Translate this page into:
Molecular characterization and identification of economically important Potyviruses in Cucurbitaceae family from Gujranwala division of Punjab, Pakistan
⁎Corresponding author. nadeem.ahmad@mnsuam.edu.pk (Nadeem Ahmed)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cucurbits belongs to Cucurbitaceae family and they are main source of energy, vitamins and minerals. The plant viruses in general and potyviruses in particular are considered to hamper the successful production of cucurbits in Punjab province of Pakistan. The present study was undertaken to determine the current status of major potyviruses infecting cucurbits in Gujranwala division of Punjab. Overall, 528 samples of cucurbit species were collected from 46 fields of six districts (Narowal, Sialkot, Gujranwala, Gujrat, Mandi Baha Uddin and Hafizabad) in Gujranwala division. All disease samples were screened for the presence of potyvirus infection by indirect plate-trapped antigen (PTA)-ELISA using “Poty-group test” kit (Bioreba AG, Switzerland). These samples were further tested for Papaya ring spot virus (PRSV), Zucchini yellow mosaic virus (ZYMV) and Watermelon mosaic virus (WMV) (Bioreba AG) using virus specific DAS-ELISA. Zucchini yellow mosaic virus (ZYMV) was also further confirmed by reverse transcriptase polymerase chain reaction (RT-PCR). ZYMV was found as the most predominant virus of cucurbits in Gujranwala division with an average incidence of 20.8%, followed by PRSV (7.2%) and WMV (1.7%). Maximum mixed double infection was observed in combination of ZYMV+PRSV (1.3%) followed by ZYMV+WMV (0.4%) and WMV+PRSV (0.8%). Overall highest disease incidence of potyviruses was in Gujranwala (40.86%) followed by Narowal (34.65%), Gujrat (28.6%), Mandi Baha Uddin (26.56%), Hafizabad (25%) and Sialkot (23.21%). Likewise, the maximum ZYMV incidence was recorded in Angel Gourd whereas no viral infection was observed in snap melon and ash gourd in Gujranwala division. All cucurbits vegetables were found to be highly prone to ZYMV followed by PRSV and WMV. The sequence of CI region of ZYMV AABRM-CI ridge gourd isolate was deposited in GenBank with an accession no. MN897101, which showed nucleotide identities from 91.501-98.401% with other isolates of ZYMV available at database. Highest nucleotide identity 98.401% was found with the sequence of ZYMV South Korean isolate BR1 (MH042024) followed by 98.301% with ZYMV isolate Y23 from Turkey (KP828425). The results of the current study would be useful in devising management strategies to mitigate potyvirus losses in cucurbits from Pakistan.
Keywords
Cucurbit
Potyviruses
ZYMV
PRSV
WMV
1 Introduction
Cucurbits (cucumber, pumpkin, watermelon, melon, squash, bottle gourd, Round Gourd, Squashes) belongs to family Cucurbitaceae and their members occupy a prominent position among vegetables crops in Pakistan. They are broadly cultivated in tropical, subtropical and temperate regions of the world.Cucurbit has high nutritional importance as it contains Vitamin B, niacin, lipid, protein, carbohydrates, Calcium and iron (Kayani et al., 2013). In the world, its area under cultivation is 50.019 thousand ha with total yield 202.39 kg/ thousand ha and production of 0.101 million tons whereas in Pakistan it is cultivated on an area of 4586 ha with 132174 kg/ha yield and 60,611 tons of production (FAOSTAT, 2017-18). Cucurbitaceae is an imperative family of vegetables that consists of 825 species from 125 genera (Rai et al., 2008). In the world, the most important summer and winter crops of family Cucurbitaceae are red pumpkin (Cucurbita maxima), cucumber (Cucumis sativus), summer squash (Cucurbita pepo), bottle gourd or loki (Lagenaria siceraria), sponge gourd (Luffa aegyptiaca), Ash gourd or white gourd (Benincasa hispida), bitter gourd (Momordica charantia), Angle gourd (Luffa acutangula), smooth gourd (Luffa cylindrica), snake or serpent gourd (Trichosanthes cucumerina) Red Gourd or halwa kaddu (Cucurbita maxima), round gourd or tinda (Praecitrullus fistulosus), water melon (Citrullus lanatus) and sweet melon (Cucumis melo L.) and Snap melon or phoot (Cucumis melo momordica).
Plant diseases are the major barrier in the successful production, consumption and export of vegetables that not only deteriorate the quality but also diminishes the yield (Yardimici and Korkmaz, 2004; Hussain et al. 2021, Qasim et al. 2021, Zaynab et al., 2019). Among those yield inhibiting or retarding factors, viral diseases possess a pivotal role in the reduction of cucurbits that are under cultivation all over the world (Islam et al. 2018). Wide range of viruses are obnoxiously affecting cucurbits cultivation in developing countries owing to abrupt variation in ecological factors and genetic diversity of the host plants. Cucurbit viruses cause significant losses to plants and 59 plant viruses has infected drastically (Lecoq and Desbie, 2012). Potyviruses severely affect cultivation of cucurbits. Potyviruses is the largest genus that belongs to family Potyviridae which contains 218 unarguable and indefinite species. It is the biggest genus currently includes nearly 111 unarguable and 86 indefinite species of virus (Berger et al., 2005). Three economically crucial and most common potyviruses are Zucchini yellow mosaic virus (ZYMV), Papaya ringspot virus (PRSV) and Watermelon mosaic virus (WMV) (Lecoq et al., 2001).
