Translate this page into:
Long non-coding RNAs act as novel therapeutic targets by regulating molecular networks associated with ischemic stroke
⁎Corresponding author at: Department of Medical Laboratory Sciences, College of Applied Medical Sciences, Majmaah University, Majmaah 11952, Saudi Arabia. m.palanisamy@mu.edu.sa (Palanisamy Manikandan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Impaired blood supply to part of the brain results in an ischemic stroke leading to dysfunction of brain tissue. Several genetic and environmental factors can contribute to stroke. Age is one of the most important risk factors for ischemic stroke. An increased incidence of stroke related mortalities is associated with aging. The pathophysiological processes triggered by stroke, such as inflammation, apoptosis, angiogenesis, and post-stroke recovery, are well described. However, the molecular mechanisms underlying disease development remain to be studied in detail. The damage and recovery process triggered by stroke is coordinately regulated by genes involved in inflammation, immune response, and angiogenesis. The transcriptional dynamics of these key pathways determine the recovery of brain tissue from damage.
The long intergenic non-coding RNAs are the key regulators of gene expression regulation. In the present study, we sought to uncover the potential lncRNAs associated with stroke and aging. In the comparison of young and old Middle cerebral artery occlusion models (MCAo) mouse models with the age-matched controls, we found an up-regulation of 27 and 89 lncRNAs in the young and old mice, respectively, after stroke induction. Similarly, we found down-regulation of 24 lncRNAs in the old mice. In our study, we also found an up-regulation of the host genes for the microRNAs miR142 and mir-675.
The potential cis-targets of the up-regulated lncRNAs are related to blood vessel morphogenesis, vascular development, and the immune system. Among the cis-targets of down-regulated lncRNAs, we find enrichment of genes involved in membrane action potential and regulation of blood circulation. Importantly, the magnitude of the cellular and molecular response to stroke correlates with differential expression of lncRNAs and the target genes. In conclusion, we demonstrate the association of lncRNAs with pathophysiological processes during stroke, such as apoptosis, angiogenesis, inflammation, blood-brain barrier breakdown, and neurogenesis.
Keywords
microRNAs
Ischemic stroke
Novel therapeutic targets
Non-coding RNAs
Cardiovascular diseases
Pathophysiology
1 Introduction
Stroke, a broad medical condition originates from the disturbance of blood supply to the brain remains a major health care challenge causing high mortality and disability rates worldwide. According to a recent report, approximately 140,000 people die every year due to stroke that corresponds to one in every 20 deaths (Yang et al., 2017). Stroke also affects the economy of the United States costing 34 billion dollars annually for treatment, medications, and loss of manpower issues (Mukundan and Seidenwurm, 2018). The prevalence of stroke in western countries is well documented which helps in planning for affordable treatment strategies. However, in Asian countries, proper data regarding the incidence and prevalence of stroke is coming up only in recent times. In the kingdom of Saudi Arabia with a population of 28 million people, the emergence of recent epidemiological data highlights that stroke is going to be a major health problem (Akala and El-Saharty, 2006). Current research data also displays the alarming situation of doubling the mortality rate in Saudi Arabia by 2030 (Tran et al., 2010). Different Mendelian stroke syndromes have been reported in patients ranging from monogenic to polygenic inheritance Genome-wide association studies (GWAS) conducted in Saudi stroke patients also reported some SNPs associated with genes such as Sortilin (SORT1) (Alharbi et al., 2018).
Recent advances in next-generation sequencing technologies, changing the traditional approach of disease phenotype with respect to gene specific mutations. New sequencing platforms uncover the role of non-coding part of the genome in disease pathology. The protein coding part of the genome constitutes only 2% while 98% of the rest of the genome was once considered to be junk DNA, that are not transcribed and translated. But the recent transcriptomic studies confirm that at least one third of the genome is transcribed into different non-translatable RNAs (Djebali et al., 2012). LncRNAs or Long non-coding RNAs are the transcripts of more than 200 nucleotides in length constitute an important member of non-protein coding RNAs. More than 100,000 lncRNAs have been annotated in mouse and human, however the degree of conservation at the primary sequence level between different species is less in comparison to protein coding RNAs (Zhao et al., 2016). The role of lncRNAs in regulating gene expression is well studied and shown to be at different levels like, transcription, RNA processing, chromatin modelling and, is very well established for some genes such as Xist (Brown and Chow, 2003). In-vitro cell culture models also claim that lncRNAs regulate multiple cellular functions such as apoptosis, cell proliferation (Lei et al., 2018; Tao et al., 2019; Yang et al., 2019).
