Molecular characterization of keratin degrading fungi isolated from semi-arid soil by PCR using ITS4 and ITS5 primers
⁎Corresponding author. drseema299@gmail.com (Seema Bhadauria)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
This study was aimed to explore molecular characters of fungal isolates and analyze the phylogenic relationship between isolates. Soil samples collected from different habitats of Jaipur, were analyzed for the prevalence of keratinophilic fungi using the hair baiting technique. The physiological conditions of soil samples were analyzed to study the effect of environmental factors on the growth and occurrence of fungi. Keratinophilic fungi were isolated and then identified on the basis of their macro and micro morphological characteristics. The identification of fungi was confirmed by DNA sequencing by PCR amplification using ITS4 and ITS5 gene primers. The fungal species were identified by the nBLAST using fungal DNA sequences against the data available in NCBI/GenBank. The isolated keratinophilic fungal species were identified as Chaetomium seminis-citrulli and Penicillium guttulosum. These two fungi were reported first time from Rajasthan, India using hair baiting technique. Phylogenic trees were constructed using MEGA7 software with the help of keratinophilic fungi reference data retrieved from NCBI GenBank data, to study the phylogenic relationship between the isolated fungi.
Keywords
Keratinophilic fungi
PCR
nBLAST
Sequencing
Phylogenic
Chaetomium and Penicillium
1 Introduction
Keratinophilic fungi are important filamentous which are capable to degrade highly stable keratinous substrates. These fungi utilize carbon and nitrogen residues and decompose keratinized materials (Kaul et al., 2013). Therefore, keratinous materials enriched soil is most appropriate for the growth and occurrence of keratinophilic fungi (Otcenasek, 1978; Mercantini et al., 1986).
Most of Keratinophilic fungal species occur in soil and are saprophytic in nature. Some of the saprophytic keratinophilic fungi are reported pathogenic in nature and cause mycotic infections in humans and animals. (Ajello, 1956; Kumari et al., 2005). Some of the keratinophilic fungi belong to a specific group called, dermatophytes, known to cause superficial infections of keratinized tissues such as skin, hair and nails in human beings. Mycotic infections are extremely contagious and reported all over the world (Hedayati et al., 2004). Therefore, identification of these fungi from the environment is epidemiologically very important.
Biological distribution of keratinophilic fungi depends on some important factors including pH, temperature, human and/or animal presence (Deshmukh and Verekar, 2006). In Rajasthan, there are high-temperature conditions. The climatic and geographic diversity of Jaipur city makes it a remarkable region to study the prevalence of keratinophilic fungi. The high temperature climate is considered as the best environmental condition for the growth and occurrence of keratinophilic fungi.
Identification of any specie on the basis of conventional morphological characters is often time-consuming and often unaffected by culture conditions (Pryce et al., 2003). Contrarily, in molecular methods, the results are unaffected by culture conditions and are more rapid than phenotypic approaches. Thus the polymerase chain reaction (PCR) based sequencing methods are used for molecular characterization (Liu et al., 2000). Analysis of fungal DNA sequence is used for fungal species identification by the amplification of the internal transcribed spacer region (ITS1-5.8S-ITS2) using PCR. The identification of the DNA sequence was implemented by comparison of desired DNA sequence data with those available from GenBank database.
The objective of this research was to report on the identification of isolated keratinophilic fungi by microscopic and molecular characterization and to study their phylogenic relationship.