Similarly, more than two hundred aphid species are recognized as the vectors of potyviruses (Edwardson and Christie, 1991). Potyviruses appear on several cucurbit crops with characteristic symptoms including mosaic, stripe, vein clearing, mottling, stunting, ringspots, vein banding, necrotic lesions and wilting (Sharma et al., 2014).
Although limited work has been done on Potyviruses of cucurbits in Pakistan (Malik et al., 2010; Ali et al., 2004) but it was restricted to some specific areas of K.P.K. and scare information is available regarding viruses of cucurbits from Punjab in general and Gujranwala division in particular. Therefore, the ideal way to combat the viral disease is use of ELISA. For this purpose, disease prevalence, incidence and distribution of major viruses is pre-requisite to detect the source in the existing germplasm of cucurbits in Pakistan.
1.1 Aims of research
The current research was conducted to find out the source of disease prevalence, incidence and distribution in cucurbits as well as genetic diversity in virulent viral strains through phylogenetic analysis for the management of viral disease under natural conditions. This disease is causing huge losses in cucurbit cultivating zones of the world.
2 Materials and methods
2.1 Survey and sample collection
Survey of major cucurbits growing areas of Gujranwala division including Narowal, Sialkot, Gujranwala, Gujrat, Mandi bahauddin and Hafizabad was conducted during 2019–2020. Samples of different cucurbits were collected from several fields (Fig. S1) in such a way that leaf samples exhibiting characteristics symptoms such as yellowing, severe mosaics, blisters, fan leaf appearance discoloration and size reduction in fruits. Each sample from different plants were kept in plastic bags in ice boxes during the survey and brought to plant virology lab in department of plant pathology MNS-University of Agriculture Multan and wash with tap water to remove superficial dust. Leaves were cleaved into two halves one is preserved at 4 °C for serological analysis and other is preserved at −20 °C for RT-PCR amplification. The infected samples were examined using PTA ELISA (Art no. 163075) and DAS ELISA (Art no, 160677) as reported by Clark and Adams (1977) for virus confirmation and incidence determination of Potyviruses in cucurbits. Basic information regarding crop e.g. variety, irrigation, agricultural practices and disease problems etc. was also collected from respective or concerned farmers/growers of cucurbit.
2.2 Serological assay
Infected samples were examined using PTA-ELISA by applying mono-clonal antibodies of potyviruses (Clark and Adams, 1977). One gram of infected samples was homogenized in 50 ml coating buffer (pH 9.6) and loaded 200 µL in each well of the ELISA plate. Plates were covered and incubated overnight at 4 °C. Polystyrene plates were given washing with phosphate buffer saline tween (PBST) three times with 3 min interval. Plants were properly dried and add 200 µL of blocking buffer in each test well, properly wrap the plates and incubated at 37 °C for one hour followed by washing and drying. The antibodies (dilute antibody IgG 1000x in conjugate buffer) were added and loaded 200 µL in each well of the ELISA plate. The plants were covered tightly and incubated in a moist chamber for two hours at 37 °C followed by washing.200 µL conjugated IgG was filled in each well of the ELISA plate and incubated at 37 °C followed by washing as described earlier. p-nitro-phenyl–phosphate tablet @ 1 mg/mL was dissolved immediately before use to prepare substrate solution. For better color development, 250 µL of substrate was added in each well of the ELISA plate followed by incubation at room temperature for 30 min in dark. Yellow colour development was assessed visually and read after 30–50 min. Reaction were visualized as strong (+++), moderate (++), mild (+) and weak (−) and by using ELISA reader intensity of development of yellow color was metered as optical density (OD405 nm).
2.3 Disease incidence
The samples exhibited ELISA positive were further interpreted to calculate disease incidence. After application of proportionate test, relative incidence was calculated in percentages (Steel and Torrie, 1980).
2.4 Indexing of viruses
The samples, showed positive response during survey, were obtained after PTA-ELISA, specific potyviruses such as Zucchini Yellow Mosaic Virus (ZYMV), Papaya Ring Spot Virus (PRSV) and Watermelon Mosaic Virus (WMV) were confirmed through DAS-ELISA as reported by the Clark and Adams (1977) by exploiting specific monoclonal antiserum. Relative occurrence in percentage of a single virus was recorded by using the following formula
2.5 Maintenance of Potyviruses on test plants through mechanical inoculation
Fresh leaf tissues from highly ELISA positive samples were standardized (1:10w/v) in 0.04 M phosphate buffer with pH 7.2 in sterilized cold pestle and mortar. Sodium diethyldithiocarbamate (Na-DIECA) was added as an additive to enhance the infectivity of inoculum and filtered through a double layer muslin cloth (Noordam, 1973). Different Test plants and cucurbits healthy plants nursery were grown in sterilized soil mixture containing of peat, clay and sand in equal ratio (1:1:1). Inoculum was applied at 2–3 leaf stage of plant. Leaves were provided with 600 mesh carborandum powder and with the help of cheese cloth or forefinger sap inoculum was also applied on the leaves. All inoculated leaves were rinsed with water after 1 to 2 min to get rid of the superfluous inoculum. All the treated plants were kept under controlled conditions for symptoms development. Characteristic viral symptoms appeared on newly emerged young leaves. Presence of potyviruses was confirmed through ELISA.
