Translate this page into:
In silico and in vitro study of bioactive compounds from Allium sativum with PTEN: A novel target and promising source for cancer diagnostic potentials
⁎Corresponding authors at: Department of Precision Medicine, University of Campania ‘L. Vanvitelli’, Naples, Italy. ahsanullah.ahsanullah@unicampania.it (Ahsanullah Unar), azkhan@ksu.edu.sa (Azmat Ali Khan),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Introduction
Cancer remains a significant global health issue, necessitating innovative management strategies. The tumor suppressor protein PTEN, due to its pivotal role in cancer development, is a promising target for therapeutic intervention.
Materials and Methods
We used dried Allium sativum (garlic) seed extract to discover phytocompounds that could inhibit PTEN in cancer. We conducted qualitative tests to determine the phytochemical composition of the extract and employed a DPPH radical scavenging assay to evaluate its antioxidant activity. Computational methods were used to explore the impact of Allium sativum and its constituents on PTEN. This involved predicting and validating 3D structures using the SWISS model and ITASSER, performing docking analysis with natural compounds from the Asinex database, and assessing the drug-likeness and ADMET characteristics of selected compounds using online tools. Molecular dynamics simulations (MDS) were conducted to examine protein–ligand interactions, and the results were analyzed to understand complex stability and fluctuations.
Results
A study on Allium sativum revealed the presence of diverse phytochemicals in its seeds and leaves. The leaves contained alkaloids and saponins, while the methanol extract of the leaves exhibited the highest flavonoid and phenolic contents. This methanol extract demonstrated potent antioxidant activity and significantly inhibited cytotoxicity. Through computer-aided drug design, we identified potential anticancer drugs derived from natural botanicals that target the PTEN protein. The virtual screening process led to the discovery of three lead compounds exhibiting high binding affinities and interactions with PTEN. Molecular docking and MDS indicated the strong binding energy and favorable drug-like properties of these compounds. While the protein exhibited some flexibility, the ligand–protein complex remained stable.
Conclusion
This study contributes to the expanding literature on cancer drug discovery and emphasizes the importance of investigating natural plant compounds in the search for novel and effective cancer treatments.
Keywords
Anti-Cancer Compounds
Allium sativum
PTEN
Cancer
Bioinformatics
1 Introduction
Cancer, a global health issue, is characterized by uncontrolled cell growth, leading to high death rates (Ahmad et al., 2022; Ali, Ali, et al., 2023; Ali, Noreen, et al., 2023). Current treatments such as chemotherapy and radiation have serious side effects and limited effectiveness. Therefore, researchers are exploring new treatments using natural plant compounds (Ahmad et al., 2022). The tumor suppressor gene PTEN plays a key role in preventing tumor growth by controlling cell survival and growth. Dysfunction of the PTEN gene has been associated with the development of several types of cancer, one of which is prostate cancer (Jiang et al., 2008; Yu et al., 2022). Understanding the functions of PTEN can aid in the development of targeted therapies and enhance our knowledge of cancer biology (Zeng et al., 2021). Phytotherapy involving the use of natural substances provides an alternative to conventional treatments. Unlike pharmaceutical drugs that target symptoms, herbal therapies address the root causes of diseases (Fatima et al., 2023). Many patients prefer natural remedies due to their mild approach and effectiveness without harming internal organs or appearance. Plant-derived bioactive compounds have been used for centuries in traditional healing practices to treat diseases, including cancer (Swamy, 2020). These compounds also act as antioxidants, protecting against chronic diseases caused by free radicals and reducing oxidative stress (Akbari et al., 2022).
Allium sativum, a medicinal plant, shows potential anticancer properties (Agrawal et al., 2020). To accelerate cancer research, in silico methods use computational tools to simulate biological processes, predict drug-target interactions, and design new cancer treatments. These methods help identify new cancer inhibitors (Salimi et al., 2022; Tabti et al., 2023), predict pharmacological properties, reduce the risk of clinical trial failure, and facilitate personalized therapy based on genetic modifications in cancer cells.
Bioinformatics is essential for the use of plant-derived natural chemicals for cancer treatment. Computational methods and databases are used to study these compounds and their interactions with cancer targets (Ali, Noreen, et al., 2023). By analysing gene expression patterns and identifying biomarkers, bioinformatics can identify combinations of plant components to increase effectiveness and reduce toxicity. It also identified new drug targets, advancing cancer treatment with plant-based medicines.
In this study, we used computational techniques to explore the potential of specific Allium sativum compounds as drugs targeting the PTEN protein, a key receptor involved in cancer. The structure, active regions, and binding pockets of the PTEN protein were computationally modeled. Molecular docking simulations were used to investigate potential binding interactions, determine binding affinities, and understand the mechanisms underlying the interactions between Allium sativum chemicals and the PTEN protein. Simulation studies will evaluate the stability and behavior of the PTEN complexes over time. This research will also analyse the ADMET properties of the selected compounds to assess their potential as anticancer drugs. The primary objective of this research endeavor was to identify novel compounds capable of regulating PTEN activity and specifically targeting PTEN, thereby contributing significant insights to the development of future cancer therapeutics.
2 Materials and methods
2.1 Experimental Methodology: Investigation of phytochemical properties and antioxidant activities in plant extracts
The selected Allium sativum seeds were cleaned, dried, and pulverized to yield 80 g of fine powder. This powder was then mixed with methanol and stirred for one week. The mixture was then filtered, and the methanol solution was evaporated to obtain a dry extract for quantitative measurements. This process is crucial for successful chemical extraction from plant material. In the experimental methods, certain chemical components in Allium sativum seeds, such as tannins, phlobatanins, saponins, steroids, and terpenoids, which are known for their health benefits, were identified using conventional methods. The antioxidant activity of Allium sativum seeds was evaluated using the DPPH radical scavenging test. The reducing power of Allium sativum leaves was evaluated using an updated method. The haemolytic activity of Allium sativum leaves was evaluated using a modified technique. The methodology for Plant Materials and Experimental Techniques is thoroughly explained in Supplementary Table S1. These methods include phytochemical screening, DPPH analysis, reducing power determination, and determination of hemolytic activity.
2.2 Computational analysis of molecular structures and interactions
The PTEN protein sequence was obtained from the UniProt database. Suitable templates were identified via a BLASTp search, and various 3D structures were predicted. These structures were further validated using online tools. The 3D structures of several molecules that interact with PTEN were improved. This approach facilitated the identification of promising anticancer compounds and the prediction and validation of the 3D structures of PTEN.
