Translate this page into:
Molecular analysis of ESBL- and AmpC-producing Enterobacteriaceae in fecal samples from broiler and backyard chickens
⁎Corresponding author at: Department of Microbiology and Biotechnology, Presidency College (Autonomous), Affiliated to University of Madras, Chennai, Tamil Nadu, India. drnarunagirinathan@gmail.com (Narasingam Arunagirinathan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
To analyse the molecular profile of antibiotic resistance in Enterobacteriaceae isolates from fecal samples of healthy broilers and backyard chickens in Tamil Nadu, India.
Methods
A total of 115 and 57 bacterial strains were isolated from fecal samples of broiler and backyard chickens, respectively using conventional bacterial culture tests. Bacterial isolates were screened for antibiotic susceptibility by Kirby-Bauer’s disk diffusion method. Colistin susceptibility was tested using the broth microdilution method. Extended-spectrum β-lactamase (ESBL), AmpC β-lactamase and metallo-β-lactamase (MBL) producers were screened by combination disk method. The drug-resistant genes’ positivity was analysed by polymerase chain reaction (PCR) method.
Results
Escherichia coli was the predominant bacterium isolated from both broiler (53%) and backyard (64.9%) chickens. Bacterial isolates from broilers and backyard chickens showed 100% resistance to ampicillin, piperacillin, and piperacillin-tazobactam. Five bacterial isolates from broiler chickens were resistant to colistin. ESBL producers were 34.7% in broilers and 19.1% in backyard chickens. AmpC production was detected in 44.3% of the isolates from broilers and 26.2% of those from backyard chickens. Molecular analysis revealed that bacterial isolates from broiler chickens were positive for drug-resistant genes such as blaTEM (25%), blaSHV (5%), blaCTX-M (4%), blaAmpC (56%), integron (39%), sul1 (30%), and sul2 (23%). Bacterial isolates from backyard chickens were positive for the blaTEM (22%), blaSHV (14%), blaAmpC (54.3%), and integron (20%) genes. None of the isolates from either broiler or backyard chickens harboured New Delhi metallo-β-lactamase (blaNDM) or colistin-resistant genes (mcr-1 to −3).
Conclusions
Bacterial isolates from broiler chickens harboured a higher number of drug resistance genes, especially blaTEM, blaCTX-M, sul1, sul2, and integron, than those from backyard chickens.
Keywords
Broiler chickens
Escherichia coli
Antimicrobial resistance
ESBL
Integron
1 Introduction
Antibiotics are extensively employed in poultry farms to enhance animal growth and treat bacterial infections in chickens. Globally the estimated usage of antibiotics in food animals was 63,151 tons in 2010 and 131,109 tons in 2013, and by 2030, it will increase to 200,235 tons. In the global consumption of antibiotics in food animals, India contributes approximately 3 % (Van Boeckel et al., 2015 & 2017). Quinolones, tetracyclines, β-lactams, polymixins and sulphonamides are major antibiotics used in poultry animals (Caneschi et al., 2023). Among the antibiotics used in poultry farms, 60 % are amoxicillin and colistin (Habiba et al., 2023).
The maximum permissible level of antibiotics in poultry animals ranges from 100 to 600 µg/kg, depending on the type of poultry animals and antibiotics (Bahmani et al., 2020). A study by the Centre for Science and Environment in India found that 40 % of chicken samples tested positive for antibiotic residues, with 17 % containing more than one antibiotic. The antibiotics detected were ciprofloxacin, chlortetracycline, oxytetracycline, enrofloxacin, doxycycline, and neomycin. Additionally, one sample contained doxycycline, enrofloxacin, and oxytetracycline, indicating the use of multiple antibiotics in poultry (Centre for Science and Environment, 2014). The primary route of antibiotic administration in poultry is through food materials. The indiscriminate use of antibiotics is a major factor contributing to bacterial antibiotic resistance in poultry farms (Kim et al., 2018). Contamination of the environment with poultry fecal material is a significant public health concern, as it can lead to the transfer of antibiotic-resistant Enterobacteriaceae from poultry fields to humans. Antibiotic residues in poultry meat raise food safety issues by causing allergic reactions and toxicity to the organs of consumers and by affecting the human gut microbiota (Zhang et al., 2021). A study reported the spread of AmpC β-lactamase and extended-spectrum β-lactamase (ESBLs) producing Escherichia coli via fecal and airborne routes in broiler farms and their surrounding areas (Laube et al., 2014). ESBL producers are showing resistance to 3rd and 4th generation cephalosporins and monobactam antibiotics. The most prominent ESBL-producing genes in bacterial pathogens are blaTEM, blaCTX-M, and blaSHV (Ewers et al., 2012; Ramesh Kumar et al., 2021). Along with ESBL production, the high positivity of tetracycline, erythromycin, sulfonamide, and chloramphenicol resistance was also reported in E. coli isolates from broiler chickens compared to layer chickens (Rahman et al., 2020). In this study, we investigated the antimicrobial resistance profile of bacterial strains isolated from fecal samples of broiler and backyard chickens.
