Translate this page into:
Molecular analysis of bedaquiline resistance and genetic variations in clinical isolates of multidrug and fluoroquinolones-resistant Mycobacterium tuberculosis
⁎Corresponding authors. usharani.unom@gmail.com (Usharani Brammacharry), ameerkhusro.sdc@saveetha.com (Ameer Khusro) armankhan0301@gmail.com (Ameer Khusro)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Mycobacterium tuberculosis developing resistance to anti-tubercular therapy leads to pioneering innovative treatment protocols featuring carefully curated combinations of potent medications. The aim of this study was to analyze the frequency and pattern of fluoroquinolone (FQ) resistant mutations among the first-line drug-resistant specimens. By employing an elective sampling strategy, we deliberately focused our investigation on a targeted subset of multidrug resistant (MDR) and FQ resistant (FQR) TB cases.
Method
Sputum samples from first-line drug-resistant patients were subjected to a GenoType MTBDRsl VER 2.0 assay to determine FQR. To investigate the effect of bedaquiline (BDQ) on these mutation-conferring genes (Rv0678, pepQ, and atpE), sanger sequencing was carried out on the MDR + FQR subset that was obtained. We have also conducted a computational analysis of the gene products of Rv0678 and pepQ with BDQ using molecular docking methods.
Results
Eighty-three samples were observed to have FQR, and the majority of these specimens were associated with naïve and treatment failure cases. Among them, 64 isolates exhibited mutations in gyrA gene, while 19 isolates showed mutations in gyrB gene. The predominant occurrence of genetic variations, particularly in D94G and A90V, was observed within the specimens displaying resistance in gyrA gene, and the same has been confirmed by PCR-based detection. And the sequence analysis of BDQ genes among MDR + FQR subset indicated that the majority of the samples did not exhibit mutations in pepQ and atpE genes. However, only 2 samples were found to have the genetic variant G337A, which involves replacing the amino acid glutamic acid (E) at codon position 113 of Rv0678 gene with lysine (K).
Conclusions
Most of the mutations were harbored in rpoB + katG + gyrA gene pattern. These findings pave the way for the development of databases and rapid diagnostic probes.
Keywords
Bedaquiline
BDQ primers
Fluoroquinolone
Mutation analysis
pepQ
Rv0678
1 Introduction
Tuberculosis (TB) is a highly contagious and lethal disease worldwide, affecting all age groups, especially those with weakened immunity (Khusro et al., 2020). It is caused by Mycobacterium tuberculosis and is latently widespread across the world's population; however, it seems to be a global burden, with India experiencing the highest incidence of TB. As per the worldwide report of WHO on 2020, India has recorded one-fourth of all TB cases worldwide (World Health Organization, 2020). HIV/AIDS patients and members of marginalized communities, and those with weakened immune systems have the greatest impact on the prevalence. It is due to a combination of factors including poor access to healthcare, overcrowding, and economic and social situations (Hargreaves et al., 2011; Khusro et al., 2018a). Approximately 90 % of adults acquire TB each year; comparatively, men are more vulnerable to this problem than women (Khusro et al., 2016; Global Tuberculosis Report, 2022).
In terms of previous TB studies, people with multidrug resistance (MDR) were reported in 132,222 individuals; among these, pre-extensive or extensively drug resistance (XDR) was reported in 25,681 individuals (Global Tuberculosis Report, 2021). In many parts of the world, the incidence of bacillary resistance is potentially expanding, with MDR- and XDR-TB being the major contributing factors (Khusro et al., 2018b).
In addition, it should be noted that M. tuberculosis remains inactive and does not replicate, making it resistant to most primary anti-tuberculosis medications (Koul et al., 2008). Currently, the majority of anti-tubercular treatments involves the use of bedaquiline (BDQ), which has been approved by the FDA. However, adult MDR-TB cases typically receive it as second-line anti-tubercular therapy (Deoghare, 2013; Khusro et al., 2018c). This disease can be prevented and treated, regardless its difficulties. Through early detection screening, timely beginning of suitable treatment, and adherence to medication regimens can considerably decrease TB-related morbidity and mortality (Khusro et al., 2016).
After being inhaled, M. tuberculosis travels to the lung's alveoli, where it comes in contact with alveolar macrophages, which are in charge of engulfing and eliminating infections. However, M. tuberculosis has developed defense against macrophage destruction, enabling it to endure and proliferate within these cells (Cohen et al., 2018). Anti-TB therapies primarily operates by interrupting vital metabolic processes of M. tuberculosis, therefore preventing the bacteria from growing and spreading (Warner, 2014).
The crucial factors that act as virulence determinants are the ESX-1 secretion system and the cell wall components, which enable M. tuberculosis to invade and maintain infection in the host cell. The TB pathogenesis involves the ESX-1 system, which secretes the proteins ESAT-6 and CFP-10, which allow the bacteria to travel across the phagosomal membrane and reach the cytoplasm of the host cell (Forrellad et al., 2013) Similarly, another key virulence determining factor is the cell wall components, which contain mycolic acids, cord factor (trehalose 6,6′-dimycolate), lipoarabinomannan, and sulfatides. These components help the bacterium develop resistance to phagocytosis, prevent the fusion of phagosomes and lysosomes, and weaken the host immune system (Rajni et al., 2011).