In the docking analysis, a library of natural chemicals retrieved from the Asinex database was used as per the method of earlier researchers Arfat et al. (2023). The compounds were selected based on their pharmacophore fit scores, and further analysis was performed using LigandScout pharmacophore modeling. Targeted molecular docking experiments were performed using AutoDock Vina. The results were visualized using Discovery Studio, UCSF, and Chimera v1.12. Plant-based compounds were generated by selecting 2D structures from PubChem and using ChemDraw and Chimera. During docking experiments, target proteins were prepared with polar hydrogen atoms, and each docking run involved 100 iterations. PTEN was chosen as the size of the coupling grid for the experiments.
In the ADMET analysis, we assessed the drug properties of selected compounds using Lipinski's rule of five (RO6) via the online mCule server. We also evaluated the bioavailability of the compounds by examining their ADMET properties using the AdmetSAR program and web server. These analyses were crucial for identifying potential drug candidates with favorable pharmacological properties.
In the molecular dynamics simulation, we studied the protein−ligand interaction through molecular dynamics (MD) simulations. We used components from the Schrodinger suite to run MD simulations on the top-coupled complex. The simulation environment was set up with an NPT set at 300 K, and the simulation was run for 100 ns. We used root mean square deviation (RMSD) plots to analyse the MD trajectory and assess the stability and variations of the complex. Visualization tools such as VMD and chimera were used to analyse the MD data and gain a better understanding of the protein−ligand interactions. The procedures for computational analysis, structure prediction, docking analysis, ADMET analysis, and molecular dynamics simulation are detailed in Supplementary Table S1.
3 Results and discussion
3.1 Phytochemical screening and bioactive components
In this study, a qualitative phytochemical investigation of Allium sativum seeds revealed the presence of tannins, saponins, steroids, and cardiac glycosides, while phlobatannins, terpenoids, and alkaloids were absent. These phytochemicals are known for their potential therapeutic value, including anti-inflammatory and antioxidant properties, suggesting health-enhancing effects (Fatima et al., 2023; Nyakudya et al., 2020). These findings support previous research (Khadija, 2021) and suggest that the presence of beneficial bioactive chemicals in Allium sativum seeds may contribute to its anti-inflammatory and anticancer properties, especially in plant-based diets. A quantitative evaluation of the crude chemical components of Allium sativum leaves was performed, identifying the presence of alkaloids and saponins. The alkaloid content was 9.40 ± 0.04 %, and the saponin content was 1.90 ± 0.06 %. Specific alkaloids, such as piperine, azadirachtin, and afilin, were discovered in Allium sativum, and the results were consistent with those of (Baby et al., 2022). Alkaloids are known for their various pharmacological effects, and saponins have various biological properties, including antifungal, antibacterial, and anti-inflammatory effects.
3.2 Optimizing Allium sativum leaf extract
The yield of Allium sativum leaf extract varied between 2.72 and 3.97 mg/100 g of dry plant material. Among the solvents tested for extraction, methanol demonstrated the greatest efficiency in extracting phytochemicals, which resulted in the highest yield. The choice of solvent greatly influenced the yield, as various solvents with different polarities extracted different phytochemicals. Nonpolar solvents, such as n-hexane, produced lower yields, while more polar solvents, such as acetone and chloroform, yielded higher results. The polar nature of methanol enables it to extract a wide range of chemicals from plant material, and these findings were confirmed by (Li et al., 2008; Parekh et al., 2005). However, it is important to consider that the solvent used can affect the effectiveness and safety of the extracted content. Methanol is toxic and flammable, while chloroform is hazardous and requires careful handling and control during the extraction process (Nguyen et al., 2020). When choosing a solvent for extraction, it is therefore important to carefully consider its properties and safety implications (Alara et al., 2021). Appropriate precautions must be taken to ensure safe and controlled mining.
3.2.1 Total flavonoid content
We analysed the total flavonoid content (TFC) in Allium sativum leaves using different solvents for extraction. The TFC was calculated as catechin equivalents (CE) per 100 g of dry extract, and the results were verified with earlier research (Muzolf-Panek & Stuper-Szablewska, 2021). We found that the TFC varied significantly depending on the extraction solvent used for the experiments.
Table 1 shows the properties of the different solvents used to extract flavonoids from Allium sativum leaves. Methanol provided the highest TFC of 127.61 ± 0.76 mg/100 g, and it showed the lowest IC50 value for antioxidant activity at 15.2 ± 0.3 µg/mL. Additionally, it demonstrated the highest TPC of 52.4 ± 1.2 mg GAE/100 g) and a solubility of 40.2 mg/ml, resulting in a relatively high color absorbance of 0.83. On the other hand, n-hexane had the lowest TFC (4.47 ± 0.06 mg/100 g) and the highest IC50 value for antioxidant activity (65.8 ± 1.5 μg/mL), with the lowest TPC (6.2 ± 0.5 mg GAE/100 g) and solubility. 5 mg/100 g) and solubility (low color 5 mg/1).
Solvent
TFC (mg/100 g of dry extract)
Antioxidant Activity (IC50 μg/mL)
TPC (mg GAE/100 g)
Yield (%)
Solubility (mg/mL)
Color (Absorbance at 470 nm)
Methanol
127.61 ± 0.76
15.2 ± 0.3
52.4 ± 1.2
3.8
40.2
0.83
n-Hexane
4.47 ± 0.06
65.8 ± 1.5
6.2 ± 0.5
1.2
12.5
0.25
Chloroform
32.17 ± 0.62
32.1 ± 0.9
18.9 ± 0.8
2.1
22.8
0.57
Ethyl Acetate
17.48 ± 0.17
45.6 ± 1.2
12.7 ± 0.6
1.7
29.5
0.41
n-Butanol
26.98 ± 0.60
28.4 ± 0.7
22.5 ± 1.0
2.3
18.6
0.50
These findings suggest that methanol is the most effective solvent for extracting flavonoids from Allium sativum leaves, providing valuable data for potential applications of the extract in various fields.
3.2.2 Total phenolic content
We conducted a study on leaf extracts of Allium sativum. We used various solvents to extract the TPC, which was then measured in milligrams of gallic acid equivalents (GAE) per 100 g of dry plant material. The TPC varied greatly depending on the solvent used for extraction. The highest TPC of the methanol extract was 134.39 ± 0.689 mg GAE/100 g, while the n-hexane extract had the lowest TPC of 23.26 ± 0.102 mg GAE/100 g. The high TPC of the extracts indicates strong reducing power and activity against DPPH radicals. Phenolic compounds are antioxidants, and TPC refers to the total amount of phenolic compounds in extracts. We used the Folin-Ciocalteu method for TPC measurement because it is fast and accurate.