2 Materials and Methods
2.1 Bacterial isolates
A total of 125 fecal samples of broiler chickens were collected from commercial broiler chicken shops at various places in Tamil Nadu, India, using pre-sterilized containers. From backyard chickens, 70 fecal samples were collected. The fecal samples of broilers from retail meat shops and backyard chickens were collected from Namakkal, Chennai, Cuddalore and Villupuram districts in Tamil Nadu (Table S1). The samples were collected by randomized selection and they were transported in the cold chain to the laboratory. For isolation of gram-negative bacterial (GNB) strains, a loop-full of fecal sample was inoculated on 5 % Sheep Blood agar and MacConkey Agar plates followed by incubation of the plates at 37 °C for 18–24 h. Each bacterial colony of the samples was taken for further analysis. Isolated colonies were characterized by the biochemical tests viz. catalase, oxidase, motility, IMViC, TSI, and urease for bacterial species identification. Isolates were further studied for antibiotic susceptibility testing and molecular analysis.
2.2 Antibiotic susceptibility test
The antibiotic susceptibility patterns of bacterial isolates from both broiler and backyard chickens were tested by Kirby–Bauer’s disc diffusion method (Bauer et al., 1996) using amikacin (30 µg), ampicillin (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), ertapenem (30 µg), fosfomycin (200 µg), meropenem (10 µg), gentamicin (10 µg), nitrofurantoin (300 µg), nalidixic acid (30 µg), piperacillin–tazobactam (100/10 µg), piperacillin (100 µg), polymyxin-B (300 units), trimethoprim–sulfamethoxazole (1.25 µg /23.75 µg) and tetracycline (30 µg) (Himedia, India). An overnight bacterial culture suspension was adjusted to 0.5 McFarland’s standard and a lawn culture was made on the Muller Hinton agar (MHA) plates. The discs containing the above-mentioned antibiotics were placed on the MHA agar surface and incubated at 37 °C for 18–24 h. The zone of inhibition around each disc was measured to interpret the results (Clinical and Laboratory Standards Institute, 2020). The standard strain Escherichia coli (25922) was used as a control.
2.3 Broth microdilution method for assessing colistin susceptibility
The susceptibility of bacterial isolates from broiler and backyard chickens to colistin was screened using the broth microdilution method. A stock of the colistin drug was made by dissolving 2 mg of colistin sulfate (MP Biomedical, France) in 1 mL of double distilled water. The colistin drug was diluted using cationic adjusted Mueller–Hinton broth (MHB)-2, and various concentrations ranging from 64 to 0.06 µg/mL were prepared. A bacterial culture suspension was made using 3–5 isolated colonies from agar plates in saline or Mueller–Hinton broth (MHB). The optical density (OD) of the liquid bacterial culture was adjusted to 0.5 McFarland standard. In the 96-well plate, 100 µL of diluted drug was added to the first well and 50 µL of MHB-2 was added to the next 9 wells in each row. 50 µL of diluted drug was transferred from 1st well to the next wells for serial dilution. Furthermore, 50 µL of culture suspension of the respective bacterial isolates was added to all the wells from 1 to 12 except for the 11th well. The 11th well was the medium control, and the 12th well was the culture control. The plate was incubated at 37 °C for 16–24 h. The minimum inhibitory concentration (MIC) of colistin was assessed by the lowest concentration that inhibited the growth of bacteria (Wiegand et al., 2008). The strain Escherichia coli (25922) was used as a control.
2.4 Identification of ESBL, MBL and AmpC production
All the bacterial isolates from broiler and backyard chickens were screened for beta-lactamase production by combination disc method (CDM). For this assay, the overnight bacterial culture suspension was adjusted to 0.5 McFarland’s standard and lawn culture was made on the MHA plate using a sterile cotton swab. For the detection of ESBL production, cefotaxime (30 μg) and cefotaxime in combination with clavulanic acid (30 μg/10 μg) and ceftazidime (30 μg) and ceftazidime in combination with clavulanic acid (30/10 μg) were used. MBL producers were detected using imipenem (10 μg) and imipenem in combination with EDTA (10/750 μg) discs, and AmpC production was detected using cefoxitin (30 µg) and cefoxitin-cloxacillin (30/200 μg) discs (Hosuru Subramanya et al., 2020).