High abundance and cell type specific expression of non-coding RNAs in nervous system and association of non-coding RNAs with neural disorders, makes them as interesting candidates to study their role in stroke (Salvatori et al., 2020). Microarray or RNASeq based studies in induced Middle cerebral artery occlusion models (MCAo) mouse models and human patients highlight the possible role of non-coding RNAs in stroke (Dykstra-Aiello et al., 2016; Zhang et al., 2016; Zhang and Wang, 2019). Aberrant expression of lncRNAs such as NEAT1, MALAT1, N1LR, ANRIL, maternally expressed gene 3 (MEG3) and H19 have been detected in animal or human models (Tao et al., 2019; Bai et al., 2014; Jin et al., 2021; Wang et al., 2019; Zhang et al., 2017). Mechanism behind the stroke induced upregulation of lncRNAs and their impact in stroke pathophysiology is not completely understood. However, some lncRNAs have been linked to stroke induced specific pathophysiological process during the onset of stroke such as excitotoxicity, apoptosis, angiogenesis, inflammation, Blood brain barrier breakdown and also in post stroke recovery like neurogenesis (Ren and Yang, 2018).
The well-known risk factors the contribute to stroke incidence are age, sex obesity, smoking, diabetes, sickle cell disease, HIV, blood pressure, and hypertension (Boehme et al., 2017). Animal models generated for stroke, mostly use young animals and males for their studies often neglecting the nonmodifiable risk factors like age and stroke. The impact of age and sex in stroke is very well documented showing an increased mortality with increasing age. Changes in expression of non-coding RNAs with respect to age especially in brain is also well documented (Pereira Fernandes et al., 2018). We used global transcriptomic profile data of ischemic challenge in young and old age groups of mice to identify stroke associated lncRNAs. To understand the influence of age on the expression of non-coding RNAs in response to ischemic stroke, we analyzed the potential cis-targets of differentially expressed lncRNAs and identified the pathways and signaling networks controlled by the lncRNAs.
2 Methods
2.1 RNASeq data mapping and analysis
We obtained the raw RNASeq data set from 3-month- and 18-month-old mice with middle cerebral artery occlusion (MCAo) (Androvic et al., 2020). Raw Fastq data were obtained from the NCBI GEO database (GSE137482). The study included 6 replicates of 3-month-old and 18-month-old MCAo ischemic stroke model and their age-matched controls. (Androvic et al., 2020). The quality of the raw Fastq files was checked using the FastQC tool. Adapters were trimmed from raw reads using Trimgalore with a maximum trim error rate of 0.1, a minimum required adapter overlap of 1 bp, and a minimum read length of 20 bp after trimming. Adapter trimmed reads were mapped to the mouse reference genome mm10 using STAR aligner (Dobin et al., 2013). For the read alignment, 10 reads were allowed for multiple alignments. The minimum number of matches for an alignment was set to 16. The ratio of mismatches observed for an alignment to mapped length was set to 0.05. The intronic gap for alignment of reads spanning more than one exon was set to a minimum of 20 bp and a maximum of 1000000 bp. The minimum overhang for spliced alignment was set to 8 bp and for annotated spliced crossover to 1 bp. Samtools was used for indexing the alignment (Li et al., 2009).
2.2 Differential expression analysis
The expression levels of lncRNAs in each sample were quantified using the cuffquant tool (Trapnell et al., 2012). The known mouse lncRNAs were obtained from the GENCODE (mm10) database (Frankish et al., 2021). The properly mapped reads were assigned to lncRNAs using cuffquant. The mean fragment length was set at 200 bp with a standard deviation of 80 bp. The maximum number of fragments in a bundle before skipping was set to 500000. The quantified unstranded reads were further normalized using the cuffnorm tool. Six replicates of each condition were used for geometric normalization of the dataset using cuffdiff (Trapnell et al., 2013). Differential expression of lncRNAs was calculated in young and old MCAo mice relative to their age-matched controls. Geometric normalization and pooled dispersion methods were used to calculate differential expression. A maximum of 5000 iterations were allowed for the MLE calculation. A cut-off value of 0.05 for false discovery rate was used to identify the significantly differentially expressed LncRNAs. To identify the most differentially expressed lncRNAs, a twofold cut-off was used.
2.3 Functional classification of LncRNAs
The potential targets of differentially expressed LncRNAs were determined using annotation of genes and LncRNAs in the UCSC genome tools (Haeussler et al., 2019). We categorized lncRNAs into two groups: gene body and intergenic RNAs. lncRNAs that occur within a protein-coding gene were grouped as gene body. These include overlapping transcripts, anti-sense transcripts, and transcripts from intronic regions. The genes overlapping to the lncRNA loci were considered to be the potential cis-targets of the lncRNAs. lncRNAs from the intergenic regions were grouped together. The immediate genes on the 5′ and 3′ sides were considered as potential cis-targets.
