Translate this page into:
Molecular typing of MRSA isolates by spa and PFGE
⁎Corresponding author. kalkharsah@iau.edu.sa (Khaled R. Alkharsah)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
Methicillin resistant S. aureus (MRSA) strains circulating among populations and crossing borders constitute a major problem for health control and require a fast and simple genotypic approach.
Methods
In the current study we compared staphylococcal protein A (spa) and pulsed field gel electrophoresis (PFGE) typing methods to genotype 106 MRSA clinical isolates.
Results
The genetic spectrum of the isolates was very diverse as revealed by the two typing approaches. In total, we identified 35 spa types in the study. The most frequently detected spa type in the study was t044 (30.18%), followed by t127 and t304 (5.6% each), t363 (4.6%), and t1200 and t002 (3.8% each). The rest of isolates were detected in low frequency and many were singletons. PFGE genotyping identified 34 pulsogroups. Most of the isolates were clustered in pulsogroup J. There was no clustering of the spa types into the pulsogroups.
Conclusions
MRSA isolates are very diverse in the region. In light of the observed MRSA diversity the spa typing could constitute a preferable approach for MRSA typing.
Keywords
MRSA
Spa typing
PFGE
Saudi Arabia
Molecular typing
1 Introduction
Methicillin resistant S. aureus (MRSA) gains tremendous attention in medical practice and microbial research because of the ascending challenge of therapy and control. MRSA causes annual deaths higher than any other infectious agent in USA (Klevens et al., 2007). In 2011 the Center for Disease Control and Prevention (CDC) estimated that more than 80,000 invasive infections and more than 11,000 deaths were caused by MRSA (Dantes et al., 2013). Furthermore, patients infected with MRSA required longer period of hospitalization and higher health care cost compared to patients infected with other staphylococcus species (Cosgrove et al., 2005). Meta-analysis study comparing MRSA infection to methicillin sensitive S. aureus (MSSA) bacteremic infection revealed that MRSA caused markedly higher mortality, which was attributed to prescribing ineffective early therapy (Cosgrove et al., 2005).
Considering the spread of MRSA strains across the borders, stringent strategies are required to control their dissemination and trace their nosocomial reservoirs including active surveillance and general infection control measures such as screening, decolonization and contact isolation. However, to ensure the effectiveness of these strategies, knowledge of the common MRSA genotypes, especially of the ones circulating in the population, is required. Several molecular typing methods employing DNA banding patterns are available such as pulsed-field gel electrophoresis (PFGE) or utilizing DNA sequencing such as the staphylococcal protein A (spa) typing and multilocus sequence typing (MLST).
Staphylococcal cassette chromosome which harbor mec (SCCmec) typing method is a polymerase chain reaction (PCR)-based and detect the type of SCCmec cassette but not its structure and is therefore less discriminative (Boye et al., 2007; Zhang et al., 2005). PFGE is a very discriminative approach for S. aureus typing because it is based on fragmentation of the bacterial chromosome using Serratia marcescens endonuclease I (SmaI) enzyme and separation of the digested DNA fragments on agarose gel using alternating pulses. However, it is laborious, time consuming and lacks the reproducibility among different laboratories in addition to its cost ineffectiveness. Furthermore, efforts to harmonize standard protocols were only successful on the national levels in some countries but not internationally (Murchan et al., 2003; Tenover et al., 1994; van Belkum et al., 1998). Sequence-based typing methods are preferable to DNA-size typing methods. MLST typing was established to analyze the allelic sequence of short DNA fragments (450–500 bp) of seven S. aureus housekeeping genes (Enright et al., 2000). This method is an excellent tool for screening and very useful in studying the molecular evolution of S. aureus. However, it is expensive, laborious, and time consuming and therefore not suitable for diagnostic purposes as for each S. aureus isolate seven PCR reactions and 14 sequencing reactions should be performed apart from the data analysis.
Spa typing method targets region X of the spa gene, which contains a tandem repeats (about 24 bp in length) (Frenay et al., 1996). Different spa types vary by the type of repeats and their copy numbers. Spa typing became very popular since it is based on sequencing of a single locus, less expensive and less time consuming than other methods. Moreover, it has more discriminative power compared to MLST (Malachowa et al., 2005). The availability of the spa types on the central spa server (http://spaserver.ridom.de) and its unified nomenclature provided another important advantage. The use of these typing tools contributes to understanding clonal diversity and transmission of MRSA in the hospital and community settings.