Key drugs have been developed to target multiple pathways within M. tuberculosis cells. These pathways include hindering the synthesis of essential cell wall components, inhibiting the onset of RNA synthesis by targeting bacterial RNA polymerase, impeding the enzymes involved in DNA replication and transcription, interfering protein synthesis by focusing on the bacterial ribosome, and disrupting the cellular ATP synthesis mechanism, which is the power hub of the cell (Kohanski et al., 2010). Interestingly, F1-F0-ATP synthase enzyme is lead factor for the ATP synthesis mechanism of cells (Zharova et al., 2023) and the c-subunit of this enzyme is targeted by the diarylquinoline inhibitor, the ATP synthase inhibitor. This inhibitor was found to possess remarkable properties in combating mycobacterial replication, as reported in previous studies (Koul et al., 2008).
The significant barrier that renders all of these main treatments useless by developing drug resistance to these anti-TB medications creates a major challenge for treatment providers and researchers. Drug resistance is generally caused by genetic changes that alter drug targets or processes involved in drug activation or metabolism (Algammal et al., 2023). Understanding the genetic basis of medication resistance in M. tuberculosis is very critical for developing effective treatment regimens (Palomino and Martin, 2014). Simultaneously, MDR has increased all over the world and is considered a public health threat. Several recent investigations have reported the emergence of MDR bacterial pathogens from different origins that increase the necessity of the proper use of antibiotics (Algammal et al., 2021; Shafiq et al., 2022). We are required to build effective diagnostic tools and strategies to combat drug-resistant TB. And this requires expanding the prevalent study and mutational analysis across the globe and enriching the TB genetic variants database. As a part of this objective, our study focuses on genetic variants that lead to BDQ resistance.
Bedaquiline was first utilized in PMDT incidents in India on March 21, 2016, according to sources (Yadav, 2016). Whole-genome sequencing revealed that drug resistance is mediated through inefficiency and an increase in the efflux mechanism of the drug target genes pepQ, Rv0678, and atpE. To fully understand the processes of resistance to BDQ, which are required to prevent the establishment of resistance, it is crucial to locate novel variants in BDQ resistance-related genes (Yadav, 2016).
Throughout this research, our focus was to analyze single nucleotide variants associated with BDQ resistance in selected clinical specimens that were not exposed to BDQ. We investigated the BDQ resistance patterns in atpE, pepQ, and Rv0678 genes among the selected MDR + FQ resistant (FQR) isolates. Further in this study, in order to better understand the effectiveness of the drug, structural predictions for pepQ and Rv0678, as well as a molecular docking study with BDQ, were conducted.
2 Materials and methods
2.1 Bacterial isolation and processing
Specimens for this study were acquired from the Intermediate Reference Laboratory in Puducherry over a period of three years. All investigations were done in a biosafety level III facility equipped with a Class II biosafety cabinet. Sputum samples from clinically confirmed MDR, rifampicin-resistant, and isoniazid-resistant patients with TB were collected. To disinfect the samples, the NALC-NaOH method was utilized. DNA was extracted from each individual sample using the GenoLyse® PCR CE/IVD Germany, following the manufacturer's protocols. The Bioline™ TB Ag MPT64TEST test is done for rapid identification of M. tuberculosis complex (Myneedu et al., 2015). Subsequently, BACTEC™ MGIT™ 960 culture tubes were employed for culturing the samples (Muthaiah et al., 2017).
2.2 MTBDRsl VER 2.0 assay
Implementing the MTBDRsl V.2 assay and interpreting it as per the directions provided by the manufacturer is crucial for accurate detection of M. tuberculosis (GenoType MTBDRsl VER 2.0 Instructions for Use). The hybridization strips clearly indicated drug resistance through the detection of specific mutant bands or the absence of wild-type (WT) bands by which drug susceptibility was confirmed (Muthaiah et al., 2017). The MDR + FQR clinical specimens from patients who had not yet received BDQ as part of their regimen for this investigation were selected.
2.3 Amplification and sequencing of targeted amplicons associated with BDQ conferring mutation
The genomic DNA from the MGIT culture of the MDR + FQR sample subset was isolated using the CTAB technique (Muthaiah et al., 2017). The extracted DNA was amplified using the existing oligo, targeting the hot spots in QRDRs (Dauendorffer et al., 2003). ReadyMix™ Taq PCR Reaction Mix from Sigma was utilized for amplifying the targets at 12.5 µl, PCR grade nuclease-free water at 7.5 µl, followed by 10 pmol of 1 µl forward and reverse primers, and 3 µl of extracted DNA, which contributes to 25 µl of total reaction volume.
The primers for the drug target genes atpE, pepQ, and Rv0678 were crafted through primer-BLAST from NCBI using reference strain NC_000962.3, as shown in Table S1 and S2 (Supplementary file). After PCR standardization, the following amplification parameters were adhered to: 5 min incubation at 94 °C, followed by 30 amplification cycles, each consisting of 30 sec at 94 °C, 45 sec at 56.2 °C (for BDQ genes), 58 °C (for gyrB), 58.6 °C (for gyrA), and 45 sec at 72 °C, and the final extension is kept for 7 min at 72 °C. After amplification, the PCR product was held at 4 °C for 15 min, and then evaluation was done by conducting an EtBr-stained agarose gel run using TAE buffer, and DNA bands were viewed using the Geldoc system (Salah Eldin et al., 2012). The QIAquick PCR Purification Kit from Qiagen was utilized to obtain the purified PCR product and the amplified product was sequenced by employing Applied Biosystems DNA sequencing equipment at ImmuGenix Biosciences Pvt. Ltd., Chennai, India. The acquired Electropherogram data and nucleotide sequences were compared to the standard strain H37Rv NC_000962.3 using the bioinformatics tools (Bioedit, BLASTn, and CLC protein workbench software) for variant analysis.
