pellet (cells) was obtained after centrifugation at 3000 rpm for 10 min. The pellets were washed with normal saline several times and freeze dried for overnight. The infrared spectra were recorded in the region of 4000 to 500 cm−1 through the FTIR spectrometer (Bruker, alpha).
2.6 Estimation of glutathione and other non-protein thiol contents
Reduced glutathione (GSH), oxidized glutathione (GSSG), and other non-protein thiols (NPSHs) contents were determined for the bacterial strain in the presence and absence of arsenite, according to procedure described in Shamim and Rehman (2015).
2.7 Quantification of antioxidant enzymes under arsenite stress
Catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), and peroxidase (POX) were quantified with As+3 stress (10 mM) and without As+3 stress. For this, cells were grown at optimum conditions for 48 h with and without arsenite stress. The cells were centrifuged at 10000 rpm (4 °C) for 5 min to collect pellets which were then washed by a phosphate buffer. Cells lysis was done by adding lysozyme and placed on ice for 10 min to homogenize. The cell's suspension was centrifuged at 16000 rpm (5 min) and the obtained supernatant was used for enzymes quantification. The protein assay kit was used to measure total protein content through Qubit fluorometer.
2.8 DNA extraction, genome sequencing and annotattion
The DNA extraction, sequencing, and genes annotation were performed according to Sher et al. (2019).
2.9 Arsenite oxidizing and removal by bacterial strain AS6
2.9.1 Arsenite oxidizing potential of Staphylococcus sp. strain AS6
The arsenic oxidizing potential of strain AS6 was determined by culturing it in 250 ml flask containing 100 ml of LB broth medium. The experiment was preceded in 3 flasks containing 250 mM As+3/l. One flask was used as control, containing arsenite with the same concentration but without bacterial culture. The arsenite oxidizing potential was determined for 96 h with a regular interval of 24 h. The broth (5 ml) was taken out from each flask after 24, 48, 72, and 96 h and was centrifuged at 3000 rpm for 5 min to collect the bacterial cells. The P S Analytical Millennium Excalibur Method (Department of ENVS, Biosciences Section Aarhus University, Denmark) was employed for arsenic estimation.
2.9.2 Arsenic removal through inactivated biomass
To obtain inactivated bacterial biomass, the bacterial strain was grown in 250 ml flask containing one liter LB medium and incubated at optimum conditions to harvest maximum growth (Tariq et al., 2019). Then the culture was centrifuged at 4000 rpm for 10 min and the pellet was washed several times with deionized water. The bacterial pellet was incubated at 70 °C to achieve powder form of cells. The procedure was repeated again and again (almost 10–15 times) to get a significant amount of bacterial biomass.
Arsenic removal experiment was run by a biosorption process with 1000 mM arsenic stress. Initially, 1 g/L bacterial biomass was mixed in 1 L of arsenic solution of 1500 ml flasks containing 1000 mM As+3 stress. The flasks were incubated at optimum conditions on the shaker for 10 h. Then after a regular interval of 2 h an aliquot of sample was drawn, filtrated (0.22 μm filter paper), and was frozen. The atomic absorption spectrophotometer was employed for arsenic determination. Finally, the amount of arsenite adsorbed in grams of biomass (q) and bioremediation efficiency (E) were calculated by given equations.
Ci and Cf represent initial and final As+3 concentration, m represents mass of biosorbent in the reaction, and V indicates volume of the mixture.
2.10 Statistical analysis
All the treatments were run in triplicate and three separate flasks were usually maintained for each treatment. For each treatment three readings were taken, their mean, and standard error of the mean were determined.