2 Materials and methods
2.1 Isolation of fungi
Thirty soil samples in sterilized polythene bags were collected from various places of Jaipur, Rajasthan, India. Soil samples were collected from dry land in the month of July with preference to human and animal presence, such as playgrounds, public parks, grazing area, barber shops from the superficial layer of soil (maximum depth of 5 cm). Soil samples were further analyzed for pH, temperature, humidity, organic matter content and moisture content of the soil using standard methods as described by Blakemore et al., 1987; Singh et al., 1999 and McLeod, 1973. Keratinophilic fungi were isolated by the hair baiting technique given by Vanbreuseghem (1952). In this technique, soil samples were filled in sterile Petri dishes and moistened with autoclaved distilled water. Small pieces of autoclaved human hairs were dispersed as a keratin source over the soil surface. These Petri dishes were incubated at room temperature and regularly observed for the fungal growth for a period of 21 days. After observing the growth, fungal mycelium was cultured on Sabouraud’s dextrose agar medium. The growing colonies were then periodically sub-cultured onto newly prepared plates of the same medium for further purification and identification. All the fungal isolates were identified morphologically by macro and micro morphological features of isolates (Sigler and Carmichael, 1976; Arx von, 1986; Cano and Guarro, 1990).
2.2 Genomic DNA extraction
The selected strains were transferred to tubes containing Sabouraud’s dextrose agar medium in slant conditions and the inoculated slants were incubated in a mycology incubator at 28 ± 2 °C for seven days of growth. Genomic DNA was extracted according to Saghai-Maroof et al. (1984) protocol with slight modification.
The fungal culture was scraped out using sterilized scalpel and transferred to a ceramic pestle containing lysis buffer (20 mM EDTA, 100 mm Tris HCl, 3,5% CTAB). To the mixture, sterile glass beads were added and the mixture was homogenized in tissue homogenizer and transferred to water bath at 65 °C for 30 min. The mixture was centrifuged at 15,000 rpm for 10 min and the supernatant was collected in a clean micro-centrifuge tube. Supernatant was mixed with RNase A and incubated at 37 °C for 20 min. Then equal volume of phenol: chloroform: Isoamyl alcohol at a ratio of 25:24:1 was mixed in centrifuge tube and centrifuged at 15,000 rpm for 10 min. The supernatant was collected and equal volume of ice-cold isopropanol was added and precipitated at −20 °C for 20 min. The sample was then centrifuged at 12,000 rpm for 10 min and the supernatant was discarded. The pellet containing DNA was washed with 70% ethanol and again centrifuged at 12,000 rpm for 5 min. The pellet was kept at room temperature for ethanol evaporation and then 1X TE buffer was added.
2.3 Quantity and quality determination
The amount of the extracted DNA was analyzed by measuring the absorbance at 270 nm using a spectrophotometer. The quality of extracted DNA was monitored by agarose gel electrophoresis in 1.2% agarose gel with ethidium bromide fluorescence. A variety of PCR based methods were utilized for downstream analysis of extracted DNA.
2.4 PCR amplification and DNA sequencing
The ITS1 –5.8S—ITS2 rDNA was amplified using primers ITS4 (forward primer) and ITS5 (reverse primer) (White et al., 1990). Amplification was achieved in a vial containing 10x buffer, MgCl2 15 mM, dNTP 0.2 mmol, Forward primer and Reverse primer 10 picomolar, Taq Polymerase 2 µl, DNA sample 50–100 ng/µl and Milli Q water. The PCR reaction was carried out using a Thermal Cycler with conditions as follows: Initiation for 10 min at 94 °C, denaturation for one minute at 94 °C, annealing for 30 s at 55 °C, extension for 1 min at 72 °C. The Thermal Cycler was run for 35 cycles and then a final extension cycle was run at 72 °C for 10 min. The PCR products were analyzed by electrophoresis on 1.2% agarose (Fig. 1). PCR products, isolated fungal DNA, were sequenced at PGIMER, Chandigarh, India. The sequence data were read by computer-based software finch TV. The sequence results were analyzed by using the web-based blasting program, basic local alignment search tool (BLAST) and data were compared with the Genebank database at the National Center for Biotechnology Information (NCBI) Nucleotide Sequence database (Altschul et al., 1990). The sequence data were submitted to Bankit for accession Number.