The aim of our study is to genotype MRSA isolates using spa typing to determine the circulating MRSA strains, which will enable a better design of surveillance protocols and control strategies. The study also aims at comparing the spa typing approach to that based on PFGE.
2 Methods:
2.1 Bacterial isolates:
MRSA isolates (106 isolates) were collected from King Fahd Hospital of the University in Alkhubar (KFHU), Saudi Arabia. All bacterial isolates were of clinical origin. Fifty one isolates were obtained from infection sites such as wound, abscess, respiratory infections, blood stream and eye infection. Fifty five isolates were obtained from carrier colonization sites such as nose, groin, or throat. Further details about the samples can be found elsewhere (Alkharsah et al., 2018). All bacterial isolates were maintained as glycerol stock and stored at −80 °C. The isolates were designated numbers M1-M114. All isolates were identified as previously described (CLSI, 2016).
We obtained the Ethical approval for the study from the Institutional Review Board at Imam Abdulrahman Bin Faisal University (IRB-2017-13-142).
2.2 Spa typing:
Extraction of DNA was performed using Qiagen DNA extraction kit following the standard protocol with addition of lysozymes (Qiagen, Germany). DNA concentration and quality assessment was done by NanoDrop (life technologies, USA). The spa1095F (5′-AAAGACGATCCTTCGGTGAGC-3′) and spa1517R (5′-GCTTTTGCAATGTCATTTACTG-3′) primers were used to amplify the target fragment of the spa gene by PCR according to the protocol previously described (Harmsen et al., 2003). The amplified DNA product was separated on 1% agarose by gel electrophoresis and the amplicons were purified by gel extraction kit from Qiagen (Qiagen, Germany). The purified DNA product was sequenced in both forward and reverse directions using the same PCR primers in combination with the big dye termination mix kit and the ABI genetic analyzer 3500 (Applied Biosystems, USA). All DNA sequences were trimmed to the required length and the full fragment sequence was obtained by aligning the forward and reverse sequences for each isolate using the DNAGear software (F et al., 2012). Spa types were allocated to each isolate according to the repeats listed in the spa typing website (http://www.spaserver.ridom.de/) using the DNAGear software (F et al., 2012). Fig. 1 illustrates the flow of spa typing method (Fig. 1).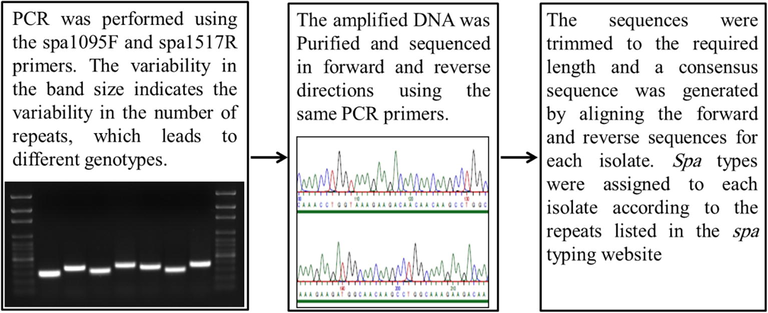
Illustration of the procedure and flow of spa typing method.