3 Results
3.1 Wastewater characteristics and heavy metal ions resistant bacterial isolation
The collected samples temperature range was 24 to 32 °C and pH ranged between 7.2 and 8.8. The light black sample color was observed and concentration of arsenic was 200 μg/ml. Wastewater samples characteristics are given in Table S1. The MIC resisted by the bacterium against arsenite and arsenate was 25 and 150 mM. Besides this, strain AS6 also showed fair resistance to other metal ions i.e. Cr (5 mM), Cd (3 mM), Pb (5 mM), Co (3 mM), Se (4 mM), Hg (2.5 mM), and Ni (5 mM) (Table S2). The resistance pattern of strain AS6 against metal ions is As+5 > As+3 > Cr+6 = Pb+2 = Ni+2 > Se+2 > Cd+2 = Co+2 > Hg+2.
3.2 Bacterial characteristics
The isolate was circular, yellow in color, non-motile, and Gram-positive, stained purple with Gram-stain. The bacterium was also positive for catalase, citrate, nitrate reduction, and Voges-Proskauer test (Table S3). The complete genome sequence of bacterial strain AS6 has been submitted to GenBank under accession number of VSRZ00000000. The bacterial strain AS6 has also been depsited to First Culture Bank of Pakistan (FCBP), University of the Punjab, Lahore, Pakistan with the accession number of FCBP-B-733.
3.3 Optimum growth conditions and growth curves
The bacterium showed optimum growth at 37 °C and pH 7 (Fig. S2a,b). In the beginning bacterial log phase was slightly delayed in arsenite stress as compared to the control i.e. medium containing no arsenite stress. The growth rate was steady although less than control and after 20 h of growth it started to decline in the presence of arsenite (Fig. 1).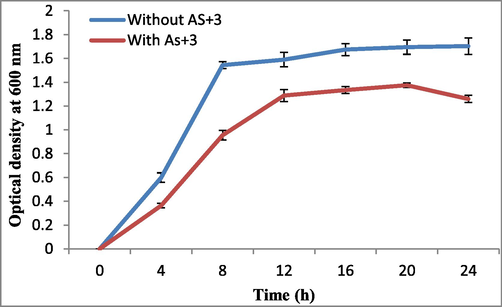
Growth curves of Staphylococcus sp. strain AS6 in the absence and presence of arsenite at 37 °C and pH 7.
3.4 SEM, EDX, and FTIR analysis