2.4 Molecular docking study
Using the technique of molecular interaction study, the current computational analysis assesses the effect of BDQ on mutation-conferring genes. We used the docking program AutoDock 4.2 to perform the docking assessment, and PreADMET and Biovia Discovery Studio visualizer were used to analyze the properties (Panikar et al., 2021).
2.5 Protein preparation
The crystallographic structure of Rv0678 and pepQ genes of M. tuberculosis H37Rv strain was derived from the Protein Data Bank site (PDB ID − 4NB5-A chain) (O’Boyle et al., 2011). We noticed that pepQ structure is not available in the PDB database. Hence, the pepQ sequence was derived from the UniProt database (UniProt ID − A0A654ZTY1) (Panikar et al., 2021).
2.6 Swiss model prediction of protein structure and PROCHECK validation
Swiss Model was used to determine the structure of pepQ using a template 5GIU. Proline dipeptidase from Deinococcus radiodurans has 39 % sequence identity in its crystal structure. Using the SAVES-PROCHECK server, Ramachandran plot prediction for pepQ and Rv0678 was performed. Almost 93.6 % and 96.7 % of the amino acid residues were present in the most favorable regions and 0.0 % was observed in the disallowed regions (Panikar et al., 2021).
2.7 Preparation for ligand
The ligand substance from the PubChem database, namely BDQ, was in the SMILES format. A converter application (open Babel) was used to create the 3D structure from the 2D structure using the SMILES structure and to carry out 3D optimization, store it in MDL-MOL File format, and convert it to PDB format (Morris et al., 2009).
2.8 Docking analysis and interpretations
A molecular interaction analysis was performed to learn the interaction of BDQ against pepQ and Rv0678 genes of M. tuberculosis, using AutoDock 4.2 (Panikar et al., 2021).
2.9 Docking visualization
The Biovia Discovery Studio visualizer was used to depict various molecular interactions, including binding energy, van der Waals interactions, electrostatic energy, including hydrogen and hydrophobic interactions.
3 Results
3.1 Evaluating specimens for potential resistance to FQ antibiotics
In this research, a total of 1158 samples displaying resistance to first-line drugs (MDR-TB, rifampicin-resistant TB, and isoniazid-resistant TB) were received for three years, of which 288 were from affected women and 870 were from affected men (Fig. S1: Supplementary material). These samples were analyzed for FQR using the MTBDRsl V.2.0 assay. The FQ prevalence study revealed that 83 (7.16 %) first-line drug-resistant samples of the population were resistant to second-line FQ drugs, and most of these samples belong to new and past-treatment failure cases.
The results of the gyrase genes mutational study are presented in Tables 1, 2, and 3. These findings indicated variation of bases like substitutions in DNA leading to subsequent changes in the associated amino acids. The study of gyrase A and B genes revealed 64 isolates with gyrA mutations and 19 isolates with gyrB mutations. According to the genetic analysis, gyrA variations are predominantly found in codon 94 and 90. Valine replaces alanine in the A90V codon, whereas in the D94G codon glycine replaces aspartic acid. Treatment failure and recurrence were associated with the D94G mutation in the majority of patients.
Type of drug resistant TB
Identified resistance pattern
No of Cases
FQR
gyrA
gyrB
gyrA and gyrB
Isoniazid- Mono-Resistant (H)
inhA, gyrA
7
25
17
0
inhA, gyrB
4
katG, gyrA
17
katG, gyrB
13
inhA, katG, gyrA
1
Rifampicin-Mono-Resistant (R)
rpoB, gyrA
10
10
1
0
rpoB, gyrB
1
rpoB, gyrA, gyrB
2
0
0
2
MDR
MDR, gyrA
27
27
1
0
MDR, gyrB
1
Total no of drug resistant-TB Cases N = 1158
Total no of TB cases resistance to second line FQ-drugs
62
19
2