2.3 Pulse field gel electrophoresis (PFGE):
All the 106 isolates were subjected to PFGE analysis. PFGE was performed as per the protocol defined by the Center for Disease Control and prevention (CDC) with few modifications (https://www.cdc.gov/pulsenet/pathogens/pfge.html). Briefly MRSA colonies from overnight cultures grown on trypticase soy agar were inoculated in 5 ml of Brain heart infusion broth and further incubated at 37 °C for 18 hrs. Cell suspension turbidity was adjusted to 0.9–1.0. Cell pellet of 200 µl of adjusted cell suspension was added to 200 µl of Tris-EDTA (TE) buffer and 4 µl of Lysostaphin in 300 µl of 1.8% Seakem agarose prepared in TE buffer. This mixture was quickly dispensed into plug mold. After 10–15 mins plugs were removed from mold and placed into a tube with 3 ml of Lysis buffer (1M tris, 0.5 M EDTA, 0.5% Brij58, 0.2% Sodium deoxycholate, 0.5% sodium laureyl sarcosine) at 37 °C overnight. Later Lysis buffer was decanted and replaced by 3 ml of ESP lysis buffer (10 mM tris, 1 mM EDTA, 1% SDS, 1 mg/ml proteinase K) at 50 °C for 4 hrs. After decanting ESP buffer, plugs were washed several times with TE buffer then stored at 4 °C. A slice of 2 × 10 mm was aseptically cut from a prepared plug and equilibrated for 30 mins in 200 µl of water buffer (buffer stock, bovine serum albumin [BSA], grade 1 water). Later water buffer was decanted and replaced by 200 µl of enzyme mixture (buffer stock, BSA, SmaI 4 µl/slice, grade 1 water) for minimum 4 hrs at 30 °C. Digested plugs were loaded into the wells of a 1% agarose gel and run in 0.5 TBE using a CHEF-DR III system (Bio-Rad Laboratories, Inc, CA, USA) according to the following parameters: 6 V, temperature 14 °C, initial switch time5 s, final switch time 40 s, included angle 120, with a run time of 21 hrs. Two controls were included in each run. The bacteriophage lambda ladder was used a PFGE marker and the NCTC 8325 strain was used a reference control. Gel was stained using ethidium bromide solution (final concentration 1 mg/ml) for 30–40 min on a rocking shaker in a covered container. Destaining was done thrice with distilled water on shaker and then the gel was visualized and captured.
Gel pictures were processed and analyzed using the software BioNumerics v6.5. Dice coefficient was employed to compare the obtained PFGE fingerprints. GelCompare and Bionumerics software v6.5 were employed to perform analysis of cluster employing the Unweighted Pair Group Method with Arithmetic Means (UPGMA). Further parameters including the band tolerance as well as optimization settings were set to 1% and 0.5%, respectively. We used the suggested recommendation by Tenover et al. to cluster the PFGE groups (Tenover et al., 1995). The similarity coefficient was set to 80% as recommended by Struelens et al. (Struelens et al., 1992).
2.4 Data analysis
SPSS version 23 software was used in the statistical analysis.
3 Results
In total, thirty-five spa types were identified in our cohort. The most common spa type in our study was t044 constituting 30.18% of the isolates (Table 1). Spa types t127 and t304 were the second most common types (5.6% each), followed by t363 (4.6%), and t1200 and t002 (3.8% each) (Table 1). The rest of the isolates were clustered either in less frequent spa types or as singletons (Table 1). Seven isolates had no matching spa type in the database. Three of the seven isolates had new repeats and four isolates had known repeats but the arrangement of the repeats didn’t match with previously identified spa types (Table 1). NR: New repeat lead to new spa type. NT: new type due to new repeat arrangement.
Isolate
PFGE group
spa type
Isolate
PFGE group
spa type
Isolate
PFGE group
spa type
M96
A
t127
M24
I
t044
M59
L
t002
M57
t044
M73
t934
M37
M
t13180
M90
t127
M70
t002
M38
t044
M92
t127
M71
t127
M79
MA
t002
M58
AA
t044
M72
t044
M81
MB
t3364
M101
B
t304
M74
t044
M86
N
t019
M108
t304
M76
t002
M87
t267
M95
t304
M14
IA