2.6 Molecular characterization of Potyviruses
2.6.1 Total RNA extraction
By applying Trizol method, total RNA was extracted. Infected fresh leaf samples (50–100 mg) were grinded by using 1 ml of Tri Reagent (Sigma) into a powder form as stated in manufacturer’s directions and transferred in an Eppendorf tube. All the wells were vortexed and kept in incubation at room temperature for 2–3 min. Following the incubation, 200 µL of chloroform/ml of Trizole reagent were added, shaken thoroughly for 15 s and incubated at room temperature for 5 min. Following the centrifugation at 14,000 RPM at 4 °C for 15 min, the uppermost aqueous phase containing RNA (supernatant) was collected and transferred into new RNA free 1.5 ml Eppendorf tube. After that, 500 µL of isopropanol was filled in each tube and incubated for 5 min at room temperature. Then centrifuged again at 14,000 RPM for 10 min at 4 °C. After centrifugation, the supernatant was taken off and 1 ml of 75% ethanol was added, vortexed shortly and centrifuged at 7500 RPM for 5 min at 4 °C. Pellet was collected, air dried for 5 min and by gentle pipetting pallet was resuspended in RNA free 50–100 µL of water. After incubating the samples at 50–60 °C for 10–15 min, samples were kept at −80 °C for future use.
2.6.2 Complementary deoxyribonucleic acid synthesis (cDNA Synthesis)
cDNA was synthesized by using downstream reverse primer specific to CI gene with the help of reverse transcriptase enzyme manufactured by thermo scientific company. In sterilized tubes on ice, master mixture of 5 µL of DEPC-containing water, 1 µL of oligo dT (20 picomol) and 5 µL of RNA (100 mg) were added in each sample followed by incubation at 65 °C for 5 min. Samples were quickly chilled on ice after incubation and then centrifuged. 4 µL of RT buffer (5x), 1 µL of dNTPs (10 Mm), 1 µL Revert Aid Reverse Transcriptase and 1 µL of Thermo Scientific Ribolock RNase inhibitor were added in the tube. Mixture was centrifuged briefly and incubated at 37 °C for 60 min. Heating was provided to the tube at 70 °C for 10 min to terminate the reaction.
2.6.3 Polymerase chain reaction (PCR)
50 µL reaction mixture was prepared by adding 35 µL nuclease free water, 5 µL PCR buffer (10x), 1 µL dNTPs (10 Mm), 3 µL MgCl2 (25 mM), 1 µL primer F(5′-GGIVVIGTIGGIWSIGGIAARTCIAC-3′) (Ha et al., 2008) and 1 µL primer R (3′-ACICCRTTYTCDATDATRTTIGTIGC-5′), 0.4 µL Taq polymerase (500uL), and 4 µL DNA template (2 µg) by providing the specific conditions for the reaction as follows, 94 °C for 3 min, 40 cycles of 94 °C for 30 s, 40 °C for 30 s and 72 °C for 60 s. The reaction was provided with incubation at 72 °C for 10 min to ensure the complete extension of amplified fragments.
2.6.4 Agarose gel electrophoresis
Amplified PCR products were run in 1% (w/v) agarose gel containing 8 µL ethidium bromide in 1x Tris-borate EDTA (TBE) buffer, mixed the amplified samples with 6X loading dye and specific marker were loaded in the gel and electrophoresis at 110 V until the bromophenol migrated approximately half of the length of the gel. DNA bands were observed under UV Transilluminator and photographs were taken.
2.7 Ligation and transformation of amplified product into E. coli
Cloning vector pGEM-T Easy was used for the ligation of amplified fragments. Reaction mixture made for ligation was composed of 3.5 μL purified PCR products, 9 μL distilled water, 4 μL ligation buffer (5X), 1.5 μL vector and 2 μL T4 DNA ligase. The incubation was followed overnight at 4 °C. Next day, tube having 100 µL of chemically competent cells strains DH5 ∝ was added with 1.5 μL of ligation reaction, and after mixing placed in ice for 20 min. In waterbath, cells were provided with heat shocks at 42 °C for 45 s and then kept on ice for 2 min. After that, mixture was placed on shaker for 1 h at 37 °C after adding 1 ml of pre-warmed LB broth at 37 °C. Centrifugation for 2 min was given to get a pellet of bacterial cells; a large portion of supernatant was discarded. White colonies were picked from these petri plates with autoclaved toothpicks and were shifted in the test tubes having 10 µL of LB broth medium, ampicillin @ 1 µL/ml of media and incubated overnight at 37 °C at 250 RPM.
2.8 Sequence comparison and phylogenetic analysis
Cylindrical inclusion (CI) gene sequence of samples obtained from Macrogen Korea, was aligned using Clustal W and MEGA 6 software was used for phylogenetic analysis. The acquired sequences of ZYMV were compared with retrieved sequences from DDBJ or GenBank data bases through Blast application (http:/www.ncbi.nlm.nih.gov/BLAST/blastn). By using the “Sequence Identity Matrix” option in BioEdit program version 7.25 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) Sequence identities were determined. Following the Neighbour-joining method, phylogenetic trees were constructed from Clustal W-aligned sequences applying MEGA version 6.0 program (Tamura et al., 2013).