Second line drug resistance TB
Gene
Band
Genetic mutation or specific genomic region investigated
Pattern and frequency of FQ mutations
FQR
(n = 83)
No. (%) detected
FQR pattern
gyrA
WT1
78
93.98 %
WTΔ1
Codon(s) 85–89
5
6.02 %
WT2
70
84.34 %
WTΔ2
Codon(s) 89–93
13
15.66 %
WT3
64
77.11 %
WTΔ3
Codon(s) 92–96
19
22.89 %
MUT1
A90V
16
19.28 %
MUT2
S91P
3
3.61 %
MUT3A
D94A
1
1.20 %
MUT3B
D94N
8
9.64 %
D94Y
MUT3C
D94G
28
33.73 %
MUT3D
D94H1
8
9.64 %
gyrB
WT1
64
77.11 %
WTΔ1
Codon(s)
19
22.89 %
536–541
(codon 497–502)
MUT1
N538D
16
84.2 %
MUT2
E540V
3
15.8 %
Mutation probe
Codon position
Nucleotide Substitution
Amino acid change
Drug Resistance Interpretation
No of cases
gyrA (n = 64)
MUT1
90
GCG to GTG
A-alanine to V-valine
LfxR, Low level MfxR
16
MUT2
91
TCG to CCG
S-serine to P-proline
LfxR, Low level MfxR
3
MUT 3A
94
GAC to GCC
D-aspartic acid to A-alanine
LfxR, Low level MfxR
1
MUT 3B
94
GAC to AAC
D-aspartic acid to N-asparagine
LfxR, High level MfxR
8
MUT 3C
94
GAC to GGC
D-aspartic acid to G-glycine
LfxR, High level MfxR
28
MUT 3D
94
GAC to CAC
D-aspartic acid to H-histidine
LfxR, High level MfxR
8
gyrB (n = 19)
MUT1
538
AAC to GAC
N-asparagine to D-aspartic acid
LfxR, Low level MfxR
16
MUT2
540
GAA to GTA
E-glutamic acid to V-valine
LfxR, Low level MfxR
3
3.2 Detection of genetic variations linked to BDQ resistance
We attempted to identify mutations predictive for potential BDQ resistance. In this study, 28 MDR-TB samples were carefully chosen from a collection of 83 samples exhibiting resistance to FQ, as shown in Fig. 1. This selective sampling strategy ensures a focused investigation into the specific subset of MDR + FQR-TB cases within the wider cohort of FQR samples, as shown in Fig. 2. Using H37Rv as a template, atpE, pepQ-S1, pepQ-S2, and Rv0678 genes were amplified. The amplification was verified by the development of clear bands at 373, 483, 735, and 572 bp (Fig. S2: Supplementary material).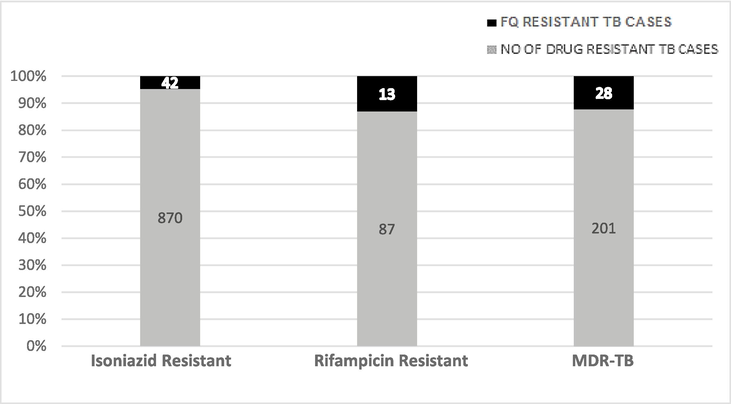
Mutational analysis of FQR among the H/R-mono-resistant and MDR-TB isolates (H-isoniazid, R-Rifampicin, MDR-Multi drug resistant).
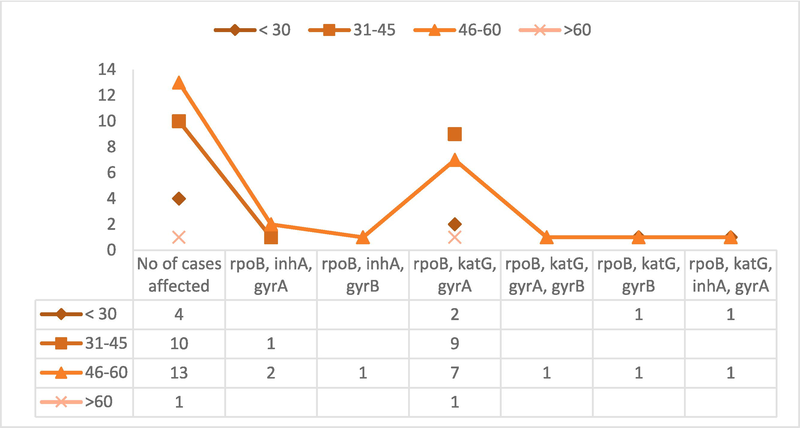
Diverse mutational pattern across age group with in MDR + FQR–TB cases.
Among the total number of cases studied, the age group of affected people was categorized as mentioned in Fig. 2. The subset's most affected age range, according to the examination of mutational trends across a range of age groups, was 45 to 60 years old. According to the mutation analysis, the combination of rpoB, katG, and gyrA gene mutations exists in almost all age groups. In particular, it became apparent that 19 of the 28 cases under investigation had mutations in each of the three genes-rpoB, katG, and gyrA.
The typical procedure involves sequencing the genomes of TB bacteria sourced from patients with no prior exposure to BDQ. Among the chosen samples, most of them lacked mutations in atpE and pepQ genes. However, in two cases, Rv0678 gene exhibited a non-synonymous mutation involving the substitution of G337A across the mutational groups of MDR + FQR subset. This mutation involves the substitution of glutamic acid (E) with lysine (K) at position 113 of Rv0678 gene (Fig. S3: Supplementary material).
3.3 Molecular docking
3D structure of target genes (pepQ and Rv0678) is shown in Fig. 3. Using AutoDock 4.2 version, the docking study on BDQ with pepQ and Rv0678 was performed. Numerous conformations of the protein-compound complex have been generated by the docking modeling, depending on the conformation with the lowest binding free energy (ΔG) and the best conformations were picked. The Biovia Discovery Studio Visualizer program is used to show the interactions between BDQ and drug target genes.
3D-structure of target genes-pepQ and Rv0678.