t044
M82
NA
t1339
M105
t304
M61
J
t13180
M85
O
t044
M18
NR1
M69
t044
M88
t044
M91
t2297
M65
t223
M89
t044
M12
C
t8400
M68
t044
M83
t044
M32
t363
M13
NT1
M1
OA
t852
M34
t363
M52
t223
M7
t362
M102
t037
M94
t044
M2
P
t037
M25
D
t1200
M45
t044
M3
t127
M28
NR2
M46
t693
M4
t044
M10
DA
t304
M43
t311
M6
NR3
M40
E
t044
M104
t1247
M5
PA
t363
M41
t657
M31
t044
M8
t1200
M42
t044
M100
t044
M75
PB
t657
M44
t044
M97
t044
M77
t1200
M39
t044
M99
t044
M54
Q
t311
M35
EA
t657
M47
NT2
M55
t037
M50
F
t021
M49
t044
M11
R
t223
M51
t8154
M17
t044
M19
t008
M9
FA
t304
M53
t309
M56
S
t932
M112
FB
t362
M66
t442
M64
t044
M63
G
t13180
M93
t044
M22
NT3
M67
t8154
M103
t044
M23
NT4
M115
t690
M29
t044
M111
T
t4573
M20
t309
M30
t363
M113
t127
M21
t690
M106
K
t044
M16
t690
M110
t044
M114
GA
t1339
M119
t004
M117
H
t224
M60
KA
t1200
M62
t224
M15
L
t363
SmaI digestion and PFGE identified the presence of 34 pulsogroups designated as A to T (Fig. 2). PFGE group J was the largest and comprised 24 of the studied isolates. Other pulsogroups (A, B, E, I, P, and S) comprised 4–7 isolates each (Fig. 2).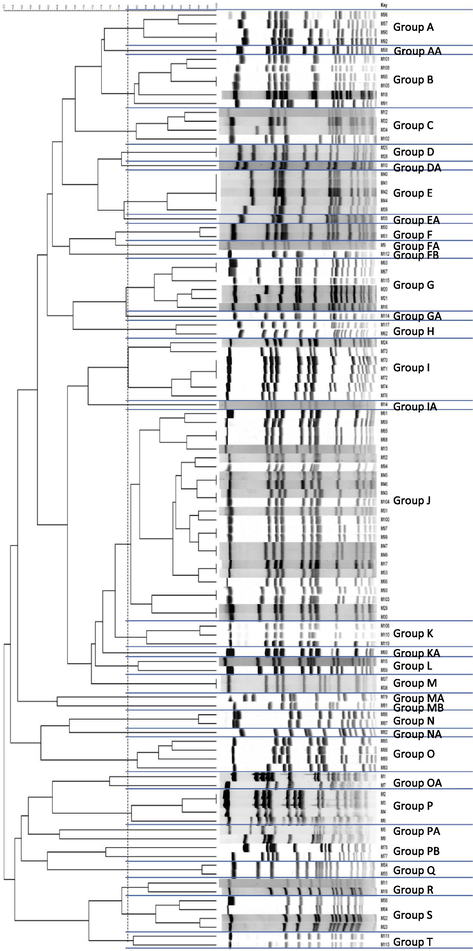
Pulsed field gel electrophoresis groups based on 80% Dice coefficient.
There was no significant clonal association between spa types and PFGE groups. There was clustering of some spa types within some of the pulsogroups. However, it was statistically non-significant, most probably due to the small numbers of isolates of such spa types. Additionally, the seven isolates that potentially represent new spa types clustered within the identified pulsogroups.
No association was found between neither of the typing methods and the clinical data nor demographic data such as gender, age group, source of isolates, or antibiotic susceptibility pattern. However, some spa types (t002 and t363) were confined to isolates recovered from a set of infection sites (wound, abscess, and endotracheal aspirate), while others were more frequently associated with isolates recovered from colonization sites (t304 and t690). There was no relation between spa types and severity of infection.
4 Discussion
Effective strategies are required to control the dissemination of MRSA worldwide such as active surveillance and institutional preventive measures, which requires insight into the molecular epidemiology of the circulating genotypes. MRSA prevalence in Saudi Arabia ranges between 12 and 49% depending on the region (Austin et al., 2003; Baddour et al., 2006; Bukharie and Abdelhadi, 2001; El Amin and Faidah, 2012). The estimated carriage rate of MRSA among health care workers is too high reaching 67% (Iyer et al., 2014) leading to an increase in the risk of hospital-acquired infections. Two studies were recently published using the spa typing approach but had either a small number of isolates or studied a selected group of patients (Abou Shady et al., 2015; Alreshidi et al., 2013). Based on that, we have initiated this study, which revealed a clear genotypic diversity within the MRSA population.
More than thirty types were identified using spa or PFGE typing approaches among 106 isolates. This genotypic diversity of MRSA strains could be attributed to the diversity in the population living in the area; people from different nationalities work and live in the region and hence importing the diversity observed. Additionally, it could be attributed to low rate of nosocomial cross-transmission due to stringent MRSA screening program adopted for more than a decade, and the selective pressure effect of antimicrobials over various circulating strains (Argudin et al., 2009).
Spa typing method relies on the PCR amplification followed by sequencing of the highly variable X fragment of the staphylococcal protein A, which makes it simple and fast compared to the MLST approach. All of the isolates, except seven, had spa types matching a known type on the database. Our isolates with new spa type were obtained from skin and nasal samples.