The enrichment of GO terms in GOdirect molecular function classification was analyzed using DAVID functional annotation tools (Haeussler et al., 2019; Jiao et al., 2012). The top 10 enriched terms associated with the cis-target genes of the up- or down-regulated lncRNAs were identified. Biological network analysis was performed using the STRING network analysis tools (Szklarczyk et al., 2019). The differentially expressed cis-target genes were subjected to interaction analysis. Available active interaction data including text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence were used to visualize the interactions. The minimum required score for interactions was set at 0.4. Known microRNA loci were extracted from mirBase, and overlap with lncRNA loci was determined using bedtools (Kozomara et al., 2019; Quinlan, 2014).
3 Results
3.1 Stroke induced lncRNA expression dynamics
To understand the role of long non-coding RNAs in the development of stroke, we analyzed the RNA expression profile of MCAo mouse model. We examined the expression of lncRNAs in young and aged MCAo mouse model and compared differential expression pattern with the age matched healthy mice. We examined the expression levels of 9855 lncRNAs annotated in the mouse Gencode database (Frankish et al., 2021). Of these, we find 6371 lncRNA genes having an expression value of above 1 in at least one sample. We also find 3492 lncRNA genes having median expression value above one. The differential gene expression analysis helped us to identify the key lncRNA genes associated with the stroke and aging.
We found differential expression of 11 lncRNAs (9 upward and 2 downward) during aging in the healthy mice (Fig. 1A). However, in the MCAo stroke mouse model, we found 36 lncRNAs differentially expressed during aging (Fig. 1B). We observed stroke-induced up- or downregulation of several lncRNAs in brain cells of MCAo mice relative to control mice. In the young mice (3 months), we found upregulation of 58 genes and downregulation of 14 genes compared with the age-matched young control mice (Fig. 1C). Similarly, we found increased levels of 89 lncRNAs and decreased levels of 24 lncRNAs in 18-month-old mice compared with controls (Fig. 1D).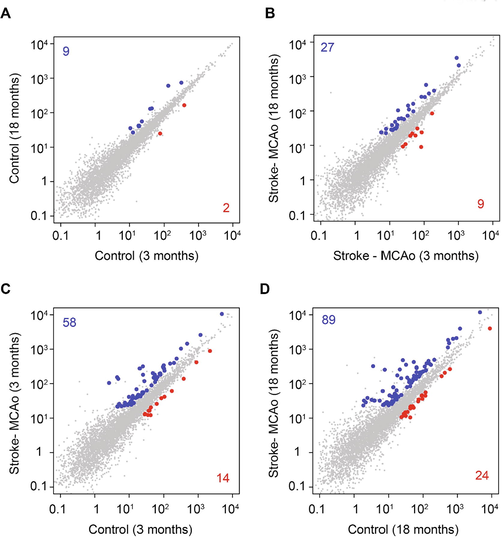
The differential expression of LncRNAs in mouse stroke model. The scatter plot showing the expression levels of LncRNAs in the comparisons of 3 months vs 18 months in control (A), MCAo (B), and age-matched comparison of control vs stroke at 3 months (C) and 18 months (D). The up- and down-regulated genes are highlighted in blue and red, respectively. The total number of up- and down-regulated genes for each comparison is also indicated. Scale – Gene expression (fpkm).
3.2 Increased risk of ischemic stroke is associated with aging
The principle component analysis of top 2000 most variable gene sets indicated a major difference between control and stroke datasets (Fig. 2A). This indicates the stroke is the major driving factor irrespective of aging in our datasets. The overlap analysis of lncRNAs associated with aging and stroke (Fig. 2B and C) indicated that the differential expression pattern in the brain is different for the aging of MCAo stroke mouse model and the control mouse model. We find an overlap of only 2 lncRNA genes commonly and differentially expressed (1 up and 1 down) upon aging in the brain of control and stroke mouse model. The comparison of the age matched stroke and control mice indicated that the aged mice are at high risk showed up regulation of 75 genes and down regulation of 22 genes. However, in the young mice we find the activation of only 52 genes and down regulation of 14 genes, indicating the severity of the disease in old mice. Interestingly, we find an overlap of 31 S induced upregulated genes between young and old mice. Only 3 genes are commonly down regulated between young and old mice upon stroke induction. Interestingly, we found 63 genes very specifically altered in old mice (44 up and 19 down) upon stroke. In the young mice we found 21 up regulated and 11 down regulated genes due to stroke.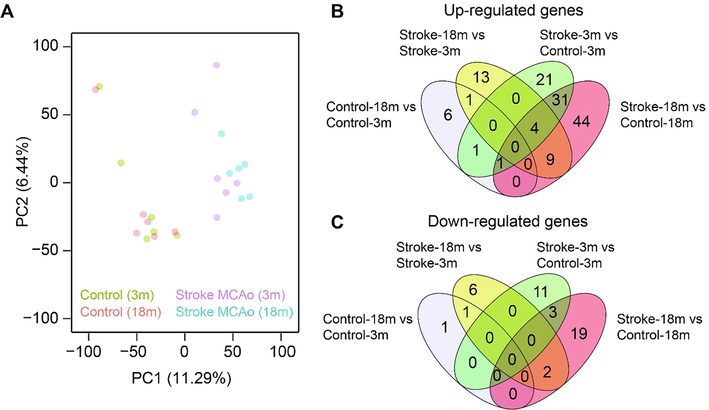
Increased alteration of LncRNAs level in aged stroke model. Principal component analysis of stroke and control RNASeq samples. Samples are color-coded according to the condition, and each dot represents a replicate of a condition (A). The overlap of up- (B) and down-regulated (C) genes in four different comparisons mentioned in Fig. 1.