3 Results
3.1 Survey and monitoring of disease
Total 46 cucurbit fields were surveyed, and 528 plant samples were randomly collected from six districts of Gujranwala division, Punjab, Pakistan (Table 1). Meanwhile, different cucurbit vegetables were surveyed, newly growing apical leaves with conspicuous characteristic symptoms as yellowing, severe mosaics, blisters, fan leaf appearance, shoe strings formation of leaves, discoloration and size reduction in fruits were observed from Red Pumpkin, Round gourd, Cucumber, Bottle gourd, Angle gourd, Ash gourd, Bitter gourd, Squashes, Red Gourd, Melon and Water-melon (Fig. 1). The highest incidence of viruses i.e. 40.86% was recorded in Gujranwala district followed by 34.65%, 28.6%, 26.56%, 25% and 23.21% in Narowal, Gujrat, Mandi Baha Uddin, Hafizabad and Sialkot districts, respectively (Table 1).
Districts
No. of fields surveyed
No. of samples collected
ELISA positive samples
Disease incidence%
Gujrat
5
115
33
28.6
Narowal
10
101
35
34.65
Gujranwala
10
93
38
40.86
Sialkot
5
56
13
23.21
Mandi Buhauddin
8
64
17
26.56
Hafizabad
8
99
25
25.0
Total
46
528
161
30.5

Cucurbit Plants (Angle Gourd, Cucumber, Bitter Gourd and Red Pumpkin) showing viral Symptoms under Field Conditions.
Some fields showed severe infections, particularly Red Pumpkin leaves were showing the symptoms of shoe-string, similar types of symptoms already reported by Ali et al. (2004). None of the districts was found to be free of virus infection. Hence three potyviruses were found more prevalent throughout the surveyed areas. Two of the three viruses were found in samples collected from all six districts while none of the samples were identified to be infected with WMV in Gujrat and Sialkot. ZYMV was identified as the most prevalent virus followed by PRSV in all the districts of Gujranwala division (Fig. 2A). The overall relative incidence of ZYMV, WMV and PRSV were 28.8%, 1.7%, and 7.2% respectively in all districts of Gujranwala division, Punjab, Pakistan (Table 2).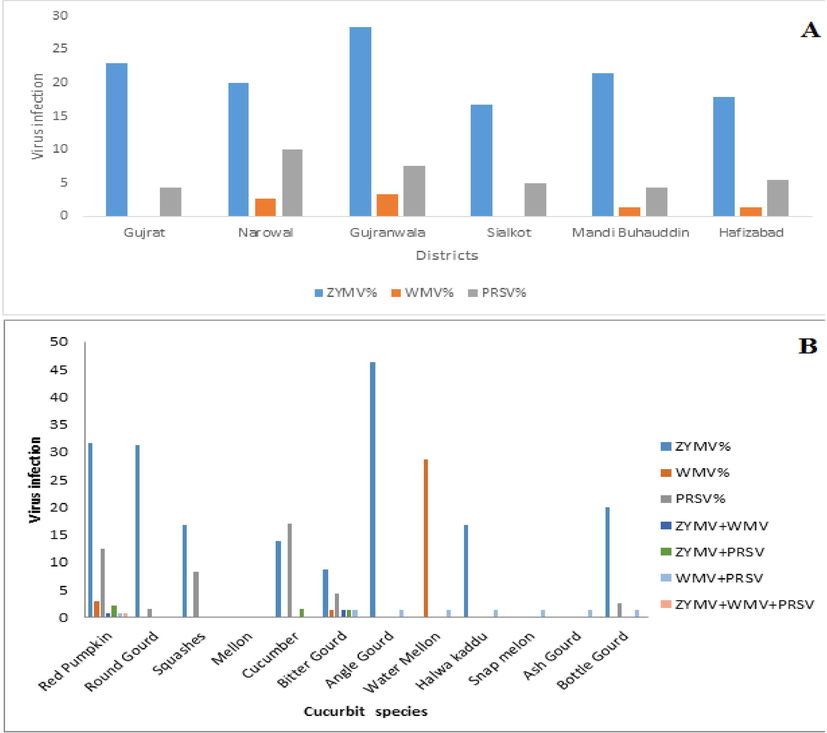
Indexing of Potyviruses in different districts (A) and cucurbit vegetables (B).
Vegetables
No. of tested samples
Poty group
ZYMV
WMV
PRSV
ZYMV + WMV
ZYMV + PRSV
WMV + PRSV
ZYMV + WMV + PRSV
Red Pumpkin
138
64
43
4
17
1
3
2
2
Round Gourd
65
21
20
0
2
0
0
0
0
Squashes
25
6
4
0
3
0
0
0
0
Mellon
36
0
0
0
0
0
0
0
0
Cucumber
66
20
9
0
11
0
2
0
0
Bitter Gourd
72
10
6
1
3
1
2
2
0
Angle Gourd
40
19
18
0
0
0
0
0
0
Water Mellon
14
4
0
4
0
0
0
0
0
Halwa kaddu
13
2
2
0
0
0
0
0
0
Snap Melon
5
0
0
0
0
0
0
0
0
Ash Gourd
12
0
0
0
0
0
0
0
0
Bottle Gourd
42
9
8
0
2
0
0
0
0
Total
528
155
110
9
38
2
7
4
2
Percent infection
29.4
20.8
1.7
7.2
0.4
1.3
0.8
0.4
Based on ELISA tests, ZYMV and PRSV were the most prevalent viruses in Gujranwala division. Their recorded incidence reached to 31.6% and 12.5% in red pumpkin, 13.8% and 15.4% in cucumber, 31.2% and 1.6% in round gourd, 16.7% and 8.3% in squashes and 8.6 and 1.3% in bitter gourd respectively. Infection rate of ZYMV in angle gourd, red Gourd and bottle gourd were 46.2%, 16.7% and 20% respectively other viruses were not found in that cucurbit species whereas WMV was predominant with an incidence of 28.6% in watermelon. WMV was also detected in red pumpkin and bitter gourd with an incidence of 2.9% and 1.4% respectively while other cucurbit species were not infected with WMV. (Table 2 and Fig. 2A).