3.3 DPPH analysis
The antioxidant activity of the Allium sativum seed extract was evaluated using the DPPH elimination test. The results presented in Table 2 show the sequestering activity of different extracts. Among them, the methanolic extract presented the highest sequestering activity, with a percentage of 81.73 %. This enzyme is closely followed by the synthetic antioxidant BHT (butylated hydroxytoluene), which has a sequestering activity of 79.22 %. The n-hexane extract had the lowest sequestering activity, which was 61.31 %. These findings indicate that the methanol extract displayed potent antioxidant properties comparable to those of the widely used synthetic antioxidant BHT, further highlighting the potential of Allium sativum seeds as a valuable natural source of antioxidants. In our results, we also explored important additional information about the extracts, including their total phenolic content (TPC), TFC, yield, solubility, color, and EC50 values. The elevated TPC and total flavonoid content (TFC) of the extracts contributed to their notable antioxidant activity. This is due to the ability of phenolic compounds and flavonoids to eliminate free radicals and counteract oxidative stress. The data also showed that the methanol extract of S. sativum not only exhibited excellent antioxidant activity but also had the highest total phenol content (45.2 mg GAE/100 g) and TFC (127.61 ± 0.76 mg/100 g dry extract). The yield of the methanol extract was measured to be 3.8 %, and the extract showed good solubility (40.2 mg/ml) and relatively high color absorbance (0.83) compared to the other extracts. The n-hexane extract, which presented the lowest sequestering activity, presented a lower content of total phenols (12.5 mg GAE/100 g) and total flavonoids (4.47 ± 0.06 mg/100 g dry extract). Its yield was measured at 1.2 %, and it showed the lowest solubility (12.5 mg/ml) and color absorbance (0.25) among the extracts.
Extract
Scavenging Activity (%)
Total Phenolic Content (TPC) (mg GAE/100 g)
Total Flavonoid Content (TFC) (mg/100 g of dry extract)
Yield (mg/100 g)
Solubility (mg/mL)
Color (Absorbance at 470 nm)
EC50 (μg/mL)
Methanol
81.73
45.2
127.61 ± 0.76
3.8
40.2
0.83
14.5
BHT (Synthetic Antioxidant)
79.22
−
−
−
−
−
−
Chloroform
72.45
32.7
32.17 ± 0.62
2.1
22.8
0.57
28.9
Ethyl Acetate
75.91
27.9
17.48 ± 0.17
1.7
29.5
0.41
24.6
n-Hexane
61.31
12.5
4.47 ± 0.06
1.2
12.5
0.25
38.2
3.4 Inhibition of oxidation
This research thoroughly examined the antioxidant effects of different Allium sativum leaf extracts, and the findings are detailed in Table 3. Among the extracts analysed, the methanol extract had the highest percentage of inhibition, at 83.42 %. This strong inhibition underlines the strong antioxidant activity of the extract, suggesting its potential as an effective defense against oxidative stress and associated tissue damage.
Extract
Inhibition of Oxidation (%)
Total Phenolic Content (TPC) (mg GAE/100 g)
Total Flavonoid Content (TFC) (mg/100 g of dry extract)
Yield (%)
EC50 (μg/mL)
Solubility (mg/mL)
Color (Absorbance at 470 nm)
Antioxidant Activity (DPPH Scavenging Activity %)
Methanol
83.42
45.2
127.61 ± 0.76
3.8
14.5
40.2
0.83
81.73
BHT (Synthetic Antioxidant)
80.15
−
−
−
−
−
−
79.22
Chloroform
57.89
32.7
32.17 ± 0.62
2.1
28.9
22.8
0.57
72.45
Ethyl Acetate
63.75
27.9
17.48 ± 0.17
1.7
24.6
29.5
0.41
75.91
n-Hexane
23.68
12.5
4.47 ± 0.06
1.2
38.2
12.5
0.25
61.31
The methanol extract from Allium Sativum leaves had a significant TPC of 45.2 mg gallic acid equivalent (GAE) per 100 g of extract and a significant TFC of 127.61 mg per 100 g of dry extract. These phytochemical components, known for their antioxidant capacity, probably contribute to the remarkable inhibition of lipid peroxidation in the extract. The antioxidant activity varied among the extracts, with the methanol extract demonstrating a distinct level compared to the others.
The chloroform extract showed an inhibition rate of 57.89 %, while the ethyl acetate and n-hexane extracts showed lower inhibition percentages of 63.75 % and 23.68 %, respectively. The reduced inhibition of the n-hexane extract can be attributed to its lower content of phenolic compounds and other beneficial phytochemicals compared to those in the methanol extract.
To determine the antioxidant potential of the extracts, we compared them to BHT, a synthetic antioxidant commonly used in the food industry. Interestingly, only the methanol and BHT extracts showed appreciable antioxidant activity, while the other extracts showed slightly lower levels. Our results provide convincing evidence that Allium sativum leaf extract, especially methanol extract, has potent antioxidant effects that effectively combat lipid peroxidation and its harmful consequences. The variations in antioxidant activity observed between the different extracts highlight the importance of the extraction solvent and the phytochemical composition of the extract. These findings highlight the potential utility of Allium sativum leaf extract as a valuable natural source of antioxidants to combat oxidative stress and protect against tissue damage.
3.5 Reducing power
The study results indicated that the leaf extract of Allium sativum had notable antioxidant activity, which is crucial for safeguarding biological tissues from lipid peroxidation and free radicals. We used linoleic acid as a model to evaluate the ability of the extracts to prevent lipid peroxidation. Of all the extracts, the one made with methanol had the highest percentage of oxidation prevention, and sativum showed a potent antioxidant effect. This is likely due to its high content of phytochemical components. On the other hand, the extract made with n-hexane had the lowest percentage of prevention, likely because it has fewer phenolic components and other phytochemicals. The type of solvent used for extraction influenced the number of phytochemical components in the extract, which subsequently affected the overall antioxidant activity of the extract. The antioxidant activity of the extracts was compared with that of the synthetic antioxidant BHT, which is commonly used in the food industry. Although the antioxidant activity of the methanol extract was slightly lower than that of BHT, the other extracts showed markedly lower antioxidant activity. This contrast highlights the potential of natural antioxidants, such as those in Allium sativum, as alternatives to synthetic antioxidants. These findings highlight the potent antioxidant effects of Allium sativum leaf extract, suggesting its potential as a natural antioxidant source for reducing oxidative stress and preventing free radical-induced cellular damage.