The SEM analysis showed that there was no considerable change in size was determined with and without arsenite in the isolated bacterial strain AS6 (Fig. 2a). The EDX results confirmed arsenite surface absorption in bacterial cells treated with arsenite (15 mM) while no surface arsenite was determined in bacterial cells without arsenite stress (Fig. 2b).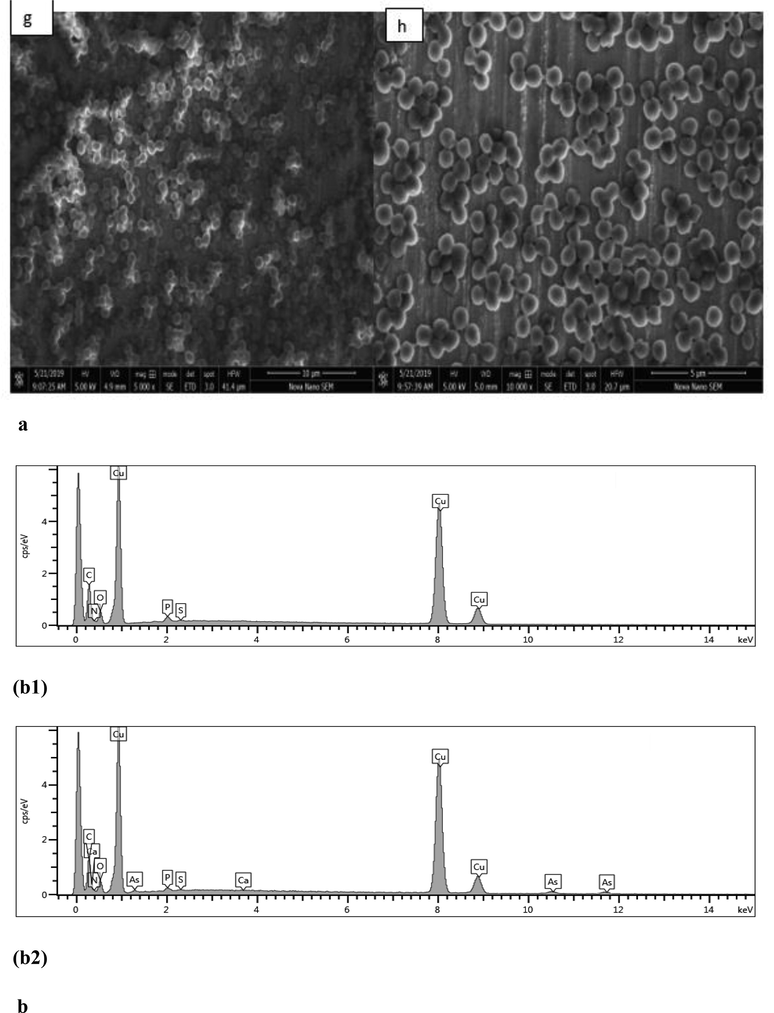
(a) Bacterial morphology under SEM; Bacterial cells in exponential growth phase without exposure to arsenite showing cocci shape morphology (g) while showing no change in bacterial cells shape with 15 mM arsenite (h), (b) EDX spectrum of bacterial cells in exponential phase without arsenite exposure (b1) and with 15 mM arsenite exposure (b2).
The FTIR analysis showed the shifting and sharpening of many peaks in bacterial strain treated with 15 mM arsenite, which could be allocated to various functional groups which might be able to adsorb or uptake As+3 into the bacterial cell. The stretching of amide and hydroxyl groups is responsible for a change in the region of 3278 to 2851 cm−1. The amide linkages from peptides and proteins are responsible for peaks shifting from 1741 to 1220 cm−1. The C-N stretching from an aliphatic amine and C-O stretching from an alcohol, carboxylic acid, and ester are behind the shift of the peaks from 1228 to 1038 cm−1 (Fig. 3).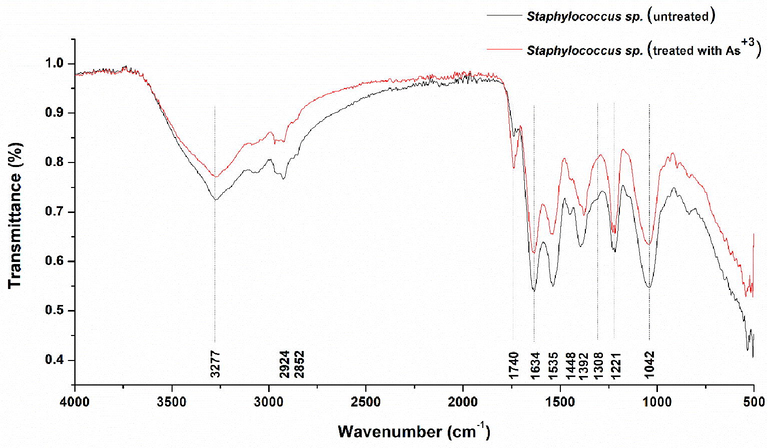
FTIR spectrum from bacterial cells of strain AS6, red line belongs to cell exposed to 15 mM arsenite and black line represents control (without arsenite).
3.5 Determination of GSH and NPSHs
GSH, GSSG, and NPSHs showed varied responses in strain AS6 in arsenite presence (15 mM). The increase (%) in GSH/GSSG ratio and NPSHs was 45.0 and 76.50%, respectively. Table S4 is showing the concentration of GSH, GSSG, total glutathione, GSH/GSSG ratio, and NPSHs in the presence and absence of arsenite.