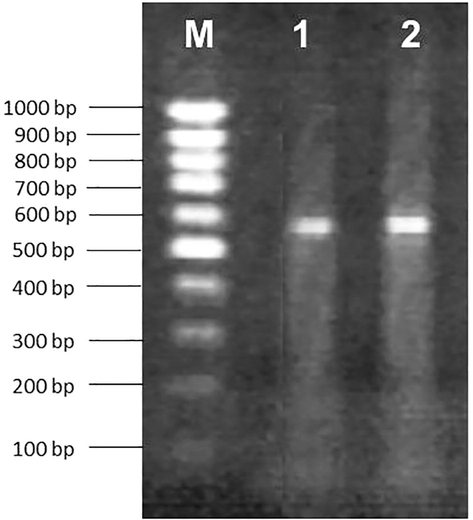
- Banding pattern of isolates resulted from PCR reactions using ITS4 and ITS5. Lane M 100 bp marker ladder, Lane 1–2 Chaetomium seminis citrulli, Penicillium guttulosum.
2.5 Phylogenetic analysis
DNA sequences were aligned with Clustal W (Version 1.74) and the alignments were corrected visually (Thompson et al., 1994). The phylogenetic tree was constructed by the neighbor-joining technique using mega7 software as described by Saito and Nei (1987).
3 Results
Out of thirty soil samples, 19 species of keratinophilic fungi were isolated. The fungi were isolated from the pH of soil ranging from 6.00 to 9.00. In the present study, most of the fungi were isolated from the pH range of 6.81–7.30, followed by 7.31–7.80 and 7.81–8.30 pH. The pH range of 6.0–6.80 and 8.30–9.0 showed a lower occurrence of fungi. The optimum temperature recorded for maximum growth of fungi was 32 °C. Most of the fungal colonies recovered from temperature range of 28 °C–38 °C. The moisture content of soil samples after collection was analyzed between 0.2 and 1.0%. The prevalence and growth of most of the fungi were in soil having a moisture content between 0.2 and 0.6%. In the present study, the keratinophilic fungi were isolated from dry soil samples having temperature above 30 °C and pH range between 6.81 and 8.30.
Out of the total 19 isolated species, two were selected for molecular characterization and phylogenic studies on the basis of their dominating occurrence. The PCR amplification of ITS region of the two isolates JECRC16 and JECRC17 yielded PCR products of 577 and 584 bp, respectively. The amplification products were sequenced for species identification. The DNA sequences of isolated strains were recited using basic local alignment search tool (BLAST) for species identification. The BLAST search showed that the sequence data of the isolated strain JECRC16 shared 99% similarity with Chaetomium seminis citrulli (JX280729) and JECRC17 shared 99% similarity with Penicillium guttulosum (HQ646592.1). Thus, the two isolated strains were identified as Chaetomium seminis citrulli and Penicillium guttulosum. DNA sequences of isolated strains were submitted in the Gen-Bank databases for the purpose of phylogenetic study (Makimura, 2001). The gene bank accession number for JECRC16 and JECRC17 are KX492890.1 and KX492891.1. Both of the fungi were reported first time from Rajasthan, India. The variable ITS regions were important tools for resolving relationships between close taxonomic relatives (Barbee et al., 1995). The NCBI GenBank data and previous studies were used for identification of the isolated strain/species (Makimura et al., 1999).
Phylogenetic trees using DNA sequences for the two isolated species were constructed with the reference strains. JECRC16 and JECRC17 (Felsenstein, 1981). The phylogenic tree shows phylogenic relationship of the isolated strains with the reference strains (Figs. 2 and 3).

- Phylogenic tree of isolated JECRC 16 specie showing their closest relationship with Chaetomium seminis citrulli (JX280729.1).
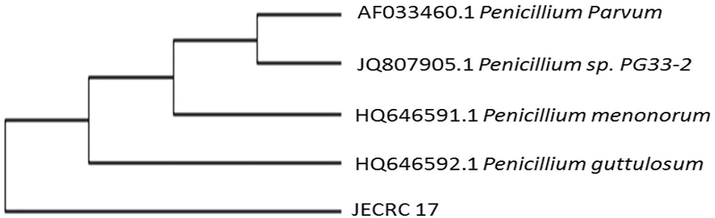
- Phylogenic tree of isolated JECRC 17 specie showing their closest relationship with Penicillium guttulosum (HQ646592.1).