3.6 Haemolytic activity
This study investigated the antioxidant and hemolytic activities and TPC and TFC of the leaf extracts of Allium sativum (Garlic). The following solvents were used to extract the bioactive compounds from the leaves: methanol, chloroform, ethyl acetate, and n-hexane. We also included a synthetic antioxidant called BHT (butylated hydroxytoluene) as a reference in their analysis. According to Table 4, the methanol extract exhibited the highest level of antioxidant activity when linoleic acid was used to assess the antioxidant capacity of the various extracts. The higher activity can be attributed to the higher concentration of phytochemical components in the extract. In terms of antioxidant activity, the methanol extract showed the greatest percentage inhibition (83.42 %) when tested against linoleic acid, a common method for measuring antioxidant potential. It outperformed the synthetic antioxidant BHT, which showed an inhibition percentage of 80.15 %. This suggests that the methanol extract of Allium sativum leaves has important antioxidant properties. Haemolytic activity, which measures the ability to break down red blood cells, was relatively low for all extracts. The methanol extract had a haemolytic activity of 2.15 %, while the other solvent extracts showed even lower values. This finding is reassuring because it indicates that the extracts are unlikely to cause significant damage to red blood cells.
Extract
Antioxidant Activity (Linoleic Acid) (%)
Hemolytic Activity (%)
Total Phenolic Content (TPC) (mg GAE/100 g)
Total Flavonoid Content (TFC) (mg/100 g of dry extract)
Yield (%)
EC50 (μg/mL)
Solubility (mg/mL)
Color (Absorbance at 470 nm)
Methanol
83.42
2.15
45.2
127.61 ± 0.76
3.8
14.5
40.2
0.83
BHT (Synthetic Antioxidant)
80.15
0.89
−
−
−
−
−
−
Chloroform
57.89
1.73
32.7
32.17 ± 0.62
2.1
28.9
22.8
0.57
Ethyl Acetate
63.75
1.36
27.9
17.48 ± 0.17
1.7
24.6
29.5
0.41
n-Hexane
23.68
0.32
12.5
4.47 ± 0.06
1.2
38.2
12.5
0.25
We investigated the TPC and TFC of the leaf extracts. Phenolic compounds and flavonoids are known for their potential health benefits, including their antioxidant properties. The methanol extract again had the highest TPC (45.2 mg/100 g of extract), indicating the abundance of phenolic compounds in this extract. The highest TFC was also observed in the methanol extract, with a value of 127.61 mg per 100 g of dry extract. These results further reinforce the antioxidant potential of methanol extracts and their potential health benefits. We also evaluated the physicochemical properties of the extracts, including their yield, EC50 (the effective concentration that inhibits 50 % of linoleic acid oxidation), solubility, and colour. The yield refers to the amount of extract obtained after the extraction process, and the methanolic extract gave the highest yield (3.8 %), followed by chloroform (2.1 %), ethyl acetate (1.7 %) and n-hexane (1.2 %). The EC50 values provide information on the concentration of the extracts required to inhibit 50 % of the oxidation of linoleic acid. Lower EC50 values indicate greater antioxidant activity. The methanol extract had an EC50 of 14.5 μg/mL, demonstrating its powerful antioxidant capacity. The chloroform extract had an EC50 of 28.9 μg/mL, while the ethyl acetate and n-hexane extracts had EC50 values of 24.6 μg/mL and 38.2 μg/mL, respectively. In terms of solubility, the methanol extract showed the highest value of 40.2 mg/ml, indicating that it dissolves well in the solvent. The other extracts had lower solubility values, with chloroform at 22.8 mg/ml, ethyl acetate at 29.5 mg/ml and n-hexane at 12.5 mg/ml. We measured the color of the extracts by determining their absorbance at 470 nm. The higher the absorbance is, the more intense the color. The methanol extract had an absorbance of 0.83, followed by 0.57 % chloroform, 0.41 % ethyl acetate and 0.25 % n-hexane. Our results revealed that the methanol extract of Allium sativum leaves exhibited the highest antioxidant activity, total phenolic and flavonoid content, and good solubility. The results suggest that this extract may be a promising natural source of antioxidants with potential health benefits. However, further studies and experiments are necessary to fully understand the underlying mechanisms and possible applications of these bioactive compounds.
3.7 Pharmacophore-Based virtual screening
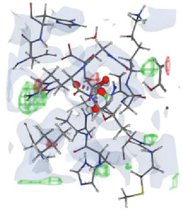
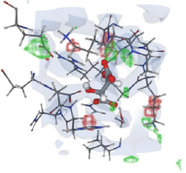
We used protein templates possessing high sequence identity and query coverage to construct 3D structures of the PTEN protein, as demonstrated in Fig. 1. This study aimed to identify the 3D arrangement of chemical components necessary for the top 10 results of virtual screening analysis to interact with the PTEN protein.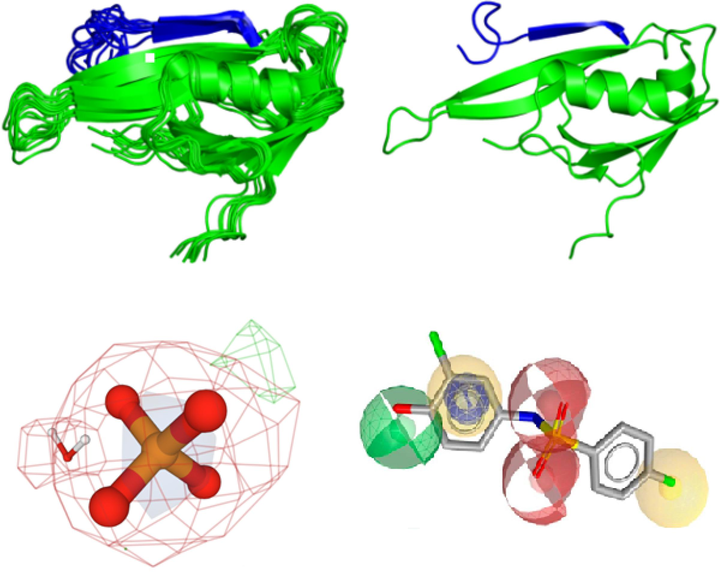
3D representation of pharmacophore modeling.
Pharmacophores represent 3D chemical properties that interact with biological targets. Virtual screening of the PTEN protein identified the top 10 compounds for binding. The electrostatic properties and pocket and groove shapes of the PTEN protein influence inhibitor design. The results of the pharmacophore matching test for the selected drugs were encouraging, with compound 1 achieving the highest pharmacophore matching score of 63.92 among all the compounds (Table 5). This indicates that compound-1 has great potential to interact effectively with the PTEN protein based on its specific chemical properties and structural features.