2.5 Molecular detection of drug resistance genes
Bacterial cultures were used for the detection of ESBL-producing genes, such as blaTEM, blaSHV & blaCTX-M; AmpC producing gene, such as blaAmpC; MBL-producing genes, such as blaNDM; sulfamethoxazole (SMX) resistance genes, such as sul1 and sul2, and integrons, by the conventional polymerase chain reaction (PCR) method. Bacterial cultures showing colistin resistance by microdilution method were also tested for colistin-resistant genes viz. mcr-1 to −3 by PCR method (Oliver et al., 2002; Mugnaioli et al., 2005; Sunde, 2005; Arabi et al., 2015; Féria et al., 2022; Nordmann et al., 2011; Lescat et al. (2018). The set of primers used for amplification of each gene is presented in Table 1. The PCR mixture had 12.5 µL of master mix (Takara, India), 0.5 µL each of forward and reverse primers of respective genes, 10.5 µL of molecular grade water, and 1 µL of bacterial DNA template, for a total reaction volume of 25 µL. PCR reactions were run on a BioRad Thermal cycler. Amplified PCR products were visualized using 1.5 % agarose gel by electrophoresis technique and the size of the product was compared with a 100 bp DNA ladder (Table 1).
Target Gene
Primer Sequence
Target Size
BlaTEM
Forward
ATGAGTATTCAACATTTCCG
867 bp
Oliver et al. (2002)
Reverse
CTGACAGTTACCAATGCTTA
BlaSHV
Forward
GGTTATGCGTTATATTCGCC
867 bp
Reverse
TTAGCGTTGCCAGTGCTC
BlaCTX-M
Forward
ATGTGCAGYACCAGTAARGT
593 bp
Mugnaioli et al. (2005)
Reverse
TGGGTRAARTARGTSACCAGA
BlaAMPC
Forward
CCCCGCTTATAGAGCAACAA
634 bp
Féria et al. (2002)
Reverse
TCAATGGTCGACTTCACACC
BlaNDM
Forward
GGTTTGGCGATCTGGTTTTC
621 bp
Nordmann et al. (2011)
Reverse
CGGAATGGCTCATCACGATC
sul1
Forward
CGGCGTGGGCTACCTGAACG
433 bp
Arabi et al. (2015)
Reverse
GCCGATCGCGTGAAGTTCCG
sul2
Forward
GCGCTCAAGGCAGATGGCATT
293 bp
Reverse
GCGTTTGATACCGGCACCCGT
Int1
Forward
GCCACTGCGCCGTTACCACC
898 bp
Sunde (2005)
Reverse
GGCCGAGCAGATCCTGCACG
mcr-1
Forward
ATGCCAGTTTCTTTCGCGTG
502 bp
Lescat et al. (2018)
Reverse
TCGGCAAATTGCGCTTTTGGC
mcr-2
Forward
GATGGCGGTCTATCCTGTAT
379 bp
Reverse
AAGGCTGACACCCCATGTCAT
mcr-3
Forward
ACCAGTAAATCTGGTGGCGT
296 bp
Reverse
AGGACAACCTCGTCATAGCA
2.6 Statistical analysis
The SPSS version 26 and R version 4.3.3 software were used for statistical analysis. This study used the t-test to find the antibiotic resistance rate between broilers and backyard chickens. The data were pre-processed and structured using SPSS software. The t-test and 95 % CI were done using R software. The p ≤ 0.05 was considered statistically significant. Significance at the 5 % level was indicated with * and at the 10 % level was indicated with **.
3 Results
A total of 125 fecal samples were collected from broiler chickens, and 115 bacterial strains were isolated from them. Escherichia coli (53 %) was the major organism isolated, followed by Proteus mirabilis (14.8 %) and Proteus vulgaris (10.4 %). From the backyard chickens, 70 fecal samples were collected, and 57 bacterial strains were isolated from them. E. coli (64.9 %) was the predominant isolated bacterium, followed by P. vulgaris (14 %) and Serratia sp. (10.5 %). Other bacterial species isolated from both broiler and backyard chickens are given in Fig. 1.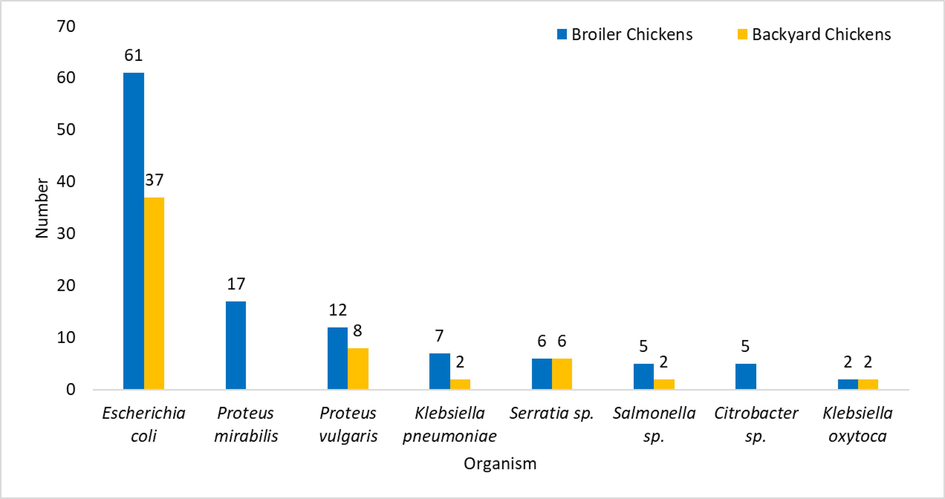
Bacterial isolates from fecal samples of broiler and backyard chickens.