The phylogeny and multiple sequence alignment analysis of isolated two strains show clear similarities with isolated fungus. Isolated JECRC16 show 100% similarity with Chaetomium seminis citrulli (JX280729.1), 99% similarity with Chaetomium sp. YZ-2015c (KP336821.1), 98% similarity with Chaetomium piluliferum (AF286406.1) and 97% similarity with Chaetomium arxii (JQ864438.1). Multiple sequence alignment results show about 100% similarity between these phylogenies strains (Fig. 4.)

- Multiple Sequence Allignment of JECRC16 ((KX492890.1), Chaetomium seminis citrulli (JX280729.1), Chaetomium sp. YZ-2015c (KP336821.1), Chaetomium piluliferum (AF286406.1) and Chaetomium arxii (JQ864438.1). showing the similarity with dark background and difference in white background.
Similarly, JECRC17 shows 100% similarity with Penicillium guttulosum (Accession No. HQ646592.1), 99% similarity with Penicillium menonorum (Accession No. HQ646591.1), 99% similarity with Penicillium sp. PG33-2 (Accession No. JQ807905.1) and 99% similarity with Penicillium parvum (Accession No. AF033460.1). Multiple sequence alignment result shows about 100% similarity between these phylogenic strains (Fig. 5).

- Multiple Sequence Allignment of JECRC17 (KX492891.1), Penicillium guttulosum (Accession No. HQ646592.1), Penicillium pimiteouiense (Accession No. AF037431.1), Penicillium turbatum (Accession No. LC178551.1) and Penicillium rubidurum (Accession No. AF033462.1. showing the similarity with dark background and difference in white background.
4 Discussion
In the present study, both of the strains were isolated from soil samples of rich organic content and animal dung. Keratinophilic fungi showed good growth at favorable temperature, pH, moisture and keratinous substrate available in the soil. Microbial population remains more stable over a period in soil with high organic matter content (De Fede et al., 2001).
The isolated fungal species Chaetomium seminis citrulli and Penicillium guttulosum were reported first time from the soil of Rajasthan, India. Chaetomium seminis-citrulli was previously recorded in USSR, Italy and Israel (Carter and Khan, 1982). In previous studies, these two species were isolated using suspension method and in current research Chaetomium seminis citrulli and Penicillium guttulosum were isolated by hair baiting technique using hair as keratinous substrate. The significance of the present study is that both of the isolates can be isolated from soil contaminated with organic and keratinous waste materials. Further, Chaetomium seminis citrulli and Penicillium guttulosum can be used in degradation of keratinous materials. The current research was focused only on isolation and identification of fungal isolates using molecular characterization. The degradation potential of these two fungi could not be included in this study.
In this study, Chaetomium seminis citrulli was isolated from semi-arid region of Rajasthan, India at 38 °C from soil. It indicates that Chaetomium seminis citrulli can be isolated high temperature climate and can tolerate temperature above 40 °C. Rodríguez et al. (2004) recorded the prevalence of Chaetomium-like species in desert soil in extremely variable temperatures and dry climate. Kumar et al., in 2017 recorded diversity of Chaetomium species near coal mines of Hazaribagh, India with their capacity to tolerate harsh environments such as high Coal concentration and extreme temperature. The natural habitats in arid areas and survival in high temperature condition is the major cause of Chaetomium infection in mammalian tissues and paying more attention around the world. These fungi can cause human infections, such as keratitis or subcutaneous infections of skin, nails, hair and have been reported by Najafzadeh et al., 2014; Mootha et al., 2012.