3.6 Antioxidant enzymes
The concentration of CAT, APX, POX, and SOD was estimated in arsenite stress (10 mM) in bacterial strain AS6 and a varied response of antioxidant enzymes was found under stress. The concentration of SOD and POX was decreased while CAT (100%) and APX (19%) activity was increased (Table S5).
3.7 Arsenic and other heavy metal genes determinants
Firstly, the genes responsible for arsenic resistance i.e. arsC, arsB, arsR, arsA, and arsD were determined in the genome of the isolated bacterium (Fig. 4; Table 1). Secondly, the genes confer resistance against other metal ions including zinc, chromium, magnesium, and cadmium were also identified (Fig. S3; Table S6).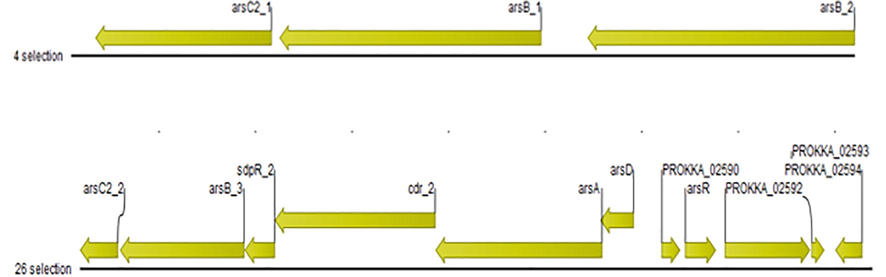
Genes responsible for arsenic resistance in Staphylococcus sp. strain AS6.
3.8 Arsenic bioremediation by bacterial strain AS6
Firstly, the strain was cultured under optimum conditions for 96 h and arsenite oxidizing potential was determined after a regular interval of 24 h up to 96 h. The metal oxidizing potential shown by Staphylococcus sp. strain AS6 was 36, 59, 78, and 91% after 24, 48, 72, and 96 h (Fig. 5a).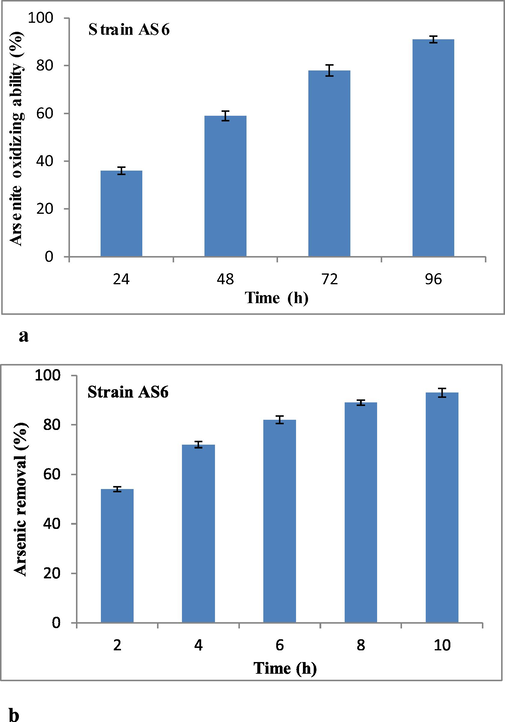
(a) Arsenite oxidizing ability of strain AS6 up to 96 h of incubation at optimum growth conditions, (b) Arsenite removal ability of strain AS6 biomass from the medium after 2, 4, 6, 8, and 10 h of contact time.
Secondly, the arsenic removal potential of bacterial biomass was estimated for 2 to 10 h from the medium containing 1000 mM arsenite stress. Bacterial biomass has shown high efficiency to remove 54, 72, 81, and 89% arsenite from the medium after 2, 4, 6, and 8 h of incubation, respectively. The strain AS6 biomass removed 93% of arsenite after 10 h of incubation showing its potential to be used in amelioration of metal contaminated sites (Fig. 5b).