3.1 Antibiotic susceptibility profile
In this study, 100 % of the E. coli isolates from broiler chickens showed resistance to ampicillin, piperacillin, and piperacillin/tazobactam followed by trimethoprim-sulfamethoxazole (29.5 %). K. pneumoniae strains exhibited 100 % resistance to piperacillin, ampicillin, and piperacillin/tazobactam and 57 % to ertapenem. K. oxytoca showed 100 % resistance to piperacillin, ampicillin, and piperacillin/tazobactam and 50 % resistance to ertapenem and gentamicin. P. mirabilis isolates showed 100 % resistance to ampicillin, piperacillin, and piperacillin/tazobactam and 52.9 % to gentamicin. P. vulgaris showed 100 % resistance to piperacillin, ampicillin, and piperacillin/tazobactam and 58.3 % to chloramphenicol. Salmonella sp. strains exhibited 100 % resistance to piperacillin, ampicillin, piperacillin/tazobactam, and polymixin B. A total of five Enterobacteriaceae isolates from broiler chickens were colistin-resistant (MIC, 16–32 µg/mL) (Table S2).
Among bacterial isolates from backyard chickens, 100 % of E. coli isolates showed resistance to ampicillin, piperacillin/tazobactam, and piperacillin. K. oxytoca showed 100 % resistance to piperacillin, ampicillin, and piperacillin/tazobactam. K. pneumoniae strains exhibited 100 % resistance to piperacillin, ampicillin, piperacillin/tazobactam, and 50 % resistance to polymyxin B. P. vulgaris strains exhibited 100 % resistance to piperacillin, piperacillin/tazobactam, and ampicillin followed by 50.0 % resistance to chloramphenicol and 25 % resistance to gentamicin and tetracycline. All the bacterial isolates from both the broiler and backyard chickens showed 100 % susceptibility to amikacin and fosfomycin (Fig. 2 and Table S3).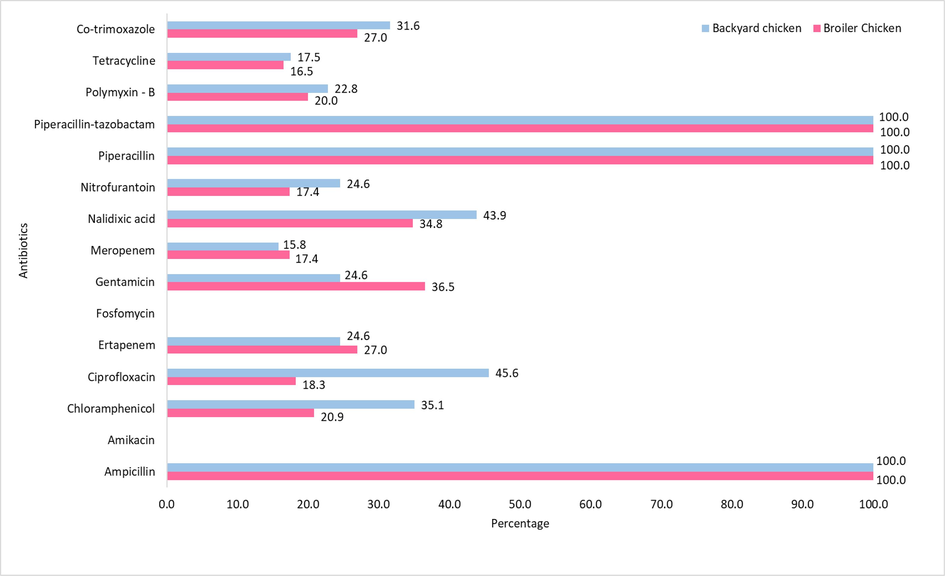
Antibiotic susceptibility of bacterial strains from broiler and backyard chickens.
It was observed that ciprofloxacin resistance was significantly higher in backyard chickens (95 % CI: 0.4421–0.0570; p = 0.0054*) than in broiler chickens. However, the gentamycin resistance was significantly higher in broiler chickens (95 % CI: 0.0164–0.3052; p = 0.0961**) than in backyard chickens.