3.3 lncRNAs regulates the tube morphogenesis and membrane potential in response to stroke-induced damages
lncRNAs function as regulatory elements in association with chromatin modifiers and regulators of gene expression (Lv et al., 2013). We investigated the potential functional role of lncRNAs by correlating them with the expression status of the nearest genes. lncRNAs can be transcribed from the intergenic regions, introns, or the opposite strand of protein-coding genes. We found common up regulation of 31 genes in both young and old mice upon stroke induction this include., Gm26719, Gm38399, AUO20206, Gm26735, Snhg18, Gm16617, Abhd11os and Gm26586 (Fig. 3). We grouped lncRNAs located within the gene body or transcribed from the intergenic regions. A total of 111 lncRNAs were upregulated and 35 lncRNAs were downregulated in the MCAo stroke model compared with age-matched controls. We found 54% of the upregulated lncRNAs and 48% of the downregulated lncRNAs within the gene body of another gene (Fig. 4A). The remaining lncRNA loci are located in the intergenic regions. lncRNAs can act as hosts for microRNAs. In our analysis, we found upregulation of mir142hg in old and H19 in young MCAo mice relative to the age-matched controls. Mir142hg is the host for mir-142a and mir-142b. H19 is the host RNA for mir-675. In the aged stroke model, we found downregulation of Gm15816, Sox2ot, and Gm27032, which are the host RNAs for mir-486a/b, mir-1897, and mir-124, respectively.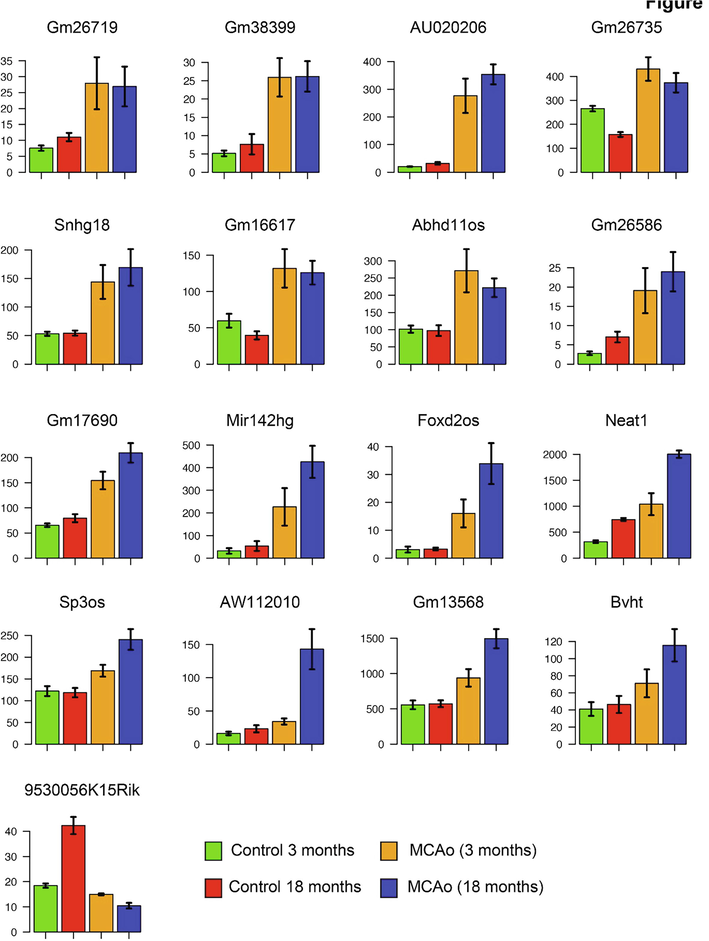
The differential expression of lncRNAs in young and old stroke models. Bar graphs show the selected lncRNAs differentially expressed in the old and/or young stroke model compared with age-matched controls. Indicated are the average expression levels of 3-month-old control mice (green), 18-month-old control mice (red), 3-month-old stroke mice (orange), and 18-month-old stroke mice (blue). The y-axis shows the expression values in fpkm.