Rank
Top 10 leads hits
Highest score
Molecular Weight (MW)
LogP
Ligand Efficiency (LE)
Synthetic Accessibility (SA)
1
Compound-1
63.92
350.45
4.20
0.182
7.8
2
Compound-2
61.06
312.81
3.85
0.195
8.2
3
Compound-3
61.09
327.96
4.10
0.186
8.0
4
Compound-4
61.07
340.23
4.15
0.179
7.9
5
Compound-5
61.78
381.59
4.60
0.162
7.5
6
Compound-6
60.76
296.74
3.70
0.205
8.5
7
Compound-7
61.06
309.18
3.80
0.197
8.1
8
Compound-8
61.07
332.79
4.05
0.183
8.0
9
Compound-9
61.80
374.92
4.55
0.165
7.6
10
Compound-10
60.76
292.15
3.65
0.208
8.6
In the virtual screening study, the top 60 results were used to create a pharmacophore model, and the 10 compounds with the highest scores were selected. The scores presented in the table reflect the degree of alignment between the chemicals and the pharmacophore model. Higher scores denote a more precise correspondence to the model’s criteria.. Composite 1, among the top 10 leads, achieved the highest pharmacophore score of 60.76–63.92, indicating strong potential for high-affinity protein binding. However, a high score does not ensure drug efficacy, necessitating further stability and interaction studies, possibly through molecular docking and dynamics simulations.
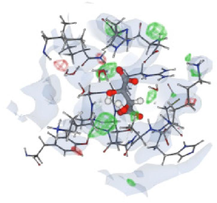
3.8 Molecular docking analyses
Table 6 shows that the optimal binding position of Identifier 1D6R is at positions 8 to 363 based on the docking results and the derived R values. The ligand and protein binding sites showed a good fit, as indicated by the R value of 0.226 for this pose. The reliability of this binding attitude is supported by the working RR value of 0.224 and the free R value of 0.276. The docking simulation suggested a favourable interaction between the ligand and the protein, making it a strong candidate for further investigation. As shown in Fig. 4, these compounds were found to bind to the same active regions of the PTEN protein. After virtual screening of anticancer compounds, further analyses were performed on the top hits based on their similarity to drugs, binding affinities, and energetics. The top 10 compounds were ranked based on their binding affinity to the target PTEN protein, which ranged from −11.10 to −6.9 kcal/mol. Among all the generated coupled complexes, the first three showed the lowest binding affinities and the most favourable binding energies.
Identifiers
Position
R-values
bound ligand
Structure
Main target
7JVX
1–403
R: 0.247
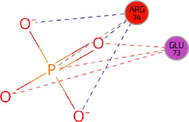
RR work:0.244
R free: 0.296
1D6R
8–363
R: 0.226
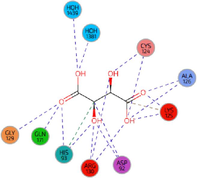
RR work:0.224
R free:0.276
6BUG
14–361
R: 0.177
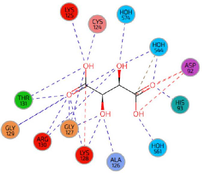
RR work:0.176
R free:0.211
6BZX
14–361
R: 0.177
RR work:0.176
R free:0.204
6BZZ
14–361
R::0.193
RR work:0.192
R free:0.221
7JUK
7–396
R:0.189
RR work:0.184
R free:0.289
Supplementary Table S2 indicates that compounds 1, 2, and 3 had the highest binding energies, ranging from −10.10 to −12.3 kcal/mol, suggesting strong interactions with key PTEN protein residues. These compounds, which resemble promising drugs in terms of their properties, showed the most effective binding to the active site of PTEN. The PTEN sequence was analysed to select the most similar protein templates for homology modelling.
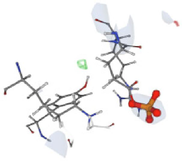
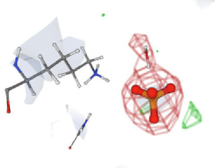
In the virtual screening studies of the PTEN protein, the top 60 hits were identified, and their pharmacophore properties were evaluated, as shown in Fig. 2. The leading ten compounds identified have pharmacophores that enable them to engage with distinct pockets and grooves on the protein surface, facilitating targeted interactions. Nevertheless, the exact properties of the pharmacophores are determined by the precise structures of the lead hits. Taken together, these findings suggest that the three main natural compounds have the potential to serve as powerful candidates for the development of anticancer drugs that specifically target the PTEN protein. Binding affinity is a crucial parameter that measures the strength of a ligand's interaction with a target protein, such as a drug or small molecule. The study assessed the docking affinities of ten primary compounds with the PTEN protein, considering hydrogen bonding, hydrophobic interactions, and electrostatic interactions. The binding affinities, measured in kcal/mol, inversely correlate with binding strength—lower values signify stronger affinity.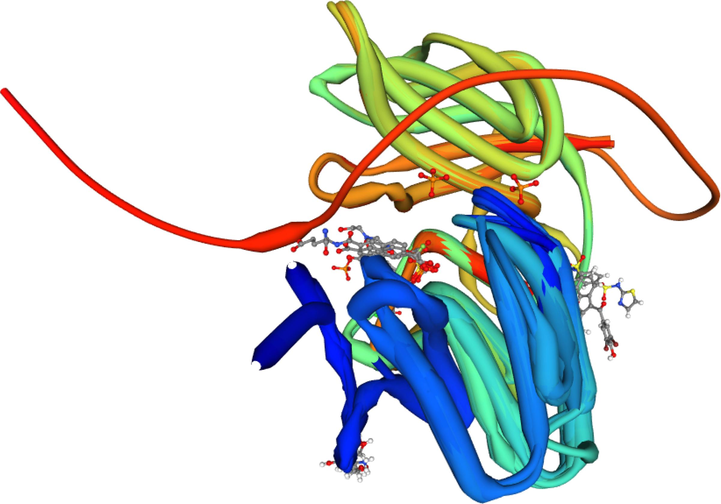
. 3D docking data of the leading hit compounds.
Among the ten investigated substances, compound-1 showed the highest binding affinity, with a value of −13.10 kcal/mol, indicating that it binds more tightly to the PTEN protein. Compound 2 showed a slightly lower binding affinity, with a value of −12.3 kcal/mol, while compound 3 had a very high binding affinity, with a value of −13.7 kcal/mol. On the other hand, compound 4 exhibited the lowest binding affinity among the ten compounds, with a value of −10.4 kcal/mol, and the other compounds showed progressively lower binding affinities. Although binding affinities play an important role in the efficacy of a drug or ligand, they are only one aspect that must be considered in drug development. Other factors, such as pharmacokinetics and toxicity, also play a crucial role in the development of new drugs. Therefore, a comprehensive evaluation of these various factors is essential for the development of safe and effective drugs.