3.2 Phenotypic detection of β-lactamase production
Phenotypic detection of beta-lactamase production revealed that 34.7 % of the bacterial isolates from broilers and 19.1 % from backyard chickens were ESBL producers. AmpC production was noted among 44.29 % and 26.2 % of the isolates from broiler and backyard chickens, respectively. None of the isolates from either the broiler or backyard chickens produced MBL (Table 2). Positivity for both ESBL and AmpC was noted for 17.4 % and 7.4 % of the Enterobacteriaceae from broiler and backyard chickens, respectively. E. coli isolates (10.4 %) from broiler chickens were the major contributors to the positivity of both ESBL and AmpC production.
S.
NoOrganism
Broiler Chicken (n = 115)
Backyard Chicken (n = 57)
ESBL
AmpC
ESBL
AmpC
n
%
n
%
n
%
n
%
1
Escherichia coli
22
19.1
23
20
6
10.5
8
14.0
2
Klebsiella pneumoniae
2
1.7
2
1.7
1
1.7
1
1.7
3
Klebsiella oxytoca
1
0.9
1
0.9
1
1.7
0
0
4
Proteus mirabilis
7
6.1
11
9.6
0
0
0
0
5
Proteus vulgaris
5
4.3
7
6.9
3
5.2
4
7.0
6
Salmonella sp.
0
0
2
1.7
0
0
0
0
7
Serratia sp.
3
2.6
2
1.7
0
0
2
3.5
8
Citrobacter sp.
0
0
3
2.6
0
0
0
0
Total
40
34.7
51
44.29
11
19.1
15
26.2
3.3 Drug-resistant genes
Molecular detection of drug-resistant genes revealed that 25 % of the isolates from broiler chickens harboured the blaTEM, 5 % blaSHV, 4 % blaCTX-M, 56 % blaAmpC and 39 % integron genes. Furthermore, 30 % of the isolates harboured sul1, and 23 % harboured sul2. None of the isolates were positive for blaNDM. All five phenotypically colistin-resistant isolates tested negative for the mcr-1 to −3 genes. In backyard chickens, 22 % of the isolates harboured blaTEM gene and other genes detected were blaSHV (14 %), blaAmpC (54.3 %) and integron (20 %) (Fig. 3). The positivity rate of blaSHV gene was significantly higher in backyard chickens (95 % CI: 0.2039–0.0232; p = 0.0941**) than in broiler chickens. The positivity rates of sul1 (95 % CI: 0.1964–0.4036; p = 0.0000*) and sul2 (95 % CI: 0.1337–0.3263; p = 0.0002*) were significantly higher in broiler chickens than in backyard chickens.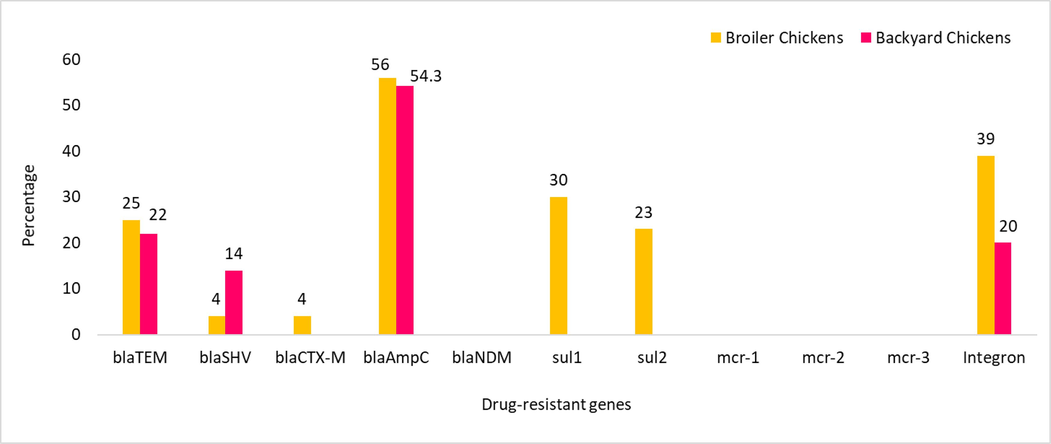
Molecular detection of antibiotic-resistant genes in bacterial strains from fecal samples of broiler and backyard chickens.
3.4 Gene sequencing
The blaTEM and blaSHVgenes were sequenced using the Sanger sequencing method. The gene sequences were deposited at NCBI (USA). Sequence accession numbers were OR492284 for the blaTEM gene and OR492283 for the blaSHVgene. BLAST analysis revealed that the blaTEM gene from E. coli and K. pneumoniae had 100 % similarity with the reference blaTEMgene. Similarly, blaSHV-1 detected in K. pneumoniae showed 99.75 % similarity with the reference gene blaSHV (Fig. S1 and S2).