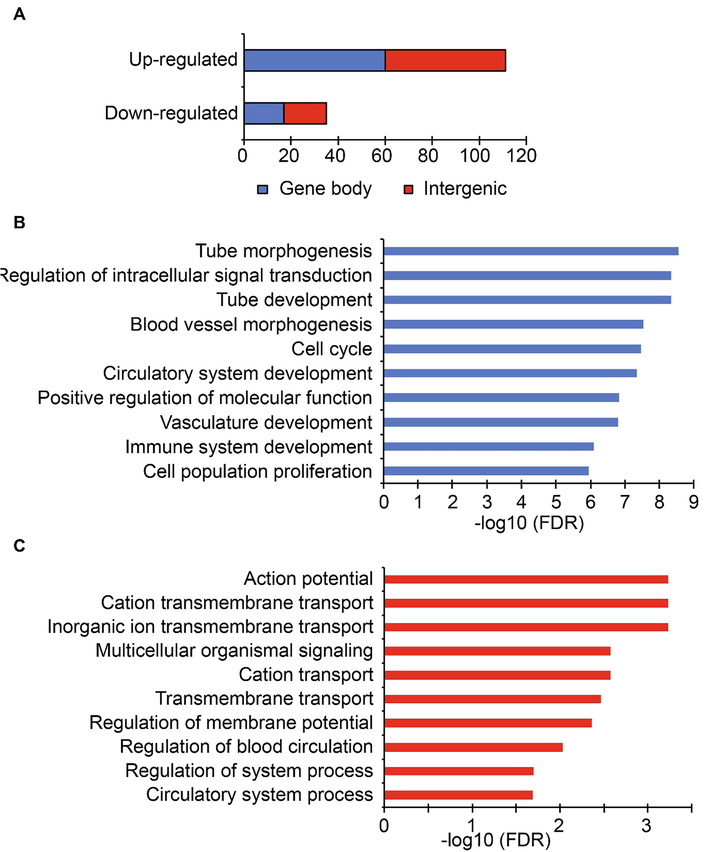
Stroke induced lncRNAs regulate angiogenesis and membrane potential. A. The distribution of cis-targets of differentially expressed lncRNAs. The x-axis indicates the total number of genes. The cis-targets are grouped according to the occurrence of the lncRNA within the gene body (blue) or intergenic regions (red). Gene ontology enrichment analysis of cis-targets of up-regulated (B) and down-regulated (C) Lnc RNAs. The top 10 overrepresented Go terms are indicated. The x-axis represents the -log10 of false discovery rate.
We examined the expression status of potential cis-targets of differentially expressed lncRNAs. We found a positive correlation between the up-regulation of lncRNAs and associated protein-coding genes. We found 24 up-regulated lncRNAs expressed from the gene body of another gene, and 11 up-regulated lncRNAs occurring in the intergenic regions showed up-regulation of their target genes. We also found 3 down regulated lncRNAs that occurred within the gene body showed down regulation of their target genes. One down-regulated lncRNA, 9530056K15Rik, showed up-regulation of its 5′ target gene Kdelr2.
Gene ontology analysis of potential cis-targets of lncRNAs revealed enrichment of genes related to tube development, tube morphogenesis, vascular development, immune development, and blood vessel morphogenesis (Fig. 4B). The cerebral vessels are very important for the function and development of the brain. After ischemic stroke, there is stimulation of neurogenesis and vascular remodeling as part of the recovery process due to decreased blood flow (24291532). Interestingly, we observe that several of these genes are regulated by stroke-induced lncRNAs. We also found enrichment of genes related to intracellular signal transduction, cell cycle, and cell proliferation. The neurodegenerative damage caused by stroke triggers the proliferation of activated endothelial cells that migrate into the newly formed blood vessels. The angiogenic and inflammatory factors expressed by the activated microglial cells promote the formation of the tube. Among the cis-targets of down-regulated lncRNAs, we find genes associated with ion transmembrane transport, regulation of membrane potential and blood circulation (Fig. 4C). The lack of blood flow due to stroke deprive oxygen and glucose supply to neurons. Due to this the neurons lose their potential to maintain the membrane potential. Subsequently, the depolarization of neurons and in appropriate firing of its action potential occurs. The genes associated with the key molecular events downstream to stroke induced damages are regulated by the lncRNAs. We find increased levels of some of the genes associated with tube development, morphogenesis, vasculature, cell cycle and proliferation (Rhoa, Casp8, Myc, Bcl2l11, Sox4, Nr2f2, Rnf213, Akap13, Ptbp1, Rps6ka1, Dock2, Gsn and Slc5a3) (Fig. 5).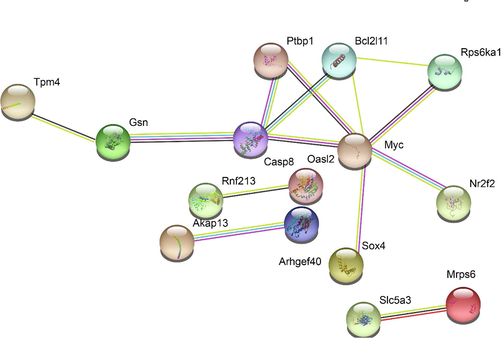
The network of differentially expressed cis-targets. STRING interaction network analysis of differentially expressed cis-targets in age-matched comparisons of stroke and control mice. Genes with at least one interaction are indicated in the figure.