Double virus infection was detected in 9 cucurbit species. The most common double infection detected was ZYMV + PRSV (1%) followed by ZYMV + WMV and WMV + PRSV which was 0.4%. Triple infection ZYMV + PRSV + WMV was found only in red pumpkin. (Table 2 and Fig. 2B).
Zucchini yellow mosaic virus (ZYMV) positive samples obtained from the survey areas were mechanically transmittable to the tested plants i.e. Cucurbita maxima, Cucumis sativus, Lagenaria siceraria, Momordica charantia, Luffa acutangula and Citrullus lanatus. The tested plants showed Leaf curling, vein clearing and mosaic like symptoms to cucumber (Cucumis sativus), red pumpkin (Cucurbita maxima) and round gourd (Citrullus lanatus) (Fig S2) whereas bottle gourd (Lagenaria siceraria) showed mild symptoms. Angle gourd (Luffa acutangulaand) smooth gourd (Luffa aegyptiaca), Cucumber and red pumpkin showed severe symptom after one week when placed in dark chamber as compare to other indicator plants which were placed under normal condition. To diagnose the virus in variety of plant species, to test the virus infectivity by local lesions on host leaves and for virus propagation, mechanical inoculation was used. Inoculation at 2–3 leaf stage expressed better results than inoculating older leaves stage.
Potassium phosphate buffer saline (PBS) was applied for maceration of the infected plant tissues. Potassium phosphate buffer destabilized the virus during extraction for that reason sodium diethyldithiocarbamate (Na-DIECA) as an additive was to enhance the infectivity in the sap.
3.2 Serological assay
Potyviruses in survey samples were identified through PTA-ELISA by using monoclonal antibodies (Clark and Adams, 1977). A high contrast yellow color developed in the wells of ELISA plate. The mean observance values at 405 nm for negative range was 0.0210 for positive control range was 1.715. Numerous plants showed ELISA positive results such as Squashes (Cucurbita pepo L.), Red pumpkin (Cucurbita maxima), Round gourd (praecitrullus lanatus var lanatus) reacted strongly positive while Bitter Gourd (Momordica charantia), bottle gourd (Lagenaria siceraria), smooth gourd (Luffa aegyptiaca Mill.) Cucumber (Cucumis sativas L.), reacted moderately positive. Angle gourd (Luffa acutangula), watermelon (Cucurbita lanatus Thunb. Mansef) and melon (Cucumis mello) showed mild positive reaction. Potyvirus was not detected in ash gourd (Beninca sahispida). ELISA is a quick, versatile and highly sensitive technique for future research of viruses aiming to the diagnosis and serological variation. ELISA comparative incidence was noted on the basis of PTA and DAS ELISA and confirmation by ELISA was considered adequate for most of the time but occasionally plants were re-checked through PCR.
3.3 Sequencing and phylogenetic analysis for ci gene of ZYMV
Sequences of cylindrical inclusion gene of Zucchini yellow mosaic virus infecting cucurbits were obtained for more precise and accurate evaluation of genetic assortment and resemblance. Cylindrical inclusion (CI) gene is regarded as most conserved component of all the Potyviruses which can be helpful for the taxonomic studies and diagnosis. Phylogenetic studies provided productive means for evaluating evolutionary hypothesis concerning genetic distinction. Sequence gaps of aligned sequences were totally removed. After running the blast and comparing with the sequences in GenBank, isolate AABRM-CI, found highest similarity percentage of nucleotide already identified from South Korean isolate BR1 (MH042024) i.e. 98.42 and lowest percentage 91.65 percent with ZYMV isolate Z-104 (MK956829) found in Italy (Table 3). The variance/resemblance percentage between species was determined at nucleotide and amino acid level (Fig S3).
Accession No.