4 Discussion
The global threat of AMR jeopardizes both human and animal health, raising concerns about the effectiveness of current antimicrobial agents used to treat bacterial infections (Tang et al., 2017). Antibiotic resistance emerges not only in pathogenic bacteria but also in the normal bacterial flora of animals and humans who are exposed to antibiotics (Hosuru Subramanya et al., 2020). This study was focussed on isolating Enterobacteriaceae from fecal samples of both broiler and backyard chickens, to analyse their molecular antimicrobial resistance profiles.
Among the broiler and backyard chicken fecal samples processed, E. coli was the predominant isolated bacterium. Besides E. coli, K. pneumoniae, P. vulgaris, Serratia sp., and Salmonella sp., were also isolated from both types of chickens. Broiler chicken-associated E. coli, Salmonella, and other Enterobacteriaceae cause enteritis in humans, and these bacteria are considered major organisms that are transmitted from poultry farms to humans (Mehdi et al., 2018). Bacterial isolates from broiler chickens exhibited resistance to tetracycline, gentamicin, nalidixic acid, nitrofurantoin, chloramphenicol, and trimethoprim-sulfamethoxazole. Bacterial isolates from backyard chickens also showed resistance to these antibiotics but at a lower rate than isolates from broiler chickens. Heightened exposure of broiler chickens to antibiotics could also explain the elevated drug resistance profile of the bacterial isolates from broilers. In a study, tetracycline and nalidixic acid resistant E. coli strains were isolated from broiler chickens (Miles et al., 2006). Backyard chickens may acquire antimicrobial-resistant bacteria from their environment, and it could potentially contribute to the spread of antibiotic resistance between animals and humans (Lepper et al., 2022). Antibiotics and drug-resistant bacteria can enter the environment through various sources such as hospitals, animal farms, and aquaculture. This environmental exposure exerting selective pressure on bacteria, leads to the development of drug resistance. Additionally, the transfer of drug-resistant genes between bacteria in the environment could increase the prevalence of multidrug–resistant bacteria. This poses a risk of transmission from the environment to animals and humans through contaminated water and food (Bobate et al., 2023).
Antibiotic resistance in GNB is mediated mainly by ESBL, AmpC, and carbapenemase production, and the prevalence of these bacteria in poultry has been extensively reported (Ghodousi et al., 2015; González-Torralba et al., 2016). In this study, ESBL production was highly noted among bacterial isolates from broiler chickens. A high percentage of AmpC production was also observed among bacterial isolates from broiler and backyard chickens. E. coli is the major organism that phenotypically produces both ESBL and AmpC enzymes. Studies have reported that broilers in Europe carried ESBL/AmpC-producing E. coli (Dierikx et al., 2013; Laube et al., 2014). A study from Greece reported that 13.6 % of isolates from broiler chickens produced ESBLs and only 2.7 % produced AmpC. However, ESBL (10.2 %) and AmpC (1 %) producers from backyard chickens were found at lower levels compared to that of broiler chickens (Xexaki et al., 2023).
A study reported that the AmpC gene was detected in > 50 % of Enterobacteriaceae isolates from both broiler and backyard chickens. They emphasized that backyard chickens also harboured AmpC-producers similar to broiler chickens (Hosuru Subramanya et al., 2020). Another study analysed the antibiotic resistance in E. coli isolates from broiler and backyard chickens with extraintestinal pathogenic E. coli from humans. They found that broiler chicken isolates shared the ESBL genes with human isolates. However, backyard chickens do not carry any one of the ESBL genes (Borges et al., 2019). In this study, ESBL genes (blaTEM and blaCTX-M) are prevalent in bacterial isolates from broiler chickens. However, the blaSHV gene was prevalent among isolates from backyard chickens. The positivity of blaSHV in bacterial isolates from backyard chickens revealed that ESBL genes might be disseminated in the environment. A study reported that most of the ESBL producers harbored blaCTX-M (13 %), and other genes detected were blaTEM (0.8 %) and blaSHV (0.52 %) (Chishimba et al., 2016). In this study, all the phenotypically colistin-resistant isolates were tested negative for mcr-1 to −3 genes. However a study reported that the mcr-1 gene was detected in 22.7 % of isolates from broiler chickens, but none of the mcr genes was detected from backyard chickens (Xexaki et al., 2023).