4 Discussion
Age is an important, non-modifiable factor in the incidence of stroke. The chances of recovery from stroke are inversely proportional with increasing age. The cellular composition of the blood-brain barrier and age-related changes in permeability are also responsible for less efficient recovery after stroke (Kaur et al., 2011). Non-coding RNAs are involved in maintaining blood-brain barrier permeability (Liu et al., 2021). Transcriptomic analysis of the widely accepted in vitro model for the blood-brain barrier, the human cerebral microvascular endothelial cell line (hCMEC/D3), identified several microRNAs, lncRNAs (Kalari et al., 2016). The association of the differentially expressed non-coding RNAs miR142, Snhg18, and H19 in our mouse model with blood-brain barrier functions suggests the possible role of these RNAs in maintaining the integrity of the blood-brain barrier after stroke.
The lncRNAs function as the cis or trans regulatory elements in the gene expression regulation. They can function as activator or repressor of the gene expression. In the mouse model of Huntington's disease, overexpression of the lncRNA Abhd11os provides neuroprotection by inhibiting the mutant huntingtin protein in the mouse striatum, whereas downregulation of Abhd11os leads to neurotoxicity (Francelle et al., 2015). Increased levels of Abhd11os were detected in the rat myocardial ischemia/reperfusion (MIRI) model. The increased level of lncRNA promoted apoptosis and prevented cell proliferation after injury. Knockdown of Abhd11os in the MIRI mouse model reduced myocardial infraction size and inhibited apoptosis (Liu et al., 2021). This suggests a possible role of Abhd11os in MCAo stroke-induced damage in our mouse model. We found increased levels of micro RNA 142 host gene (mir142hg) upon stroke in adult mice. The inflammation associated microRNA miR142 is elevated upon traumatic brain injury. The miR142 activates astrocyte and induce brain inflammation in response to traumatic brain injury (Korotkov et al., 2020). This indicates the involvement of mir142 in stroke induced damages in adult mice. In the tMCAo model, an increased level of the circular RNA HECD1 was detected, which inhibits the function of miR142 by acting like a sponge (Han et al., 2018). The circular RNA HECD1 has high therapeutic potential for controlling mir142-induced damages after stroke.
We find adjacent opposite strand lncRNA transcripts for the transcription factors Foxd2 Sox4 and Sp3. FoxD2-os is highly upregulated in adult mice upon stroke. In human brain malignancy glioma, an increased level of FOXD2-AS1 lncRNA correlated with poor patient outcome (Shangguan et al., 2019). Though we do not find a significant homology between mouse Foxd2-os and human FoxD2-AS1, the available evidences suggest an important role of this RNA in disease development. Snhg18 is upregulated after stroke in both old and young mice. Snhg18 is involved in the pathogenesis and progression of gliomas. It promotes glioma cell invasion and migration by inhibiting nucleocytoplasmic transport of α enolase (Zheng et al., 2019). Elevated levels of Gm38399 were observed in young and adult mice after stroke induction. Regular consumption of green or black tea may reduce the risk of stroke (Arab et al., 2009). Elevated levels of lncRNA Gm38399 could be controlled by green tea administration in an obese mouse model (Li et al., 2019). This suggests that Gm38399 may be a therapeutic target to reduce stroke risk. We found increased levels of AU020206 in both young and old mice after stroke-induced damage. An increased level of AU020206 was observed after sciatic nerve injury in a mouse model (Barrette et al., 2010). AU020206 is one of the possible candidate genes involved in regeneration by eliciting a safe immune response that may lead to rapid clearance of axon and myelin remnants after injury. Altered expression of GM17690 was detected in brain microvascular endothelial cells after oxygen deprivation in an in-vitro model of cerebral ischemic stroke (Zhang et al., 2016). We found up-regulation of AW112010 specifically in the aged mice after stroke induction. AW112010 functions as a pro-inflammatory factor in T cells. In conjunction with KDM5A, it is involved in the regulation of IL10 gene expression (Yang et al., 2020). Increased levels of AW112010 were observed in microglial cells in the in-vitro OGD stroke model (Zhang et al., 2019). We found an increased level of Nuclear enriched abundant transcript 1 (NEAT1) upon stroke induction in the aged mice. The lncRNA was significantly upregulated in patients with ischemic stroke (Zhang and Wang, 2019). In an in vitro stroke model with oxygen-glucose deprivation (OGD)/reoxygenation, NEAT1 reduced stroke-induced damages by downregulating AKT/STAT3 pathway activity (Ni et al., 2020). We found upregulation of H19 in young animals. H19 promotes neuroinflammation in stroke patients and has been proposed as a potential biomarker for stroke (Wang et al., 2017). Polymorphisms associated with the H19 locus are associated with altered susceptibility to stroke (Huang et al., 2019).