Country Name
Host Name
Isolate Name
Year
Amino Acids similarity %
MH042024
South Korea
Cucurbita pepo
ZYMV
2016
98.42
KP828425
Turkey
Cucurbita pepo
ZYMV
2012
98.28
MN296124
China
Cucumis melo
ZYMV
2019
97.84
MK124612
USA
Cucurbita moschata
ZYMV
2016
97.7
AY279000
South Korea
Cucurbita moschata
ZYMV
2006
97.7
KX664482
China
Cucurbita pepo
ZYMV
2016
93.82
KX421104
China
Sesamum indicum
ZYMV
2016
93.25
MK033874
China
Luffa aegyptiaca
ZYMV
2018
93.1
AB369279
South Korea
Cucumis melo
ZYMV
2007
92.96
KX249747
China
Luffa aegyptiaca
ZYMV
2017
92.39
AY278998
South Korea
Cucurbita moschata
ZYMV
2006
92.39
AB020477
Japan
Cucurbita pepo
ZYMV
1999
91.94
MN598576
Australia
Curcumis melo
ZYMV
2017
91.80
KJ875865
USA
Cucurbita pepo
ZYMV
2011
91.80
KP828404
Turkey
Cucurbita moschata
ZYMV
2014
91.8
KJ923769
USA
Cucurbita pepo
ZYMV
2011
91.8
KJ923767
USA
Cucurbita pepo
ZYMV
2011
91.8
MT161993
Iraq
Cucumis sativus
ZYMV
2018
92.02
AJ307036
China
Cucumis sativus
ZYMV
2002
91.80
MK956829
Italy
Cucurbita pepo
ZYMV
2019
91.65
The isolate AABRM-CI displayed that nucleotide resemblance ranged from 91.501-98.401 percent with other isolates of ZYMV available in GeneBank. The highest nucleotide identity 98.401% was found with the sequence of ZYMV South Korean isolate BR1 (MH042024) from Cucurbita pepo followed by 98.31 percent with Cucurbita pepo isolate Y23 from Turkey (KP828425) downloaded from GenBank (Table 3) and maximum amino acid based similarity percentage 98.28 was observed with Turkish isolate (AKV89043) (Table 4).
Accession No.
Country Name
Host Name
Isolate Name
Year
Nucleotides similarity %
AKV89043
Turkey
Cucurbita pepo
ZYMV
2015
98.28
AKV89037
Turkey
Cucurbita pepo
ZYMV
2015
98.28
AKV89062
Turkey
Cucumis melo
ZYMV
2015
98.28
AKV89057
Turkey
Cucurbita moscheta
ZYMV
2015
98.28
AKV89042
Turkey
Cucurbita pepo
ZYMV
200
97.84
QRG28834
Iraq
Cucumis sativus
ZYMV
2020
98.69
AKV89061
Turkey
Cucurbita pepo
ZYMV
2015
97.84
AKV89054
Turkey
Cucurbita moscheta
ZYMV
2015
97.84
To regulate the relationship of AABRM-CI isolate with isolates from other countries, phylogenetic tree was constructed (Fig. 3A) based on nucleotide a sequences of CI coding region. Phylogenetic tree revealed that ZYMV isolates grouped into two main clusters. The cluster I was further divided into two sub-clusters A and B. The isolate AABRM-CI present in the sub-cluster A forming close relationship with the isolates MH042024, KP828425, AY279000 from South Korea, Turkey and South Korea respectively. while the sub-cluster B grouped the isolates from China, South Korea and USA.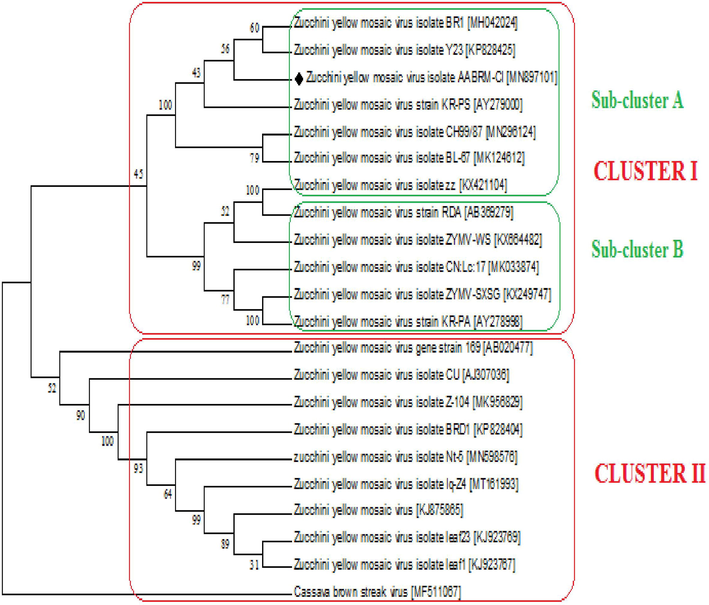
Tree showing Phylogenetic relationship of ZYMV isolates on the basis of nucleotides with other ZYMV reported isolates.
The results of current research depicted that molecular level characterization and diagnosis is quite accurate and very precise. Cylindrical inclusion region of Potyviruses is considered as conserved region (Adams et al., 2005) and now extensively used for the characterization of Potyviruses.