In this study, sul1 and sul2 genes for sulfamethoxazole resistance were detected among the broiler chickens but none among the backyard chickens. In a study from Bangladesh, 37.2 % of the Salmonella sp. strains isolated from broiler chicken fecal samples harboured the sul1 gene (Das et al., 2022). These findings revealed that sulfamthoxazole drug usage might be high in poultry farms. Integrons are widespread in bacterial isolates from chicken droppings. The integron has gene cassettes that capture the ESBL and other β-lactam antibiotic resistance genes and transfer them to other organisms (Langata et al., 2019). In our study, 39 % of bacterial isolates from broiler chickens harboured the integron gene. However, only 20 % of bacterial isolates from backyard chickens harboured integron. Gene cassettes of Integrons had cephalosporins, carbapenems, sulfamethoxazole, trimethoprim, and tetracycline antibiotic-resistant genes (Ramesh Kumar et al., 2017; Girlich et al., 2020). The positivity of integrons in poultry droppings could influence the transmission of ESBL/AmpC genes to susceptible bacterial species in the environment, and these genes can further be transmitted to humans. Racewicz et al. (2022) identified 75 % of MDR bacteria from broiler chicken harbored integrons, of which class 1 was the majorly detected one than class 2 integron. In the class 1 integron, aadA1 and dfrA1-aadA1 gene cassettes were detected. Class 2 integron had no gene cassettes. In this study, the integron was not studied for gene cassettes.
Antimicrobial stewardship in poultry is crucially needed to combat the high prevalence of antibiotic-resistant bacteria. The emergence of AMR bacteria in poultry is also threatening food safety since antibiotics cause other health issues (Patel et al., 2020). The findings of this study highlight the importance of implementing alternative strategies and judicious antibiotic use in poultry production. To control the AMR spread in poultry, biosecurity should be improved. Vaccine introduction, use of alternatives of antibiotics as growth promoters, and reduced antibiotic use are major strategies that should be implemented in poultry farms (Van Boeckel et al., 2017).
The limitation of this study is the smaller sample size from both broiler and backyard chickens and a lack of bacterial sequence typing. Sequence typing could identify the prevalent sequence types of bacteria in poultry farms and backyard chickens. Additionally, environmental samples from areas where backyard chickens consume food and water were not analysed, which could provide more insights into the environmental acquisition of antimicrobial-resistant bacteria in backyard chickens. Another limitation of this study is not detecting gene cassettes found on integrons that would provide more light on drug-resistant genes.
5 Conclusion
In this study, E. coli was the predominant bacterium isolated from both broiler and backyard chickens. Broiler chicken fecal isolates exhibited higher levels of antibiotic resistance compared to backyard chickens. Nonetheless, antibiotic resistance was still evident in backyard chicken isolates, suggesting possible environmental acquisition. The prevalence of ESBL/AmpC-producing bacteria in poultry is a serious public health concern. Strengthening surveillance efforts is crucial to assess the emergence of antimicrobial-resistant bacteria in poultry farms and their potential impact on the surrounding environment and human health.
CRediT authorship contribution statement
Balasubramanian Senthamilselvan: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. Marimuthu Ragavan Rameshkumar: Conceptualization, Data curation, Validation, Writing – original draft, Writing – review & editing. Aridass Dhanasezhian: Data curation, Resources, Validation. Sarva Kamalakar: Formal analysis, Investigation. Selvaraj Sivakumar: Formal analysis, Investigation. Arunagirinathan Nishanth: Data curation, Writing – original draft. Hissah Abdulrahman Alodaini: Funding acquisition, Resources, Writing – review & editing. Ashraf Atef Hatamleh: Funding acquisition, Resources, Writing – review & editing. Narasingam Arunagirinathan: Conceptualization, Data curation, Methodology, Supervision, Validation, Writing – review & editing.
Acknowledgments
The authors extend their appreciation to the Researchers supporting project number (RSP2024R479), King Saud University, Riyadh, Saudi Arabia. We also thank Dr. A. Amalraj, Professor of Biostatistics, Meenakshi Academy of Higher Education and Research, Chennai, India, and Dr. N. Viswanathan, Associate Professor and Head, Department of Statistics, Presidency College, Chennai, India for their support in statistical analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Monitoring and risk assessment of tetracycline residues in foods of animal origin. Food Sci. Biotechnol.. 2020;29:441-448.
- [CrossRef] [Google Scholar]
- The emergence of environmental antibiotic resistance: mechanism, monitoring and management. Environ. Adv.. 2023;13:100409
- [CrossRef] [Google Scholar]
- The use of antibiotics and antimicrobial resistance in veterinary medicine, a complex phenomenon: a narrative review. Antibiotics. 2023;12:487.
- [CrossRef] [Google Scholar]
- Detection of extended-spectrum beta-lactamase-producing Escherichia coli in market-ready chickens in Zambia. Int. J. Microbiol.. 2016;2016:1-5.
- [CrossRef] [Google Scholar]
- Clinical and Laboratory Standards Institute, 2020. Performance standards for antimicrobial susceptibility testing. 28th Informational supplement. CLSI document M100-S25.
- Antimicrobial resistance profiling and burden of resistance genes in zoonotic Salmonella isolated from broiler chicken. Vet. Med. Sci. 2022;8:237-244.