As an immediate response to stroke, membrane depolarization occurs in brain tissue due to the lack of blood supply and decreased oxygen levels. We found enrichment of genes related to ion transmembrane transport and regulation of membrane potential as cis-targets of downregulated lncRNAs. Angiogenesis occurs in response to stroke-induced damage. Angiogenesis is stimulated by hypoxic conditions with the upregulation of hypoxic factors such as HIF-1α and VEGF (Greenberg, 1998). However, hypoxia-induced upregulation of HIF-1α is reduced in aged mice (Benderro and Lamanna, 2011). The expression of HIF-1α is regulated by the lncRNA NEAT1 (Zheng et al., 2018). Also, NEAT1 level is controlled by HIF1-α (Kong et al., 2019). We found upregulation of NEAT1 in 18-month-old mice, suggesting a possible age-dependent regulation of angiogenesis. LncRNA H19 promote invasion and angiogenesis in glioblastoma cells (Jiang et al., 2016). In addition, we found several genes associated with blood vessel morphogenesis and development as the cis-targets of upregulated lncRNAs. We also found a positive correlation between differential expression of lncRNAs and associated protein-coding genes indicating a prominent role of lncRNAs in regulating several molecular signaling network in response to stroke.
5 Conclusions
Our study illustrates the stroke-induced lncRNA transcriptome and its association with aging. Differentially expressed lncRNAs are predominantly associated with genes involved in blood vessel morphogenesis, vascular development, and the immune system. The magnitude of the cellular and molecular response to stroke correlates with differential expression of lncRNAs and target genes. We show that lncRNAs are associated with pathophysiological processes during stroke, such as apoptosis, angiogenesis, inflammation, and neurogenesis. The identified lncRNAs could serve as a potential therapeutic target to reduce the risk of stroke.
Acknowledgements
The authors extend their appreciations to the deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (lFP-2020-39).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Vital Signs: Recent Trends in Stroke Death Rates – United States, 2000–2015. MMWR Morb Mortal Wkly Rep. 2017;66(35):933-939.
- [Google Scholar]
- Economic and Societal Aspects of Stroke Management. Neuroimaging Clin N Am. 2018;28(4):683-689.
- [Google Scholar]
- Public-health challenges in the Middle East and North Africa. Lancet. 2006;367(9515):961-964.
- [Google Scholar]
- The epidemiology of stroke in the Middle East and North Africa. J Neurol Sci. 2010;295(1-2):38-40.
- [Google Scholar]
- Molecular genetic studies in Saudi population; identified variants from GWAS and meta-analysis in stroke. Saudi J Biol Sci. 2018;25(1):83-89.
- [Google Scholar]
- NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44(D1):D203-D208.
- [Google Scholar]
- Beyond sense: the role of antisense RNA in controlling Xist expression. Semin Cell Dev Biol. 2003;14(6):341-347.
- [Google Scholar]
- Long non-coding RNA ABHD11-AS1 promotes colorectal cancer development through regulation of miR-133a/SOX4 axis. Biosci Rep. 2018;38
- [Google Scholar]
- LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol Reprod Dev. 2019;86(6):686-695.
- [Google Scholar]
- LncRNA: Shedding light on mechanisms and opportunities in fibrosis and aging. Aging Res Rev. 2019;52:17-31.
- [Google Scholar]
- Non-coding RNAs in Nervous System Development and Disease. Front Cell Dev Biol. 2020;8:273.
- [Google Scholar]
- Altered Expression of Long Noncoding RNAs in Blood After Ischemic Stroke and Proximity to Putative Stroke Risk Loci. Stroke. 2016;47(12):2896-2903.
- [Google Scholar]
- Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol. 2016;277:162-170.
- [Google Scholar]
- Long Non-coding RNA in CNS Injuries: A New Target for Therapeutic Intervention. Mol Ther Nucleic Acids. 2019;17:754-766.
- [Google Scholar]
- Regulation of CARD8 Expression by ANRIL and Association of CARD8 Single Nucleotide Polymorphism rs2043211 (p.C10X) With Ischemic Stroke. Stroke. 2014;45(2):383-388.
- [Google Scholar]
- Transcriptome-Wide Analysis to Identify the Inflammatory Role of lncRNA Neat1 in Experimental Ischemic Stroke. J Inflamm Res. 2021;Volume 14:2667-2680.
- [Google Scholar]
- Long noncoding RNA H19 prevents neurogenesis in ischemic stroke through p53/Notch1 pathway. Brain Res Bull. 2019;150:111-117.
- [Google Scholar]
- Long Noncoding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J Neurosci. 2017;37(7):1797-1806.