Penicillium guttulosum in this study was isolated from soil with pH of 6.5 and containing high organic matter. Coutinho et al. (2010) isolated 13 species of Penicillium from the semi – arid soil of Petrolina, Brazil containing high organic material content. Vico et al. (2014) used species specific primer for identification of Penicillium species. Similarly, Suhail et al. (2006) isolated 10 species of Penicillium from the Indus river bank at Kotri, Pakistan on Rose Bengal-Streptomycin agar medium.
Identification and characterization of filamentous fungi in clinical laboratories are based on microscopic examination of sporulating mycelium. Fungi of the class, that have absence of spores generate difficulties in identification of genera and species by microscopic method (Pounder et al., 2007). For identification of isolated fungus, DNA sequencing technique was preferred because Penicillium sp. cannot be identified morphologically because of a large number of fungal species in Penicillium genus (Pitt, 1979). Therefore, DNA sequencing of the isolates was performed for conformation of species. Phylogenic tree was constructed to study the phylogenic relationship of the isolated species and multiple species sequences were aligned to study base pairs of the DNA sequence. According to present study, Chaetomium seminis citrulli is phylogenetically related to Chaetomium sp. YZ-2015c (KP336821.1), Chaetomium piluliferum (AF286406.1) and Chaetomium arxii (JQ864438.1).Zhanga et al. (2017) studied on Chaetomium species and concluded that C. uniseriatum is phylogenetically related to C. crispatum, C. acropullum, and C. seminis-citrulli. The genetic diversity of some Penicillium species using random amplified polymorphic DNA (RAPD) was reported by Tiwari et al. (2011) and a similar study was conducted by Josephine and Gnanadoss, in 2014.
5 Conclusion
The present study highlights the isolation and identification of two fungi Chaetomium seminis-citrulli and Penicillium guttulosum. The difference in the prevalence of different species of keratinophilic fungi may be due to tolerance to various environmental factors such as pH, temperature, moisture content and organic carbon. This study represents a combined approach to assess the genetic diversity in environmentally important fungal isolates. The NCBI GenBank data have provided the major source for identification of fungi.
Acknowledgement
The authors extend their appreciation to JECRC University, Jaipur, and PGIMER, Chandigarh, India for providing necessary research facilities and support for completion of this work.
References
- Soil as natural reservoir for human pathogenic fungi. Science. 1956;123(3203):876-879.
- [Google Scholar]
- Is Penicillium monophyletic? an evaluation of phylogeny in the family Trichomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87(2):210-222.
- [Google Scholar]
- Blakemore, L.C., Searle, P.L., Daly, B.K., 1987. Methods for Chemical Analysis of Soils. New Zealand Soil. Bureau Scientific Report.10A.
- New and interesting Chaetomium species from East Africa. Can. J. Bot.. 1982;60(7):1253-1262.
- [Google Scholar]
- Filamentous fungi isolated from the rhizosphere of melon plants (Cucumis melo L. cv. Gold Mine) cultivated in soil with organic amendments. Acta Bot. Brasilica. 2010;24(1):292-298.
- [Google Scholar]
- Characterization of dilution enrichment cultures obtained from size-fractionated soil bacteria by BIOLOGR community-level physiological profiles and restriction analysis of 16S rDNA genes. Soil Biol. Biochem.. 2001;33(11):1555-1563.
- [Google Scholar]
- The occurrence of dermatophytes and other keratinophilic fungi from the soils of Himachal Pradesh (India) Czech Mycol.. 2006;58(1/2):117-124.
- [Google Scholar]
- Evolutionary trees from DNA Sequences: a maximum likelihood approach. J. Mol. Evol.. 1981;17(6):368-376.
- [Google Scholar]
- A survey on the pathogenic fungi in soil samples of potted plants from Sari hospitals. Iran J. Hosp. Infect.. 2004;58(1):59-62.
- [Google Scholar]
- Screening, molecular characterization and phylogenetic analysis of Penicillium sp producing high levels of protease. Int. J. Curr. Sci. 2014:10-17.