- [CrossRef] [Google Scholar]
- Extended-spectrum-β -lactamase- and AmpC- β-lactamase-producing Escherichia coli in dutch broilers and broiler farmers. J. Antimicrob. Chemother.. 2013;68:60-67.
- [CrossRef] [Google Scholar]
- Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect.. 2012;18:646-655.
- [CrossRef] [Google Scholar]
- Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother.. 2002;49:77-85.
- [CrossRef] [Google Scholar]
- Extended-spectrum ß -lactamase, AmpC-producing, and fluoroquinolone-resistant Escherichia coli in retail broiler chicken meat. Italy. Foodborne Pathog. Dis.. 2015;12:619-625.
- [CrossRef] [Google Scholar]
- Genetics of acquired antibiotic resistance genes in Proteus spp. Front. Microbiol.. 2020;11:256.
- [CrossRef] [Google Scholar]
- Survey of carbapenemase-producing enterobacteriaceae in companion dogs in Madrid. Spain. Antimicrob. Agents Chemother.. 2016;60:2499-2501.
- [CrossRef] [Google Scholar]
- Use of antibiotics in poultry and poultry farmers- a cross-sectional survey in Pakistan. Front. Public Health. 2023;11:1154668.
- [CrossRef] [Google Scholar]
- Detection and characterization of ESBL-producing enterobacteriaceae from the gut of healthy chickens, Gallus gallus domesticus in rural Nepal: dominance of CTX-M-15-non-ST131 Escherichia coli clones. PLoS ONE. 2020;15:e0227725.
- [Google Scholar]
- Comparison of the loads and antibiotic-resistance profiles of enterococcus species from conventional and organic chicken carcasses in South Korea. Poult. Sci.. 2018;97:271-278.
- [CrossRef] [Google Scholar]
- Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi. Kenya. BMC Res. Notes. 2019;12:22.
- [CrossRef] [Google Scholar]
- Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet. Microbiol.. 2014;172:519-527.
- [CrossRef] [Google Scholar]
- The role of the environment in dynamics of antibiotic resistance in humans and animals: a modelling study. Antibiotics. 2022;11:1361.
- [CrossRef] [Google Scholar]
- Use of antibiotics in broiler production: global impacts and alternatives. Anim. Nutr.. 2018;4:170-178.
- [CrossRef] [Google Scholar]
- Antimicrobial resistance of Escherichia coli isolates from broiler chickens and humans. BMC Vet. Res.. 2006;2:7.
- [CrossRef] [Google Scholar]
- Dissemination of CTX-M-type extended-spectrum β-lactamase genes to unusual hosts. J. Clin. Microbiol.. 2005;43:4183-4185.
- [CrossRef] [Google Scholar]
- Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother.. 2002;46:3829-3836.
- [CrossRef] [Google Scholar]
- Antibiotic stewardship in food-producing animals: challenges, progress, and opportunities. Clin. Ther.. 2020;42:1649-1658.
- [CrossRef] [Google Scholar]
- Prevalence and characterisation of antimicrobial resistance genes and class 1 and 2 integrons in multiresistant Escherichia coli isolated from poultry production. Sci. Rep.. 2022;12:6062.
- [CrossRef] [Google Scholar]
- Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep.. 2020;10:21999.
- [CrossRef] [Google Scholar]
- Dissemination of trimethoprim–sulfamethoxazole drug resistance genes associated with class 1 and class 2 integrons among gram-negative bacteria from HIV patients in South India. Microb. Drug Resist.. 2017;23:602-608.
- [CrossRef] [Google Scholar]
- Occurrence of extended-spectrum β-lactamase, AmpC, and carbapenemase-producing genes in gram-negative bacterial isolates from human immunodeficiency virus infected patients. J. Infect. Public Health. 2021;14:1881-1886.
- [CrossRef] [Google Scholar]
- Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of norwegian origin. J. Antimicrob. Chemother.. 2005;56:1019-1024.
- [CrossRef] [Google Scholar]
- Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet. Health. 2017;1:e316-e327.
- [CrossRef] [Google Scholar]
- Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. u.s.a.. 2015;112:5649-5654.
- [CrossRef] [Google Scholar]
- Reducing antimicrobial use in food animals. Science. 2017;357:1350-1352.
- [CrossRef] [Google Scholar]
- Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc.. 2008;3:163-175.
- [CrossRef] [Google Scholar]
- Prevalence of antibiotic resistant E. coli strains isolated from farmed broilers and hens in Greece, based on phenotypic and molecular analyses. Sustainability. 2023;15:9421.
- [CrossRef] [Google Scholar]
- Antibiotic residues in cattle and sheep meat and human exposure assessment in southern Xinjiang. China. Food Sci. Nutr.. 2021;9:6152-6161.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103191.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