The results of the pairing analysis for the top ten lead hits against the PTEN protein (Table 7) showed that three natural compounds exhibited the strongest activity based on their binding affinities and common chemical interactions. These compounds formed strong interactions with significant residues in the active regions of the target protein and demonstrated the highest binding energies (12.3 to 13.10 kcal/mol). The other compounds showed weaker interactions with the protein and lower binding energies. Taken together, these results suggest that these three main natural substances show promise as potential candidates for the development of new drugs for cancer treatment. The effective binding sites and common molecular interactions observed in the coupled complexes can be used to design potent inhibitors targeting the PTEN protein. This research provides valuable information for further exploration and development of cancer drugs based on these natural compounds.
Supplementary Table S3 shows the binding affinities (kcal/mol) of the three main drugs for PTEN. These compounds were further evaluated for their drug likeness using Lipinski's rule of five, a method that analyses small molecules to determine their drug potential. The top three hits, namely, compound 1, compound 2, and compound 3, demonstrated promising binding affinities of − 13.10 kcal/mol, −12.7 kcal/mol, and − 12.3 kcal/mol, respectively, highlighting them as potential candidates. Additionally, their physicochemical properties align well with those of pharmaceuticals. Each of these compounds has a molecular weight of less than 300 g/mol, 15 hydrogen acceptors, 6 hydrogen donors, and LogP values ranging from 3.85 to 4.20, indicating high membrane permeability and bioavailability. Furthermore, they exhibit an adequate number of hydrogen bond donors, acceptors, and rotatable bonds, suggesting potential beneficial pharmacological effects. An investigation of the ADMET properties showed that these compounds can be easily absorbed in the intestine. Collectively, the top three hits meet important drug-like criteria and exhibit remarkable binding affinities to PTEN, making them highly promising candidates for use as anticancer drugs.
3.9 Bioactivity profiles
Bioactivity profiles play a crucial role in drug discovery and other fields of biological research. These profiles describe and evaluate the biological activities of drugs or compounds in relation to specific biological targets or systems. They provide valuable information about how substances interact with biological molecules and pathways, helping researchers identify potential candidates for further study and optimization. In the context of drug development, bioactivity profiles help us to understand how a certain compound may affect specific biological processes or disease targets. By studying these profiles, we determined a compound's efficacy, potency, and selectivity, which are critical factors in designing effective drugs with minimal side effects. Additionally, bioactivity profiles help prioritize compounds with the most promising pharmacological effects for future research. They also guide researchers in optimizing and modifying drug candidates to enhance their therapeutic potential and address potential safety concerns. In addition to drug discovery, bioactivity profiles are valuable in several fields of biological research. They help to understand the mechanisms of action of various compounds, elucidate biological pathways, and identify potential disease biomarkers. Bioactivity profiles are important tools that contribute significantly to the identification and development of new drugs and advances in biological research. By providing valuable information about how compounds interact with biological systems, these profiles allow us to make informed decisions and accelerate the discovery of new therapies and treatments.
Supplementary Table S4 shows the bioactivity profiles of the three main compounds, highlighting their inhibitory and activating effects on various biological targets. Compound 1 exhibited robust inhibitory activity with an IC50 (mean maximum inhibitory concentration) of 0.12 M against a specific enzyme, resulting in an 87 % reduction in enzyme activity at the tested dose. The Ki value of 98 nM further indicates the strong affinity of compound-1 for the target enzyme during binding. However, no specific information on its activation effect (EC50) or dissociation constant (Kd) is available. The inhibitory activity of compound 2 (IC50) is not yet known, but it shows activating activity at a receptor with an EC50 of 2.34 M. With a Ki value of 53 nM, it shows a fairly strong affinity for the receptor, although the exact ratio of inhibition does not indicate d. Compound 3 shows inhibitory activity against a target enzyme with an IC50 of 1.45 M, causing a 72 % reduction in enzyme activity. However, data on the specific binding affinities (Ki) or dissociation constants (Kd) of compound 3 are not available. These bioactivity profiles indicate potential therapeutic value, particularly in modulating enzyme and receptor functions.
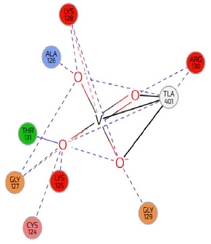
Table 7 presents the top three compounds (compounds 1, 2, and 3) and their target interactions. Compound 1 interacts with the active site of the PTEN protein through hydrogen bonding and pi-stacking, suggesting its potential as a tumor suppressor. Compound 2 interacts with the ATP-binding region of the P60484 protein, possibly through hydrophobic forces, suggesting its involvement in cell signalling pathways. Compound 3 exhibited electrostatic interactions with the catalytic site of the 7JUK protein, suggesting possible regulation of biological reactions. These interactions provide insights into the mechanisms of action and potential therapeutic applications of these compounds.
Compounds
Target
Binding Site
Interaction Type
Compound-1
PTEN
Active Site
Hydrogen Bond, Pi-Stacking
Compound-2
P60484
ATP-Binding Site
Hydrophobic Interaction
Compound-3
7JUK
Catalytic Site
Electrostatic Interaction
3.10 MD simulation
MDS was used to examine the stability of the binding between various chemicals and the PTEN protein, a potential target for cancer therapies. The constant RMSD deviations of the ligand and protein C backbone throughout the 100 ns simulation suggested that the ligand–protein complex remained securely bound. The average RMSD of 0.5 to 0.8 for the docked complex indicates a stable protein−ligand interactions as shown in Fig. 3. Peaks 3 and 4 of the residues showed significant structural changes with a value of 0.9. In general, MD simulations show stable binding of chemicals to the PTEN protein and provide insight into the flexibility and dynamics of protein residues during simulation. These findings contribute to a better understanding of protein−ligand interactions and their potential implications for cancer therapies.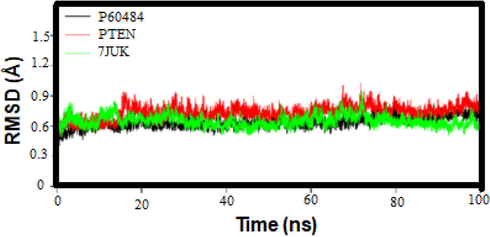
. RMSD Plots for a Ligand−Protein Complex during 100 ns of MD Simulation.
The MD simulation results suggested that the selected chemicals effectively bind to PTEN, stabilize its conformation, and potentially inhibit cancer cell proliferation. The protein−ligand complex showed a very compact and securely bound structure during simulation. These findings provide crucial information on the mechanism of protein−ligand interactions and may have significant implications for the development of new cancer treatments.