4 Discussion
In this study, strain AS6 has MIC 25 mM against arsenite (a toxic form of arsenic) while the MIC of arsenite in Klebsiella pneumonia is 21 mM (Mujawar et al., 2019). Bacillus cereus has 40 mM MIC against arsenite (Naureen and Rehman, 2016). The MIC of strain AS6 for arsenate was 150 mM while another study from this laboratory reported that the MIC for arsenate in Brevibacterium sp. strain CS2 and Micrococcus luteus strain AS2 is 275 and 280 mM, respectively (Sher et al., 2019). Manzoor et al. (2019) reported that Pseudomonas sp. strain PG-12 showed resistance to various metal ions besides arsenic. The strain PG-12 resisted Cd and Pb up to 10 and 0.6 mM. Elahi and Rehman (2019) reported that the multiple metal tolerance in S. sciuri A-HS1 is 18.5 mM Pb, 2.5 mM Cu, 3 mM Cd, and 25 mM Cr.
The strain AS6 showed peaks under EDX in As+3 stress (15 mM) while no peaks were found in samples without As+3. The EDX spectrum confirmed that As+3 adsorbed on the bacterial cell surface while similar peaks were also found in strain RJB-2 and K. pneumoniae strain SSSW7 (Mujawar et al., 2019). The FTIR analysis of strain AS6 under As+3 stress shows that there is some sort of interaction between As+3 and functional groups (carboxyl, hydroxyl, and amino group) present on the surface of bacterial cell wall. Many studies reported that similar functional groups interact with arsenite in bacterial strains including Arthrobactor sp., Bacillus aryabhattai, and E. coli (Prasad et al., 2013; Singh et al., 2016). Arsenic produces ROS being oxidizing agent and antioxidant enzymes protect the cells against ROS (Hughes et al., 2011; Jha et al., 2015). In the present study, it was noted that CAT production against ROS was significant as compared to the APX. Another study reported that Enterobactor sp. MUM2 increased CAT activity 4.6 folds in arsenite presence (9 mM) as compared to the non-stressed bacterial culture and no considerable change was determined in APX and POX activities (Jobby et al., 2016).
In the current study, the genes confer resistance against arsenic i.e. arsC, arsB, arsR, arsA, and arsD were found in the bacterial genome and play roles in the reduction of arsenic, arsenite and arsenate transport, arsenate efflux, and overall regulate arsenic resistance operon (Table 1). Butcher et al. (2000) reported that the genes e.g. arsC, arsB, arsH, and a putative arsR are responsible to confer resistance in T. ferrooxidans against arsenite. Jia et al. (2019) reported that two ars operons inside Bacillus strain PVR-YHB1-1 are present: 1st operon is arsRacr3arsCDA and 2nd operon is arsRKacr3arsC. Cai et al. (2009) reported that the genes related to arsenic resistance i.e. aoxB, acr3, and arsB are commonly found in arsenic resistant bacterial genera including Agrobacterium, Pseudomonas, and Achromobacter. czcD_1 gene with putative function to transport inorganic ion transport and metabolism and cadA_1 with putative function to play a role in translocating cadmium and other heavy metal divalent ions are also present in the isolated strain AS6 (Table S6).
The strain AS6 was checked for its ability to oxidize arsenic for 96 h with the interval of 24 h and bacterium oxidized 91% arsenite after 96 h. Another study reported that one bacterial strain, Thermus HR13, was able to oxidize arsenite 100% within 16 h of incubation (Gihring and Banfield, 2001). Research showed that Stenotrophomonas panacihumi was capable to oxidize 500 μM As+3 within 12 h of incubation (Bahar et al., 2012). The bacterial inactivated biomass was also used to remove arsenite from the medium for 10 h with regular interval of 2 h and strain AS6 efficiently removed 93% arsenite after 10 h while P. aeruginosa strain ATCC27853 has removal ability of 90.72% after 30 min and 98% after 2 h of incubation (Tariq et al., 2019). Multiple metal tolerance, high arsenite oxidation potential (91%), and efficient arsenite removal ability (93%/10 h) make this bacterium indispensable for metal removal strategies.