- [Google Scholar]
- Pathophysiology of Long Non-coding RNAs in Ischemic Stroke. Front Mol Neurosci. 2018;11:96.
- [Google Scholar]
- Pereira Fernandes D, Bitar M, Jacobs FMJ, Barry G: Long Non-Coding RNAs in Neuronal Aging. Noncoding RNA. 2018. 4.
- Decoding the Transcriptional Response to Ischemic Stroke in Young and Aged Mouse Brain. Cell Rep. 2020;31(11)
- [Google Scholar]
- Genome Project Data Processing S: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078-2079.
- [Google Scholar]
- Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562-578.
- [Google Scholar]
- Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46-53.
- [Google Scholar]
- The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853-D858.
- [Google Scholar]
- DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805-1806.
- [Google Scholar]
- STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613.
- [Google Scholar]
- miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155-D162.
- [Google Scholar]
- BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11 12:11-34.
- [Google Scholar]
- Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 2013;41:10044-10061.
- [Google Scholar]
- Quantitative MRI reveals the elderly ischemic brain is susceptible to increased early blood-brain barrier permeability following tissue plasminogen activator related to claudin 5 and occludin disassembly. J Cereb Blood Flow Metab. 2011;31(9):1874-1885.
- [Google Scholar]
- Involvement of noncoding RNA in blood-brain barrier integrity in central nervous system disease. Noncoding RNA Res. 2021;6(3):130-138.
- [Google Scholar]
- BBBomics-Human Blood Brain Barrier Transcriptomics Hub. Front Neurosci. 2016;10:71.
- [Google Scholar]
- Striatal long noncoding RNA Abhd11os is neuroprotective against an N-terminal fragment of mutant huntingtin in vivo. Neurobiol Aging. 2015;36(1601):e1607-e1616.
- [Google Scholar]
- Knockdown of lncRNA Abhd11os attenuates myocardial ischemia/reperfusion injury by inhibiting apoptosis in cardiomyocytes. J Cardiovasc Pharmacol 2021
- [Google Scholar]
- Korotkov A, Puhakka N, Gupta SD, Vuokila N, Broekaart DWM, Anink JJ, Heiskanen M, Karttunen J, van Scheppingen J, Huitinga I, et al: Increased expression of miR142 and miR155 in glial and immune cells after traumatic brain injury may contribute to neuroinflammation via astrocyte activation. Brain Pathol 2020, 30:897-912.
- Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14(7):1164-1184.
- [Google Scholar]
- FoxD2-AS1 is a prognostic factor in glioma and promotes temozolomide resistance in a O(6)-methylguanine-DNA methyltransferase-dependent manner. Korean J Physiol Pharmacol. 2019;23:475-482.
- [Google Scholar]
- Long Noncoding Ribonucleic Acid SNHG18 Promotes Glioma Cell Motility via Disruption of alpha-Enolase Nucleocytoplasmic Transport. Front Genet. 2019;10:1140.
- [Google Scholar]
- Green and black tea consumption and risk of stroke: a meta-analysis. Stroke. 2009;40(5):1786-1792.
- [Google Scholar]
- lncRNA profiling to elucidate the metabolic mechanism of green tea extract on weight loss in mice. Tropical Journal of Pharmaceutical Research. 2019;18:1733-1738.
- [Google Scholar]
- Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun. 2010;24(8):1254-1267.
- [Google Scholar]
- Long Noncoding RNA AW112010 Promotes the Differentiation of Inflammatory T Cells by Suppressing IL-10 Expression through Histone Demethylation. J Immunol. 2020;205(4):987-993.
- [Google Scholar]
- LncRNA-1810034E14Rik reduces microglia activation in experimental ischemic stroke. J Neuroinflammation. 2019;16:75.
- [Google Scholar]
- Knockdown lncRNA NEAT1 regulates the activation of microglia and reduces AKT signaling and neuronal apoptosis after cerebral ischemic reperfusion. Sci Rep. 2020;10:19658.
- [Google Scholar]
- Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke. 2017;48(8):2211-2221.
- [Google Scholar]
- Association of long noncoding RNA H19 polymorphisms with the susceptibility and clinical features of ischemic stroke in southern Chinese Han population. Metab Brain Dis. 2019;34(4):1011-1021.
- [Google Scholar]
- Hypoxia-induced angiogenesis is delayed in aging mouse brain. Brain Res. 2011;1389:50-60.
- [Google Scholar]
- HIF-2alpha activated lncRNA NEAT1 promotes hepatocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem. 2018;119:3247-3256.
- [Google Scholar]
- Overexpression of HIF-2alpha-Dependent NEAT1 Promotes the Progression of Non-Small Cell Lung Cancer through miR-101-3p/SOX9/Wnt/beta-Catenin Signal Pathway. Cell Physiol Biochem. 2019;52:368-381.
- [Google Scholar]
- Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;2016(1):129-136.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102119.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