Figs. 4 and 5 show solid ribbon model conformation of pepQ and Rv0678 genes. The essential residues are indicated as red lines, and the compound was depicted as blue ball and stick. It also showed the interactions of BDQ at the active binding sites.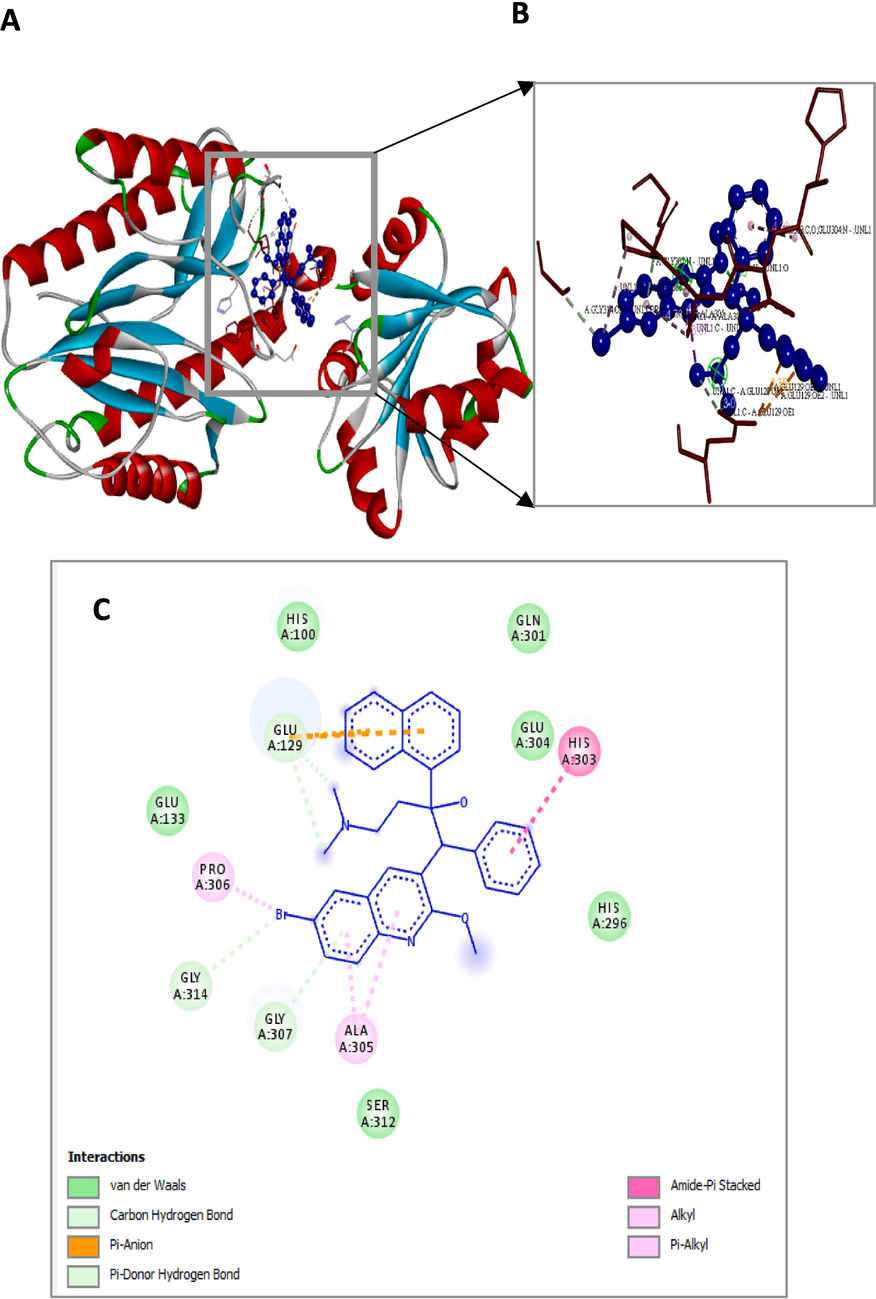
Bedaquiline (blue colour ball and stick model) interactions with pepQ represented in (A) solid ribbon model with; (B) active site amino acid residues amino acid residues represented in red colour line model; (C) hydrophobic interactions (alkyl and pi-alkyl are shown in light pink colour dotted lines; Amide-pi-Anion are shown in orange), Carbon hydrogen bonds are shown in (light blue colour circles). van der Waals interactions (light green colour circles).
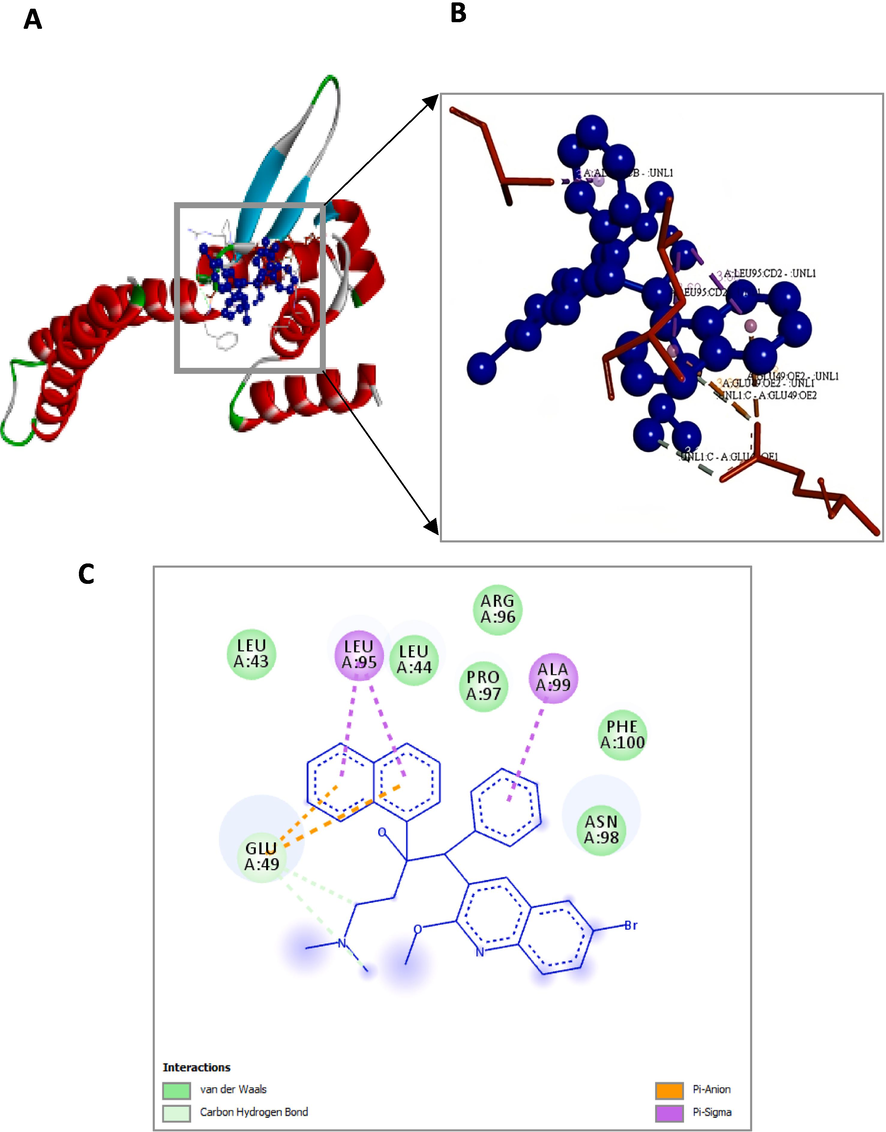
Bedaquiline (blue colour ball and stick model) interactions with Rv0678 represented in (A) solid ribbon model with; (B) active site amino acid residues amino acid residues represented in red colour line model; (C) Pi-Anion are shown in orange dotted lines and Pi-sigma are shown in purple dotted lines), Carbon hydrogen bonds are shown in (light blue colour circles). van der Waals interactions (light green colour circles).