5 Conclusions
In conclusion, the isolated bacterial strain AS6 has high resistance against As and other heavy metal ions i.e. Zn, Cd, Hg, Ni, Co, and Cr. The analysis of EDX and FTIR has confirmed the interaction of arsenite with the outer surface of bacterium. Antioxidant enzymes showed varied response and CAT activity was almost doubled in arsenite stress as compared to the non-stressed cells. The genes responsible to confer resistance against arsenic as well as other metal ions are present in the bacterial genome. The arsenite oxidizing potential of strain AS6 was 36, 59, 78, and 91% after 24, 48, 72, and 96 h and bacterial inactivated biomass has removed 54, 72, 81, 89, and 93% arsenite from the medium after 2, 4, 6, 8, and 10 h of incubation. Several industries in Pakistan release arsenic containing wastes into open land to corrupt the environment. So the isolated bacterial strain AS6 can be employed for the treatment of wastewater containing toxic metal ions. Further research work is needed to explore its molecular biology and investigate arsenite oxidizing potential of strain AS6 from the real wastewater so that it can become an attractive environmental tool for green chemistry.
Acknowledgements
Prof. Lars Hestbjerg Hansen and Tue Kjærgaard Nielsen, Department of Environmental Science, Aarhus University, Denmark, are highly acknowledged for providing research facilities. Higher Education Commission (HEC), Pakistan is also gratefully acknowledged for providing funds to the first author to visit Denmark under IRSIP program.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Arsenic bioremediation potential of a new arsenite-oxidizing bacterium Stenotrophomonas sp. MM-7 isolated from soil. Biodegradation. 2012;23:803-812.
- [Google Scholar]
- The chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol.. 2000;66:1826-1833.
- [Google Scholar]
- Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol.. 2009;9:4.
- [Google Scholar]
- Microbiology: A Laboratory Manual (sixth ed.). Benjamin Cummings, San Francisco: Pearson Education; 2001.
- Characterization of siderophore producing arsenic-resistant Staphylococcus sp. strain TA6 isolated from contaminated groundwater of Jorhat, Assam and its possible role in arsenic geocycle. BMC Microbiol.. 2018;18(1):104.
- [Google Scholar]
- Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol. Rep.. 2016;10:1-7.
- [Google Scholar]
- Multiple metal resistance and Cr6+ reduction by bacterium, Staphylococcus sciuri A-HS1, isolated from untreated tannery effluent. J. King Saud Uni. Sci.. 2019;31:1005-1013.
- [Google Scholar]
- Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett.. 2001;204:335-340.
- [Google Scholar]
- Arsenic methylation by a novel ArsM As(III) S-adenosylmethionine methyltransferase that requires only two conserved cysteine residues. Mol. Microbiol.. 2018;107:265-276.
- [Google Scholar]
- Arsenic exposure and toxicology: a historical perspective. Toxicol. Sci.. 2011;123(2):305-332.
- [Google Scholar]
- Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature. 2004;430(6995):68-84.
- [Google Scholar]
- Differential expression of antioxidant enzymes during degradation of azo dye reactive black 8 in hairy roots of Physalis minima L. Int. J. Phytoremediat.. 2015;17(4):305-312.
- [Google Scholar]
- Efficient arsenate reduction by As-resistant bacterium Bacillus sp. strain PVR-YHB1-1: Characterization and genome analysis. Chemosphere. 2019;218:1061-1070.
- [Google Scholar]
- Differential expression of antioxidant enzymes under arsenic stress in Enterobacter sp. Environ. Prog. Sustain. Energy. 2016;35:1642-1645.
- [Google Scholar]
- Cadmium resistance and uptake by bacterium, Salmonella enterica 43C, isolated from industrial effluent. AMB Express. 2016;6(1):1-16.