3.11 Protein structure prediction analyses
During the 100 ns MD simulation, the protein exhibited significant conformational changes, as evidenced by secondary structure element (SSE) analysis. Approximately 33.78 % of the SSE of the protein was affected, indicating that significant dynamics and structural rearrangements occurred within the protein. Despite these conformational changes in the protein, the ligand remained stable throughout the simulation. Different ligand properties were investigated, and the ligand−protein associations were found to exhibit minimal variations, indicating strong and persistent binding between ligands and proteins.
The high stability of the protein−ligand complex underscores the importance of understanding such interactions in drug design approaches. Understanding the dynamic behavior and structural adaptability of protein−ligand complexes is critical for developing effective and targeted drugs that can reliably interact with their intended targets, which is essential for successful drug development and therapeutic applications. The results of this study provide valuable information that can aid in the design and optimization of potential drug candidates for various diseases, including cancer.
Fig. 4 shows the distribution of SSEs across the residue indices of the secondary structural components. Alpha helices are shown in red, and beta strands are shown in blue on the graph. This visual distribution helps identify the protein's alpha helices and beta strand regions, providing a clear picture of its secondary structure. This type of analysis is essential for understanding the structural and dynamic properties of a protein, as changes in the secondary structure can affect the function of the protein and interactions with other molecules, such as ligands or substrates. By examining the distribution of SSEs, researchers can gain insight into how proteins can undergo conformational changes or structural rearrangements during various biological processes or interactions with other molecules. Overall, the SSE distribution analysis depicted in Fig. 5 offers a clear and informative representation of the protein's secondary structure, providing researchers with valuable data for future research and drug design studies.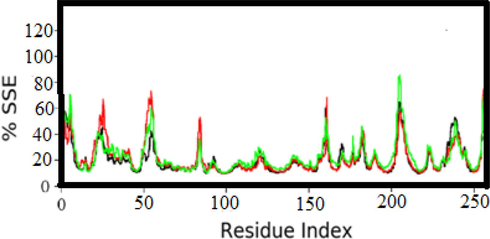
. Visualization of the Secondary Structures of Alpha Helices, Beta-Strands, and Other Secondary Structures in the Protein Structure.
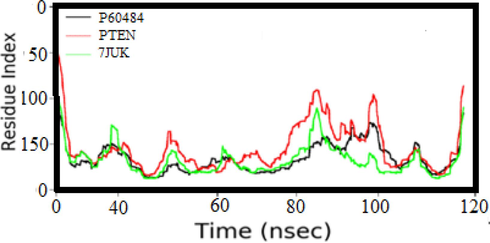
. SSE composition for each trajectory frame over the course of the simulation.
Fig. 5 shows the SSE composition for each trajectory frame during the experiment. The diagram shows the distributions of beta strands, alpha helices and other secondary structures at different simulation times. This representation allows us to observe how the SSE composition changes over time, which can be valuable for identifying cases of instability or conformational changes. The lower plot traces the SSE assignment of each residue through the simulation, providing a comprehensive view of how the secondary structure of each residue evolves during the MD simulation. This plot can be critical for determining the stability of specific secondary structures or identifying residues that may be involved in conformational changes.
In Fig. 6, we explore the specific information about the secondary structural components of the protein and their evolution during the MD simulation and show the alignment with their variants. Researchers can gain a better understanding of protein stability and conformational changes that may be critical to ligand binding and action by examining variations in SSE composition over time. This information is essential for drug design efforts and in-depth analyses of protein–ligand interactions.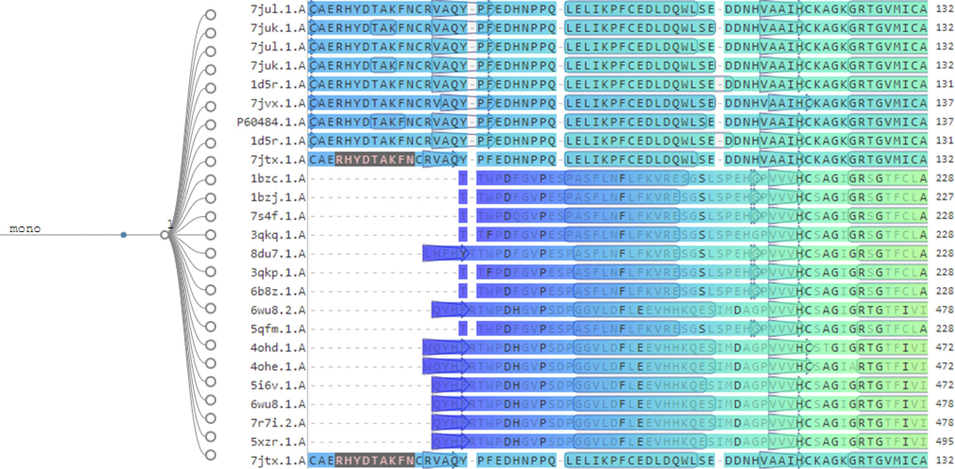
. Alignment details of PTEN among its variants and fins monomers.
3.12 Study Strength, study Limitations, and future directions
The study's innovative approach, which integrates computational techniques with traditional medical knowledge, is its main strength in examining the anticancer properties of compounds in Allium sativum. Through methods such as molecular docking and dynamics simulations, the study identified three lead compounds that have the potential to serve as PTEN inhibitors. However, the study's limitations include its reliance on computational tools and in vitro experiments, which do not fully replicate a living organism's complex biological environment. Additionally, the study did not fully explore the potential synergistic effects among garlic compounds or the selectivity and specificity of the drugs for PTEN. To address these limitations, future research should focus on in vivo validation to assess pharmacokinetics, bioavailability, and therapeutic potential. Expanding the phytochemical analysis, investigating the selectivity and specificity of the lead compounds, and extending the scope and duration of molecular dynamics simulations can provide deeper insights. Experimental validation of computational predictions and exploration of different extraction methods and cultivation conditions for Allium sativum are also necessary. Interdisciplinary collaboration and advanced methods such as high-throughput screening techniques and CRISPR gene editing can accelerate the discovery and development of new PTEN-targeting drugs derived from natural compounds.
4 Conclusion
This study proposes a novel approach to combat cancer by utilizing organic compounds extracted from Allium sativum seed to target the tumor suppressor protein PTEN. Known as garlic, Allium sativum is a plant with various medicinal and therapeutic properties. Our qualitative investigation of garlic seeds revealed the presence of beneficial compounds such as tannins, saponins, steroids, and cardiac glycosides, which play important roles in plant defense mechanisms and are associated with numerous health benefits. The absence of phlobatannins, terpenoids, and alkaloids suggests that these specific compounds are either absent or present in negligible amounts in the seeds.