- [Google Scholar]
- Polymerase chain reaction: restriction fragment length polymorphism differentiates the environmental and clinically important fungal isolates. Nat. Acad. Sci. Lett.. 2013;36(2):139-146.
- [Google Scholar]
- Isolation and identification of fungi from coal mines near Hazaribagh and their Diversity Study. J. Cell Sci Apo.. 2017;1(1)
- [Google Scholar]
- Prevelance of non keratinophilic fungi in the soil. Indian J. Med. Microbiol.. 2005;23(2):44-145.
- [Google Scholar]
- Application of PCR to the identification of dermatophytic fungi. J. Med. Microbiol.. 2000;49(6):493-497.
- [Google Scholar]
- Species identification system for dermatophytes based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1. Nippon Ishinkin Gakkai Zasshi. 2001;42(2):61-67.
- [Google Scholar]
- Phylogenetic classification of Trichophyton mentagrophytes complex strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 region. J. Clin. Microbial.. 1999;36(9):2629-2633.
- [Google Scholar]
- Studies on wet oxidation procedures for the determination of organic carbon in soils. CSIRO Division of Soils, Notes on Soil Techniques, 73–79.
- Isolation of keratinophilic fungi from floors in Rome (Italy) kindergarten and secondary schools. Mycopathologia. 1986;94(2):109-115.
- [Google Scholar]
- Identification problems with sterile fungi, illustrated by a keratitis due to a nonsporulating chaetomium-like species. Med. Mycol.. 2012;50(4):361-367.
- [Google Scholar]
- Implantation phaeohypho mycosis caused by a non-sporulating Chaetomium species. J. Mycol. Med.. 2014;24(2):161-165.
- [Google Scholar]
- The Genus Penicillium and its Teleomorphic States Eupenicillium and Talaromyces. London, UK: Academic Press; 1979.
- Discovering potential pathogens among fungi identified as non-sporulating molds. J. Clin. Microbiol.. 2007;45(2):568-571.
- [Google Scholar]
- Rapid identification of fungi by sequencing the ITS1 and ITS2 region using an automated capillary electrophoresis system. J. Mycol.. 2003;41(4):369-381.
- [Google Scholar]
- Ribosomal DNA spacer-length polymorphism in barley: mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. USA. 1984;81:8014-8018.
- [Google Scholar]
- The neighbor-joining method for reconstructing phylogenetic tree. Mol. Biol. Evol.. 1987;4(4):406-425.
- [Google Scholar]
- Taxonomy of Malabranchea and some other hyphomycetes with arthroconidia. Mycotaxon.. 1976;4:349-488.
- [Google Scholar]
- Singh, D., Chhonkar, P.K., Pande, R.N. 1999. Soil Reaction in Soil, Plant, Water analysis Method Manual, IARI, ICAR, New Delhi, 1: 4.2 (b) 11–13.
- Isolation and identification of Penicillium spp., from the river indus bed at Kotri. Pak. J. Bot.. 2006;38(4):1289.
- [Google Scholar]
- CLUSTAL W. improving the sensitivity of progressive multiple sequence alignments through sequence weighting. Position specific gap penalties, and weight matric choice. Nucleic Acid Res.. 1994;22(22):2673-2680.
- [Google Scholar]
- Morphological and molecular study of different penicillium species. Middle East J. Sci. Res.. 2011;7(2):203-210.
- [Google Scholar]
- Technique biologique pour l'isolement des dermatophytes du sol. Ann. Soc. Belge. Med. Trop.. 1952;32(2):173-178.
- [Google Scholar]
- Identification of Penicillium expansum causing postharvest blue mold decay of apple fruit. Pesticidi i Fitomedicina.. 2014;29(4):257-266.
- [Google Scholar]
- Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. California. USA: Academic Press., San Diego. 1990;18(1):315-322.
- [Google Scholar]
- Polyphasic characterisation of Chaetomium species from soil and compost revealed high number of undescribed species. Fungal Biol.. 2017;121(1):21-43.
- [Google Scholar]