3.4 Binding interaction of BDQ with pepQ and Rv0678
The interaction details of BDQ with the crucial residues of two drug target genes are shown in Table 4. The computational analysis of pepQ to BDQ had shown that it formed two hydrogen bonds (GLY314 and GLU129) with pepQ; six hydrophobic bonds formed with GLU129, GLY307, HIS303, GLU304, PRO306, and ALA305; and two electrostatic interactions with GLU129 (OE1) and GLU129 (OE2), as shown in Fig. 4, with the interaction energy of −6.07 kcal/mol and an inhibitory constant of 35.46 µM (Table 5). These amino acid residues were critical in the establishment of the hydrophobic interactions between the BDQ and pepQ compounds.
Conformation
Hydrogen-bonding
Donor-Acceptor linkage of Amino acid
Hydrophobic interactions
Electrostatic interaction
(Atom⋯Ligand atom)
Distance (Å)
(Atom⋯Ligand atom)
Distance (Å)
(Atom⋯Ligand atom)
Distance (Å)
PepQ +
BDQCA-BR
3.22955
GLU129 (OE2)
3.86883
OE1-C
3.02404
OE1-C
3.00781
GLY307(N)
3.42698
GLU129(OE2)
3.34607
HIS303 (C,O)
4.19601
GLU304 (N)
4.19601
PRO306 (BR)
5.46667
ALA305
4.28584
ALA305
4.72995
Rv0678 + BDQ
OE2- C
3.19971
LEU95 (CD2)
3.59752
GLU49(OE2)
3.65846
OE1- C
3.34369
LEU95 (CD2)
3.683
GLU49(OE2)
3.17952
ALA99 (CB)
3.97165
Conformation
Binding Energy
Ligand
efficiency
Inhibitory
constant,
Ki (μm)
Intermolecular energy
vdW + H
bond + desolv Energy
Electrostatic energy
Torsional
energy
Total internal Unbound
BDQ + pepQ
−6.07
−0.16
35.46
−8.76
−7.51
−1.25
2.68
−3.2
BDQ + Rv0678
−5.72
−0.15
63.68
−8.41
−7.65
−0.76
2.68
−2.79
The binding energy and inhibition constant used in the molecular interaction analysis of Rv0678 with BDQ are −5.72 kcal/mol and 63.68 M, respectively. Fig. 5 represents that the Rv0678 with BDQ conformation depicting two of hydrophobic bonds with LEU95 and ALA99 amino acids, were crucial in establishing the hydrophobic contacts between the BDQ. There were two pi-sigma interactions with LEU95 and GLU49, two hydrogen bonds with GLU49 (OE1 and OE2), and two electrostatic interactions with GLU49 residues.
The least energy values along with the inhibition constants, torsional, intermolecular, electrostatic, and unbound energy as well as the ref RMS value of BDQ with pepQ and Rv0678 are shown in Table 5. The units of measurement for all energy are kcal/mol.
4 Discussion
The efficacy of BDQ in treating TB appears to be reducing in current times, primarily due to the emergence of drug resistance. Longer regimens have no control over alterations in the patients' treatment plans (Singhal et al., 2016; Wu et al., 2021).
FQR occurs primarily due to inappropriate regimen concordance and mutations in the QRDR of gyrA, specifically in the amino acid 74–113 coding region (Kabir et al., 2020). The drug efflux process pumps the drug out of the cell, thus leading to FQR (Singhal et al., 2016).
It was observed that 83/1158 isolates (7.1 %) were FQR. Among all samples examined, INH resistance was found in 75 % of samples (870/1158). Out of 83 FQR isolates, 64 was gyrA mutants and 45 (72.5 %) resistant isolates exhibited mutations at codon 94, resulting in an alteration of the amino acid from Asp to Gly. The remaining 16 isolates (25.8 %) possessed mutations in codon A90V (GCG to GTG), whereas 3 (4.8 %) possessed mutations in codon S91P (TCG to CCG). These findings align with the research conducted by Kabir et al. (2020) and Singhal et al. (2016). Out of 19 gyrase B mutants, 16 resistant isolates (84.2 %) had mutations at codon 538 (asparagine to aspartic acid), while the other 3 (15.8 %) had a mutation at codon 540 (glutamic acid to valine).