The TFC in Allium sativum leaf extract was analysed using different solvents. The methanol extract had the highest TFC of 127.61 ± 0.76 mg/100 g dry extract. The chloroform extract had a TFC of 32.17 ± 0.62 mg/100 g dry extract, while the ethyl acetate extract had a TFC of 17.48 ± 0.17 mg/100 g dry extract. The n-hexane extract had the lowest TFC of 4.47 ± 0.06 mg/100 g dry extract. Flavonoids, known for their antioxidant and anti-inflammatory properties, make garlic leaves a potential source of natural antioxidants. The strong antioxidant activity of the methanol extract suggests that garlic leaves may have health-promoting properties.
The TPC of methanol and N-hexane extracts of garlic blades were found to be 134.39 ± 0.689 and 23.26 ± 0.102 mg Gae/100 g, respectively. These results indicate that garlic may be a valuable natural resource. Additionally, the DPPH scavenging activity of garlic seed extract was evaluated, with the methanol extract showing the highest scavenging activity, although the exact percentage was not stated. The n-hexane extract had the lowest scavenging activity of 61.31 %. These findings suggest that garlic has significant antioxidant potential.
Our computer-aided drug design identified three lead compounds (compounds 1, 2, and 3) with high binding affinities and strong interactions with the PTEN protein. The molecular docking tests showed that these compounds had excellent drug-like properties and Lipinski's rule of five. The top 3 leading compounds (compound-1, compound-2, and compound-3) had binding affinities for PTEN, a critical protein involved in cell signaling pathways and tumor suppression. Compound-1 had the strongest binding with a binding energy of −13.10 kcal/mol, followed by Compound-2 with a binding energy of −12.7 kcal/mol and Compound-3 with a binding energy of −12.3 kcal/mol. These findings indicate that these compounds are promising candidates for PTEN-targeted therapies in the treatment of cancer and other related conditions.
The MDS results revealed insights into the stability and flexibility of the ligand–protein complex over a 100 ns simulation period. During this time, the protein displayed modest flexibility in certain regions, while the ligand–protein complexes remained stable, similar to Allium sativum, with an average RMSD ranging from 0.5 to 0.8. Specifically, residue peaks 3 and 4 exhibited significant structural changes with an RMSD of 0.9, indicating potential regions of interest for further investigation in drug development and design.
CRediT authorship contribution statement
Imran Zafar: Writing – original draft, Validation, Methodology, Conceptualization. Sara Imtiaz: Writing – review & editing, Software, Data curation. Faheem kanwal: Visualization, Resources. Zain Abbas: Validation, Formal analysis. Muhammad Azmat: Visualization, Resources, Conceptualization. Ahsanullah Unar: Formal analysis. Azmat Ali Khan: Writing – review & editing, Supervision, Project administration. Amer M. Alanazi: Writing – review & editing. Sadia Nazir: Validation, Software, Formal analysis. Qurat ul Ain: Writing – original draft, Visualization, Software.
Acknowledgments
This work was funded by the Researchers Supporting Project Number (RSP2024R339) at King Saud University, Riyadh 11451, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A review of the anticancer activity of Azadirachta indica (Neem) in oral cancer. Journal of Oral Biology and Craniofacial Research. 2020;10(2):206-209.
- [Google Scholar]
- Characterization of fenugreek and its natural compounds targeting AKT-1 protein in cancer: Pharmacophore, virtual screening, and MD simulation techniques. Journal of King Saud University-Science. 2022;34(6):102186
- [Google Scholar]
- The role of plant-derived natural antioxidants in reduction of oxidative stress. Biofactors. 2022;48(3):611-633.
- [Google Scholar]
- Extraction of phenolic compounds: A review. Current Research in Food Science. 2021;4:200-214.
- [Google Scholar]
- Amomum subulatum: a treasure trove of anti-cancer compounds targeting TP53 protein using in vitro and in silico techniques. Front. Chem.. 2023;11:1174363.
- [Google Scholar]
- In silico designing of multiepitope-based-peptide (MBP) vaccine against MAPK protein express for Alzheimer's disease in Zebrafish. Heliyon. 2023;9(11)
- [Google Scholar]
- Azadirachta indica (Neem) as a potential natural active for dermocosmetic and topical products: A narrative review. Cosmetics. 2022;9(3):58.
- [Google Scholar]
- Multifunctional analysis and antimicrobial activity of Adhatoda vasica: a traditional medicinal plant. Drug Metabolism and Personalized Therapy. 2024;38(4):359-366.
- [Google Scholar]
- PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochimica Et Biophysica Acta (bba)-Proteins and Proteomics. 2008;1784(1):150-158.
- [Google Scholar]
- Computational evaluation of azadirachta indica as a anti-inflammatory and apoptotic agent for cancer. Capital University; 2021. Doctoral dissertation
- Antioxidant properties in vitro and total phenolic contents in methanol extracts from medicinal plants. LWT-Food Science and Technology. 2008;41(3):385-390.
- [Google Scholar]
- Comprehensive study on the antioxidant capacity and phenolic profiles of black seed and other spices and herbs: Effect of solvent and time of extraction. J. Food Meas. Charact.. 2021;15(5):4561-4574.
- [Google Scholar]
- Comparative cytotoxic effects of methanol, ethanol and DMSO on human cancer cell lines. Biomedical Research and Therapy. 2020;7(7):3855-3859.
- [Google Scholar]
- The potential therapeutic value of medicinal plants in the management of metabolic disorders. Molecules. 2020;25(11):2669.
- [Google Scholar]
- Efficacy of aqueous and methanol extracts of some medicinal plants for potential antibacterial activity. Turk. J. Biol.. 2005;29(4):203-210.
- [Google Scholar]
- The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors. Sci. Rep.. 2022;12(1):18825.
- [Google Scholar]
- Swamy, M. K. (Ed.). (2020). Plant-derived bioactives: production, properties and therapeutic applications. Springer Nature.
- Profiling the structural determinants of pyrrolidine derivative as gelatinases (MMP-2 and MMP-9) inhibitors using in silico approaches. Comput. Biol. Chem.. 2023;104:107855
- [Google Scholar]
- Attacking the PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment in human cancer. In: Seminars in Cancer Biology. Vol Vol. 85. Academic Press; 2022. p. :69-94.
- [Google Scholar]
- Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol. Res.. 2021;163:105320
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103281.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