- [Google Scholar]
- Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol.. 2010;1:53-55.
- [Google Scholar]
- Prospects of genetic engineering utilizing potential genes for regulating arsenic accumulation in plants. Chemosphere. 2018;211:397-406.
- [Google Scholar]
- Arsenic resistance operon structure in Leptospirillum ferriphilum and proteomic response to arsenic stress. Bioresour. Technol.. 2010;101(24):9811-9814.
- [Google Scholar]
- Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PloS One. 2014;3:e99236
- [Google Scholar]
- Arsenic: In search of an antidote to a global poison. Environ. Health Perspect.. 2005;113(6):A378-A386.
- [Google Scholar]
- Conventional as well as emerging arsenic removal technologies—a critical review. Water Air Soil Pollut.. 2017;10:381.
- [Google Scholar]
- Arsenite biotransformation and bioaccumulation by Klebsiella pneumoniae strain SSSW7 possessing arsenite oxidase (aioA) gene. BioMetals. 2019;32:65-76.
- [Google Scholar]
- Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol.. 2003;1:135-141.
- [Google Scholar]
- Arsenite oxidizing multiple metal resistant bacteria isolated from industrial effluent: their potential use in wastewater treatment. World J. Microbiol. Biotechnol.. 2016;32:133-144.
- [Google Scholar]
- Arsenic removal technologies for drinking water treatment. Rev. Environ. Sci. Biotechnol.. 2004;1:43-53.
- [Google Scholar]
- Biosorption of arsenite (As+3) and arsenate (As+5) from aqueous solution by Arthrobacter sp. biomass. Environ. Technol.. 2013;19:2701-2708.
- [Google Scholar]
- Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA. 2006;7:2075-2080.
- [Google Scholar]
- Micro-colonization of arsenic-resistant Staphylococcus sp. As-3 on arsenopyrite (FeAsS) drives arsenic mobilization under anoxic sub-surface mimicking conditions. Sci. Total Environ.. 2019;669:527-539.
- [Google Scholar]
- Antioxidative enzyme profiling and biosorption ability of Cupriavidus metallidurans CH34 and Pseudomonas putida mt2 under cadmium stress. J. Basic Microbiol.. 2015;55:374-381.
- [Google Scholar]
- Sher, S., Hussain, S.Z., Rehman, A., 2020. Phenotypic and genomic analysis of multiple heavy metal–resistant Micrococcus luteus strain AS2 isolated from industrial waste water and its potential use in arsenic bioremediation. Appl. Microbial. Biotechnol. 104, 2243-2254.
- Use of heavy metals resistant bacteria-a strategy for arsenic bioremediation. Appl. Microbial. Biotechnol.. 2019;1:1-15.
- [Google Scholar]
- Complete genome sequences of highly arsenite-resistant bacteria Brevibacterium sp. strain CS2 and Micrococcus luteus AS2. Microbiol. Resour. Announc.. 2019;31:e00531-e619.
- [Google Scholar]
- Arsenic mediated modifications in Bacillus aryabhattai and their biotechnological applications for arsenic bioremediation. Chemosphere. 2016;164:524-534.
- [Google Scholar]
- Contamination of drinking-water by arsenic in Bangladesh: a public health emergency. Bull. World Health Organ.. 2000;78:1093-1103.
- [Google Scholar]
- Isolation and characterization of Staphylococcus sp. strain NBRIEAG-8 from arsenic contaminated site of West Bengal. Appl. Microbiol. Biotechnol.. 2012;95:1275-1291.
- [Google Scholar]
- Biosorption of arsenic through bacteria isolated from Pakistan. Int. Microbiol.. 2019;1:59-68.
- [Google Scholar]
- Transcriptomic analysis reveals adaptive responses of an enterobacteriaceae strain LSJC7 to arsenic exposure. Front. Microbial.. 2016;7:636.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.08.012.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