Spa type t044 (30.18%), which was the most common in this study, was previously reported as being the second most common after type t037 in Riyadh (Alreshidi et al., 2013). This indicated the high prevalence of type t044 in Saudi Arabia with some regional variation. However, in another study from Buraydah city in Al-Qassim district near to Riyadh none of the two former common spa types were detected, which could be attributed to the low sample size (15 MRSA isolate) (Abou Shady et al., 2015).
Type t044 represents the so called ST80 MRSA, which is very common in the Middle East. It was detected in other neighboring countries at variable frequencies such as Jordan (Al-Bakri et al., 2013; Bazzoun et al., 2014); however, it was only reported in adults and healthcare workers but not in children (Aqel et al., 2015). This was in contrast to our results, as type t044 was detected in all age groups in our study. It was also reported from Lebanon (Harastani et al., 2014), United Arab Emirate (Sonnevend et al., 2012), Kuwait (Udo and Al-Sweih, 2017), Iran (Goudarzi et al., 2016), and in one but not all studies from Tunisia (Ben Nejma et al., 2013; Ben Said et al., 2016; Kechrid et al., 2011), and widely disseminated in Europe, Australia (Larsson et al., 2014), and USA (Fluit et al., 2015). No correlation between type t044 and the source of the isolate was reported. However, some spa types were confined to isolates obtained from a specific site of infection; for example t002 and t363 were isolated from wound, abscess, and endotracheal aspirate. However, the small number of isolates (5 each) precludes firm conclusion. There was no relation between spa types and severity of infection.
Three new spa types were reported, and four isolates had known repeats but the repeat sequence did not match any previously identified spa types on database. The X region of the spa protein is very polymorphic, with 761 repeats being so far registered on the Ridom spa server. The random arrangement of these repeats generated 7643 spa types. New spa types are generated by the rearrangement of the repeats in a recombination event or by generation of new repeats due to DNA polymerase error, deletion, or duplication events.
Despite its limitations, spa typing has shown, in this work, its utility to study local MRSA epidemiology and to help in clustering isolates. It also showed a better discriminative power. The lack of association between the spa type and PFGE pattern could be attributed to the large genetic diversity of the studied isolates.
5 Conclusion
The data presented showed high diversity of MRSA in our population providing a baseline for further molecular characterization in the Region. Spa-typing can be considered a practical approach to investigate and manage MRSA-linked outbreaks because of its simplicity, relative low cost, high throughput and discriminative power besides the standardized nomenclature and portability into an international database. Infrequently when misclassification or non-typeability is a concern, a sophisticated typing tool, such as PFGE in this study, is an informative supplementary addition.
Acknowledgements
The authors are grateful to Mr. Melchor Jimenez, Mr. Nestor Recella, Ms. Janaica Yu Logan, and the technical staff in the diagnostic microbiology laboratory (KFHU) for their technical assistance.
This Article contains the results and findings of the research project funded by King Abdulaziz City for Science and Technology (KACST) Grant No. LGP-36-108.
Conflict of interest
The authors have no conflict of interest to declare.
Author contribution
KRA: overall design and plan of the work and raising fund.
SR: PFGE experimental work.
AA: spa data analysis.
AD: spa experimental design.
AH: PFGE experimental design.
ST: Analysis of PFGE data.
All authors read and edited the manuscript.
Funding
This work was supported by King Abdulaziz City for Science and Technology (KACST) [Grant number LGP-36-108].
References
- Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz. J. Infect Dis.. 2015;19:68-76.
- [Google Scholar]
- The epidemiology and molecular characterization of methicillin-resistant staphylococci sampled from a healthy Jordanian population. Epidemiol. Infect. 2013;141:2384-2391.
- [Google Scholar]
- Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann. Clin. Microbiol. Antimicrob.. 2018;17:7.
- [Google Scholar]
- Genetic variation among methicillin-resistant Staphylococcus aureus isolates from cancer patients in Saudi Arabia. Eur. J. Clin. Microbiol. Infect. Dis.. 2013;32:755-761.
- [Google Scholar]
- Molecular epidemiology of nasal isolates of methicillin-resistant Staphylococcus aureus from Jordan. J. Infect. Public Health. 2015;8:90-97.