The most prevalent mutation, S315T, located in the active site of the katG enzyme, leads to reduced binding affinity to isoniazid and induces a high degree of drug resistance (Rabha et al., 2019). There were 201 MDR and 87 Rifampicin-monoresistant samples. Mutations alter the conformation of the drug binding site and cause resistance to all FQ (Singhal et al., 2016; Kabir et al., 2020).
Since levofloxacin is resistant to the A90V mutation, moxifloxacin can be used at a higher dose. However, neither levofloxacin nor moxifloxacin work if the D94G mutation is present. These results agree with the majority of the earlier investigations (Singhal et al., 2016; Kabir et al., 2020).
A high frequency of A90V mutations was observed in newly identified incidents of TB isolates with FQR; hence, moxifloxacin is still the treatment of choice at higher dosages for these mutations. On the other hand, the D94G mutation is frequently present in cases of therapy failure and recurrence.
Among these 83 isolates, a substitution of Ala with Val was observed in major isolates when the MUT1 probe was present. Our most common pattern was the presence of the MUT3C probe and the lack of WT3, a mutation of Asp to Gly (Kabir et al., 2020).
We found a significant association between MDR-TB and the gyrA mutations among the MDR + FQR subset (Dreyer et al., 2022). Among the selected subset, the age range of 45 to 60 years exhibited the most pronounced prevalence of mutations. The mutations were identified in the genes rpoB, katG, and gyrA. The age group between 31 to 45 years showed a similar level of impact compared to other age groups. In the age range of 45 to 60 years, 46.4 % of the group was affected, while in the 31 to 45 years range, the affected percentage was 36 %.
Variations in Rv0678, atpE, and pepQ play a role as potential determinants of BDQ resistance. Overexpression of the MmpL5 efflux pump gene may result from a mutation in the Rv0678 transcriptional regulatory gene (Hartkoorn et al., 2014). In two distinct samples, the Rv0678 gene displayed nonsynonymous mutations, including the nucleotide substitution of G337A. This mutation alters glutamic acid (E) to lysine (K) at codon position 113 of the Rv0678 gene, generating elevated BDQ resistance. And the pattern was consistent with that reported by the CRyPTIC collection and Sonnenkalb et al. (2023).
Based on the computational analysis results indicating a binding energy of −6.07 kcal/mol and −5.72 kcal/mol and an inhibition constant of 35.46 µM and 63.68 µM in the molecular interaction analysis of pepQ and Rv0678 with BDQ, it can be concluded that there is a favorable interaction between these two genes and BDQ. And this suggests that pepQ has a slightly higher binding affinity for BDQ than Rv0678 does. However, both interactions show that the corresponding proteins and BDQ bind well, implying that BDQ could potentially be effective against targets associated with TB.
We are seeking out the new mutation in the FQR specimens to check whether there is any prior mutation in the drug target genes for BDQ (Deshkar and Shirure, 2022). With enhanced research and development, TB eradication objectives can be achieved by 2030 or 2035 (Global Tuberculosis Report, 2022).
5 Conclusions
The findings revealed a strong occurrence of gyrA gene mutations among the MDR-TB specimens. Nearly all (96.4 %) of the MDR-TB samples exhibited gyrA mutations, while only 3.6 % displayed gyrB mutation among the MDR + FQR subset. The mutation analysis suggests that the highest prevalence of mutations occurred within 45 to 60-year-old age group. Moreover, each case of the MDR + FQR subset in all age categories harbored mutations in rpoB + katG + gyrA gene group. Among the MDR + FQR subset, an E113K mutation in the Rv0678 gene was found, which is potentially linked to BDQ resistance. Investigating unique BDQ mutations in MDR + FQR samples aims to identify variants in drug target genes, facilitating the development of improved probes for rapid diagnosis in TB suspects. The computational study demonstrated a negative binding energy of -6.07 kcal/mol and −5.72 kcal/mol, implying that the interaction between pepQ and BDQ is energetically more beneficial than Rv0678, indicating the potential efficacy of BDQ drug against these genes. These findings provide valuable insights into the potential effectiveness of BDQ as a therapeutic agent targeting pepQ and Rv0678 in the treatment of TB. However, these experimental validations are required to create a database of potential variations to determine the association between effective drug concentrations for treating patients. It will also help us understand how these point mutations alter protein structure and drug binding effectiveness. Thus, consistent surveillance and analysis of resistance patterns play a pivotal role in shaping effective strategies for treating TB.
CRediT authorship contribution statement
Vijayalakshmi Jawaharlal Nehru: Conceptualization, Investigation, Methodology, Writing – original draft, Software. Usharani Brammacharry: Data curation, Project administration, Supervision, Visualization. S.R. Sri Ramkumar: Resources, Visualization. Ameer Khusro: Writing – review & editing, Methodology, Data curation, Conceptualization. Maria Jose Vadakunnel: Data curation, Visualization. Shoba Gunasekaran: Validation, Visualization. Esther David: Resources, Software, Validation. Veeraraghavan Vishnu Priya: Formal analysis, Visualization. Reem M. Aljowaie: Formal analysis, Funding acquisition. Saeedah Musaed Almutairi: Visualization, Funding acquisition.
Acknowledgement
We feel grateful for the guidance and facilities provided by Dr. M. Muthuraj, Head of the Department of Microbiology of the Intermediate Reference Laboratory, Puducherry. This study was supported by grants from - The Indian Council of Medical Research- Ad-Hoc project. The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R418), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol.. 2021;21
- [Google Scholar]
- Editorial: Emerging multidrug-resistant bacterial pathogens “superbugs”: A rising public health threat. Front. Microbiol. 2023:14.