4 Discussion
This study demonstrated the pervasive occurrence of ZYMV infecting cucurbits crops in Gujranwala division of Pakistan. It implies that this disease has been existing in the field since it has been identified for a long time (Ashfaq et al., 2015; Ashfaq and Ahsan, 2017). Although data regarding yield losses were unavailable, the higher disease incidence in this region was a cause of concern as premature crop defoliation was attributed to the disease, which ultimately had an adverse effect on fruit size, yield and quality of crop. The occurrence of lesions and mosaic pattern on the leaves had a negative impact on the marketability and edibility of leafy vegetables because leaves with lesions were discarded (Zitter and Murphy, 2009). In addition to direct losses, the virus weakens plants, making them liable to other pest and diseases (Sharma, 2014). Most farmers are unaware of the symptoms of the virus infection and therefore do not recognize the disease, which means less or no adoption of control measures. Farmers need to be educated about the disease through extension programs; electronic and print media campaigns so that they know about it and adopt control measures. Virus characterization at a molecular level helps in the proper understanding of genetic composition, recombination, variation, and exact taxonomic status (Ahmad and Ashfaq, 2018). It also aids in predicting new strain evolution and its adaptation to new hosts and geographical conditions. The phylogenetic analysis revealed that determination of genetic diversity among ZYMV isolates is a productive step in virus managementand ZYMV AABRM-CI ridge gourd isolate showed close relationship with ZYMV South Korean isolate BR1 (MH042024) and ZYMV isolate Y23 from Turkey (KP828425) (Lin et al., 2003; Pratap et al., 2012). The molecular sequencing results of present study concluded that isolates of ZYMV from western Iran and northwestern Iran have the same origin as reported for Pakistani ZYMV AABRM-CI ridge gourd isolate. Similarly, the molecular divergence of ZYMV isolates in Czech and Slovakia Republic was evaluated from recovered isolates between 2001 and 2006 from variety of cucurbit hosts. Sequence analysis illustrated a great level of nucleotide resemblance between the isolates, characterized previously by Tobias and Palkovics (2003). The close genetic association resembling the ZYMV isolates with the central European region proposes that either they possibly have a common ancestor or have been developed from specific infection point (Svoboda et al., 2002). The phylogeny of the isolates as documented in the agreement reported by Glasa et al. (2007), where a genetic association relating the ZYMV isolates with the central European region (Slovakia and Czech Republic) was observed. According to Tymchyshyn et al. (2018) the phylogenetic studies of ZYMV among cucurbits is a key in controlling this catastrophic crop disease. The phylogenetic tree suggested that ZYMV isolates may be act as potent viral reservoirs and partly accounting for the geographic distribution of the virus (Simmons et al., 2011). Wang et al. (2020) revealed that phylogenetic analyses showed accuracy to understand the genetic diversification of ZYMV, and provided a theoretical base for developing preventive and management strategies. Sequences of the ZYMV gene available from NCBI databases revealed different groups of this virus isolates worldwide (Coutts et al., 2011) showing different geographical distributions (Zhao et al., 2003; Simmons et al., 2008). Similarly, Nasr-Eldin et al. (2016) concluded that identification and phylogenetic analysis of ZYMV in Egypt, determined the genetic diversity of ZYMV, and is one of the main dangerous virus infecting the major cucurbit crops.
5 Conclusion
Information generated from this study may prove fundamental for long term management of these notorious viruses. A comprehensive survey of this dangerous virus should be conducted to identify the infection in other division as well as in other vegetables. Moreover, the escalating host range of this virus is alarming and shows a great threat to crops, which should be tackled using active viral diagnostic and management approaches.
Acknowledgement
The authors gratefully acknowledge the financial support of Higher Education Commission of Pakistan for providing funding through NRPU project No. 8162 to conduct this research. The authors are also thankful to King Saud University for their support through Researchers Supporting Project Number (RSP-2021/5) King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant. Pathol.. 2005;6:471-487.
- [Google Scholar]
- Genetic diversity and recombination analysis based on capsid protein gene of Chilli veinal mottle virus isolates from Pakistan. Eur. J. Plant Pathol.. 2018;151:891-900.
- [Google Scholar]
- Identification and molecular characterization of viruses infecting cucurbits in Pakistan. J. Phytopathol.. 2004;152:677-682.
- [Google Scholar]
- First Report of Zucchini yellow mosaic virus in Round Gourd (Praecitrullus fistulosus) in Pakistan. Plant Dis.. 2017;101:265.
- [Google Scholar]
- First report of Zucchini yellow mosaic virus in ridge gourd in Pakistan. Plant Dis.. 2015;99:1870.
- [Google Scholar]
- Berger, P. H., Adams, M. J., Barnett, O. W., Brunt, A. A., Hammond J., Hill, J. H., Jordan, R. L., Kashiwazaki, S., Rybicki E., Spence, N., Stenger, D. C., Ohki, S. T., Uyeda, I., Van Jaayen, A., Valkonen, J., Vetten H. J., 2005. Potyviridae. In Fauquet, C. M., Mayo, M, A., Maniloff, J., Desselberger U., Ball, L. A., (eds) Virus taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London.
- Characteristics of micro plate method of Enzyme Linked Immunosorbent Assay for the detection of plant viruses. J. Gen. Virol.. 1977;34:475-482.
- [Google Scholar]
- Coutts, B. A., Kehoe M. A., Webster, C. G., Wylie, S. J., Jones, A. L., 2011. Zucchini yellow mosaic virus biological properties, detection procedures and comparison of coat protein gene sequences. Arch. Virol. 156, 2119-31.
- Edwardson, J. R., Christie, R. G., 1991. The potyvirus group. Florida agricultural experiment station monograph series 16.
- FAOSTAT, 2017-18. Food and agriculture organizations of United Nation. Vialedelle terme di caracalla 00153 Rome, Italy. http://www.fao.org/faostat/en/#home.