- [Google Scholar]
- Clonal complexes and diversity of exotoxin gene profiles in methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from patients in a Spanish hospital. J. Clin. Microbiol.. 2009;47:2097-2105.
- [Google Scholar]
- MRSA prevalence in a teaching hospital in Western Saudi Arabia. Saudi Med. J.. 2003;24:1313-1316.
- [Google Scholar]
- Trends in antibiotic susceptibility patterns and epidemiology of MRSA isolates from several hospitals in Riyadh, Saudi Arabia. Ann. Clin. Microbiol. Antimicrob.. 2006;5:30.
- [Google Scholar]
- Molecular typing of staphylococcus aureus collected from a major hospital in amman, Jordan. J. Infect. Dev. Ctries. 2014;8:441-447.
- [Google Scholar]
- Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn. Microbiol. Infect. Dis.. 2013;77:20-24.
- [Google Scholar]
- Genetic characterization and antimicrobial resistance of Staphylococcus aureus isolated from bovine milk in Tunisia. Lett. Appl. Microbiol.. 2016;63:473-481.
- [Google Scholar]
- A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect.. 2007;13:725-727.
- [Google Scholar]
- The epidemiology of methicillin-resistant Staphylococcus aureus at a Saudi university hospital. Microb. Drug. Resist.. 2001;7:413-416.
- [Google Scholar]
- CLSI, 2016. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. CLSI document M100-S26. Wayne, PA.
- The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol.. 2005;26:166-174.
- [Google Scholar]
- National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Int. Med.. 2013;173:1970-1978.
- [Google Scholar]
- Methicillin-resistant Staphylococcus aureus in the western region of Saudi Arabia: prevalence and antibiotic susceptibility pattern. Ann. Saudi Med.. 2012;32:513-516.
- [Google Scholar]
- Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol.. 2000;38:1008-1015.
- [Google Scholar]
- DNAGear–a free software for spa type identification in Staphylococcus aureus. BMC Res. Notes. 2012;5:642.
- [Google Scholar]
- Comparison of an ST80 MRSA strain from the USA with European ST80 strains. J. Antimicrob. Chemother.. 2015;70:664-669.
- [Google Scholar]
- Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis.. 1996;15:60-64.
- [Google Scholar]
- Spa typing of staphylococcus aureus strains isolated from clinical specimens of patients with nosocomial infections in Tehran, Iran. Jundishapur J. Microbiol.. 2016;9:e35685
- [Google Scholar]
- Molecular characteristics of Staphylococcus aureus isolated from a major hospital in Lebanon. Int. J. Infect. Dis.. 2014;19:33-38.
- [Google Scholar]
- Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol.. 2003;41:5442-5448.
- [Google Scholar]
- High incidence rate of methicillin-resistant Staphylococcus aureus (MRSA) among healthcare workers in Saudi Arabia. J. Infect. Dev. Ctries. 2014;8:372-378.
- [Google Scholar]
- Molecular analysis of community-acquired methicillin-susceptible and resistant Staphylococcus aureus isolates recovered from bacteraemic and osteomyelitis infections in children from Tunisia. Clin. Microbiol. Infect.. 2011;17:1020-1026.
- [Google Scholar]
- Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763-1771.
- [Google Scholar]
- Epidemiology of MRSA in southern Sweden: strong relation to foreign country of origin, health care abroad and foreign travel. Eur. J. Clin. Microbiol. Infect. Dis.. 2014;33:61-68.
- [Google Scholar]
- Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol.. 2005;43:3095-3100.
- [Google Scholar]
- Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol.. 2003;41:1574-1585.
- [Google Scholar]
- Change in meticillin-resistant Staphylococcus aureus clones at a tertiary care hospital in the United Arab Emirates over a 5-year period. J. Clin. Pathol.. 2012;65:178-182.
- [Google Scholar]
- Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol.. 1992;30:2599-2605.
- [Google Scholar]
- Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol.. 1994;32:407-415.
- [Google Scholar]
- Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol.. 1995;33:2233-2239.
- [Google Scholar]
- Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS One. 2017;12:e0179563
- [Google Scholar]
- Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol.. 1998;36:1653-1659.
- [Google Scholar]
- Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol.. 2005;43:5026-5033.
- [Google Scholar]