- [Google Scholar]
- Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. 2018;24:439-446.e4.
- [Google Scholar]
- Identification of Mycobacterial species by PCR sequencing of quinolone resistance-determining regions of DNA gyrase genes. J. Clin. Microbiol. 2003;41:1311-1315.
- [Google Scholar]
- Bedaquiline: A new drug approved for treatment of multidrug-resistant tuberculosis. Indian J. Pharmacol. 2013;45:536.
- [Google Scholar]
- Bedaquiline: A novel diarylquinoline for multidrug-resistant pulmonary tuberculosis. Cureus 2022
- [Google Scholar]
- High fluoroquinolone resistance proportions among multidrug-resistant tuberculosis driven by dominant L2 Mycobacterium tuberculosis clones in the Mumbai Metropolitan Region. Genome Med.. 2022;14(1):95.
- [Google Scholar]
- Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4:3-66.
- [Google Scholar]
- GenoType MTBDRsl VER 2.0 Instructions for Use. (n.d.). www.hain-lifescience.com/products/msds.html.
- Global Tuberculosis Report 2021. (2021). http://apps.who.int/bookorders.
- Global Tuberculosis Report 2022. (2022a). http://apps.who.int/bookorders.
- The social determinants of tuberculosis: From evidence to action. Am. J. Public Health. 2011;101:654-662.
- [Google Scholar]
- Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014;58:2979-2981.
- [Google Scholar]
- Fluoroquinolone resistance and mutational profile of gyrA in pulmonary MDR tuberculosis patients. BMC Pulm. Med. 2020:20.
- [Google Scholar]
- Anti-tubercular peptides: A quest of future therapeutic weapon to combat tuberculosis. Asian Pac. J. Trop. Med. 2016;9:1023-1034.
- [Google Scholar]
- Venom as therapeutic weapon to combat dreadful diseases of 21st century: A systematic review on cancer, TB, and HIV/AIDS. Microb. Pathog. 2018;125:96-107.
- [Google Scholar]
- Neoteric advancement in TB drugs and an overview on the anti-tubercular role of peptides through computational approaches. Microb. Pathog. 2018;114:80-89.
- [Google Scholar]
- Anti-tubercular and probiotic properties of coagulase-negative staphylococci isolated from Koozh, a traditional fermented food of South India. Microb. Pathog. 2018;114:239-250.
- [Google Scholar]
- Computational modelling and docking insight of bacterial peptide as ideal anti-tubercular and anticancer agents. Biocatal. Agric. Biotechnol. 2020;26:101644
- [CrossRef] [Google Scholar]
- How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 2010;8:423-435.
- [Google Scholar]
- Diarylquinolines are bactericidal for dormant Mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 2008;283:25273-25280.
- [Google Scholar]
- AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785-2791.
- [Google Scholar]
- Prevalence of mutations in genes associated with rifampicin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Tuberc. Other Mycobact. Dis. 2017;8:19-25.
- [Google Scholar]
- Utility of MPT64 antigen detection for rapid confirmation of Mycobacterium tuberculosis complex. J. Glob. Infect. Dis. 2015;7:66.
- [Google Scholar]
- Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics. 2014;3:317-340.
- [Google Scholar]
- Essential oils as an effective alternative for the treatment of COVID-19: Molecular interaction analysis of protease (Mpro) with pharmacokinetics and toxicological properties. J. Infect. Public Health. 2021;14:601-610.
- [Google Scholar]
- Structural basis for isoniazid resistance in KatG double mutants of Mycobacterium tuberculosis. Microb. Pathog. 2019;129:152-160.
- [Google Scholar]
- Biosynthesis and virulent behavior of lipids produced by Mycobacterium tuberculosis: LAM and cord factor: An overview. Biotechnol. Res. Int. 2011;2011:1-7.
- [Google Scholar]
- Detection of fluoroquinolone resistance in Mycobacterium tuberculosis clinical isolates as determined by gyrA/B gene mutation by using PCR technique. Egypt J. Chest Dis. Tuberc. 2012;61:349-353.
- [Google Scholar]
- Coexistence of blaNDM–5 and tet(X4) in international high-risk Escherichia coli clone ST648 of human origin in China. Front. Microbiol. 2022:13.
- [Google Scholar]
- Sequence analysis of fluoroquinolone resistance-associated genes gyra and gyrb in clinical Mycobacterium tuberculosis isolates from patients suspected of having multidrug-resistant tuberculosis in New Delhi, India. J. Clin. Microbiol. 2016;54:2298-2305.
- [Google Scholar]
- Bedaquiline and clofazimine resistance in Mycobacterium tuberculosis: an in-vitro and in-silico data analysis. Lancet Microbe. 2023;4:e358-e368.
- [Google Scholar]
- Mycobacterium tuberculosis Metabolism. Cold Spring Harb Perspect. Med. 2014;5:a021121-a.
- [Google Scholar]
- World Health Organization, 2020. Global tuberculosis report 2020.
- Primary Bedaquiline resistance among cases of drug-resistant tuberculosis in Taiwan. Front. Microbiol. 2021:12.
- [Google Scholar]
- Bedaquiline: A novel antitubercular agent for the treatment of multidrug- resistant tuberculosis. J. Clin. Diagn. Res. 2016;10:FM01-2.
- [Google Scholar]
- F1·Fo ATP Synthase/ATPase: Contemporary view on unidirectional catalysis. Int. J. Mol. Sci. 2023;24:5417.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103226.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