- Glasa, M., Svoboda, J., Novakova, S., 2007. Analysis of the molecular and biological variability of Zucchini yellow mosaic virus isolates from Slovakia and Czech Republic. Virus Genes. 35, 415–421. https://doi: 10.1007/s11262-007-0101-4.
- Identification and sequence analysis of potyviruses infecting crops in Vietnam. Arch. Virol. 2008;153(1):45-60.
- [Google Scholar]
- A basic helix-loop-helix transcription factor CabHLH113 positively regulate pepper immunity against Ralstonia solanacearum. Microb. Pathog.. 2021;156:104909
- [Google Scholar]
- Bemisia tabaci mediated facilitation in diversity of begomoviruses: evidence from recent molecular studies. Microb. Pathog.. 2018;123:162-168.
- [Google Scholar]
- Principles and Procedures of Statistics: A Biometrical Approach. McGraw-Hill Book Company, New York 1980
- [Google Scholar]
- Distribution, variability and overwintering of Zucchini yellow mosaic virus in the Czech Republic. Plant. Protect. Sci.. 2002;38:125-130.
- [Google Scholar]
- Infestation and assessment of root-knot nematodes (Meloidogyne spp.) associated with cucumber in the Pothowar region of Pakistan. Crop. Prot.. 2013;47:49-54.
- [Google Scholar]
- Viruses of cucurbit crops in the Mediterranean region: an ever-changing picture. Adv. Virus. Res.. 2012;84:67-126.
- [Google Scholar]
- Biological and molecular characterization of Morrocan watermelon mosaic virus and a potyvirus isolate from eastern Sudan. Plant. Dis.. 2001;85:547-552.
- [Google Scholar]
- Genetic diversity and biological variation among California isolates of Cucumber mosaic virus. J. Gen. Virol.. 2003;84:249-258.
- [CrossRef] [Google Scholar]
- Severe disease of melon in North West frontier province is associated with simultaneous infection of two RNA viruses. Pak. J. Bot.. 2010;42:361-367.
- [Google Scholar]
- Characterization and phylogenetic analysis of Zucchini yellow mosaic virus infecting Cucurbita pepo in Egypt. Am. J. Sci.. 2016;12(3):93-104.
- [Google Scholar]
- Identification of Plant Viruses. Methods and Experiments. Wageningen: Centre for Agricultural Publishing and Documentation (Pudoc); 1973. p. :207.
- Biological and molecular characterization of Cucumber mosaic virus isolate causing shoestring disease of tomato in India which has closer affinity to European or East Asian isolates of CMV. Indian. J. Virol.. 2012;23(1):57-63.
- [CrossRef] [Google Scholar]
- Host-pathogen interaction between Asian citrus psyllid and entomopathogenic fungus (Cordyceps fumosorosea) is regulated by modulations in gene expression, enzymatic activity and HLB-bacterial population of the host. Comp. Biochem. Physiol. C: Toxicol. Pharmacol.. 2021;248:109112
- [Google Scholar]
- Rai, M., Pandey S., Kumar, S., 2008. Cucurbit research in India: a retrospect. Pitrat M. (ed): Cucurbitaceae 2008, Proceedings of the IXth EUCARPIA meeting on genetics and breeding of Cucurbitaceae, Avignon (France), May 21-24th, 2008, pp. 285–293.
- Current status of Potyvirus in India. Arch. Phytopathol. Plant. Protect.. 2014;47:906-918.
- [Google Scholar]
- Survival, Detection and Management of Cucumber Mosaic Virus (CMV) And Zucchini Yellow Mosaic Virus (ZYMV) Infecting Cucurbits in Punjab. Ludhiana: Punjab Agricultural University; 2014. (Unpublished) thesis
- Rapid evolutionary dynamics of zucchini yellow mosaic virus. J. Gen. Virol.. 2008;89:1081-1085.
- [Google Scholar]
- Experimental verification of seed transmission of Zucchini yellow mosaic virus. Plant. Dis.. 2011;95:751-754.
- [Google Scholar]
- Characterization of Hungarian isolates of zucchini yellow mosaic virus (ZYMV, potyvirus) transmitted by seeds of Cucurbita pepo var Styriaca. Pest Manage. Sci.: Formerly Pesticide Sci.. 2003;59(4):493-497.
- [Google Scholar]
- Phylogenetic analysis of see-transmitted isolate of Zucchini yellow mosaic virus. Bull. Taras Shevchenko Natl. Univ. Kyiv-Biol.. 2018;74(2):46-50.
- [Google Scholar]
- Wang, D. F., Wang, J. R., Cui, L. Y., Wang, S. T., Niu, Y., 2020. Molecular identification and phylogeny of cucumber mosaic virus and zucchini yellow mosaic virus co-infecting Luffa cylindrica L. in Shanxi, China. J. Plant. Pathol. 1–11.
- Studies on spread and identification of Zucchini yellow mosaic virus disease in the North-West Mediterranean of Turkey by biological indexing and Double stranded RNA analysis. J. Plant. Pathol.. 2004;3:1-4.
- [Google Scholar]
- Molecular analysis of Zucchini yellow mosaic virus isolates from Hangzhou, China. J. Phytopathol.. 2003;151:307-311.
- [Google Scholar]
- Role of primary metabolites in plant defense against pathogens. Microb. Pathog.. 2019;137:103728
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101642.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







