Identification and mutational analyses of KpsF gene containing PxIxIT motif in Aspergillus fumigatus
⁎Corresponding author at: Department of Dermatology and Venerology, Second Hospital of Shanxi Medical University, No.382 Wuyi Road, Taiyuan 030001, Shanxi, China. shanximayan88@163.com (Yan Ma)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Invasive Aspergillosis (IA) has increased significantly in recent years in which about 90% is caused by Aspergillus fumigatus, and the mortality rate is on the rise. Its treatment has become a common problem clinically. Calcineurin (CaN) plays an important role in the morphogenesis and toxicity of Aspergillus fumigatus, making it an attractive antifungal target. Although the Calcineurin inhibitors FK506 and cyclosporine A have successfully applied in modern transplantation medicine, the systemic toxic and side effects of the two drugs have greatly limited their clinical antifungal applications. Previous studies have shown that CaN of the Aspergillus fumigatus is localized at the top and division of hyphae, and interactions of CaN with cbpA are closely related to the PxIxIT motif. The functioning of CaN is closely related to the PxIxIT motif. Calcineurin substrates or target proteins such as Crz1, SIm1, SIm2, Hph1, and CbpA all contain similar PxIxIT motifs, and similar motifs are present in the Aspergillus fumigatus KpsF gene sequence. We can find KpsF gene in the Aspergillus fumigatus genome through bioinformatics methods, which contains a similar motif in its sequence. Therefore, we can speculate that this motif may interact with Calcineurin, further illustrate the specific mechanism of pathogenesis of CaN in Aspergillus fumigatus, which will help us to have a deeper understanding of the pathogenesis of invasive aspergillosis and provide new ideas for us. By completing the construction of the Aspergillus fumigatus KpsF gene-deficient strain and over-exp strain, we found that the KpsF gene over-exp strain of Aspergillus fumigatus is significantly different from that of the control strain in morphology, and is different from the cbpA in calcium ion regulation. In the low concentration of Ca2+, KpsF may be involved in the negative feedback regulation of Calcineurin, and this negative feedback regulation is inhibited under the condition of high concentration Ca2+.
Keywords
Aspergillus fumigatus
PxIxIT gene
KpsF gene
Calcineurin
Gene mutation
1 Introduction
Aspergillus fumigatus is a saprophytic fungus widely existed in the natural environment. Its conidia are small in volume and widely existed in nature. The human body inhales hundreds of Aspergillus fumigatus conidia every day, which can reach the end of the bronchus and alveoli through the nasal cavity and trachea. In the case of a healthy immune function, non-specific and specific immune responses can effectively remove these spores. However, with the increase in population with immune deficiency or impairment in recent years, the application of immunosuppressive therapy. The most common infections are patients with solid organs, hematopoietic stem cell transplantation and hematologic malignancies (Igbalajobi et al., 2017).
Calcineurin (CaN) is the target protein of immunosuppressive agent cyclosporin A and FK506 commonly used clinically (Schneider et al., 2016), and is a Ca2+/calmodulin (CaM)-dependent serine/threonine protein phosphatase. It consists of two subunits: catalytic subunit CnaA and regulatory subunit CnaB. Previous studies have suggested that the CnaA subunit has a catalytic region at the amino terminal, which interacts with the phosphorylation of the substrate, and contains a helical region connected to CnaB at the carboxy terminal. CnaB contains four EF-calcium linkage motifs, which are interconnected with CnaA. Calcineurin is widely existed in eukaryotic organisms and is relatively conserved evolutionarily (Juvvadi et al., 2011), which recognizes various substrates and controls various conductive pathways of eukaryotic cells in developmental and physiological functions. It was found that the inhibitory effect of RCNAs on Calcineurin requires PxIxIT and LxVP motifs. The PxIxIT motif was first discovered in NFAT and is highly conserved (Nargesi and Rezaie, 2018). It is located at the N-terminal of nuclear transcription factors, and Huiming Li et al. Previous studies have shown that the homologous gene CbpA of RCAN1 in Aspergillus fumigatus is very important for hyphal growth and calcium ion homeostasis.
We find that, through bioinformatics research, the gene KpsF is present in the genome of Aspergillus fumigatus, and the motif of similar sequences is existed in the sequence (PVIAIT). Therefore, we speculate that the gene may be new substrate of Calcineurinor undiscovered substrate. How does it interact with Calcineurin, participate in the physiological and biochemical functions of Aspergillus fumigatus, and whether it has an effect on the virulence of Aspergillus fumigatus? There questions are still waiting for us to study further.
2 Materials and methods
2.1 Strains, media, and culture conditions
The A. fumigatusakuBKU80 and akuBKU80 pyrG− uracil/uridine auxotrophic strains were used for deletion analyses, and the A. fumigatus akuBKU80 strain was used as the wild-type reference strain. Cultures were grown on glucose minimal medium (GMM) supplemented with 5 mM uracil and 5 mM uridine (GMM plus UU) at 37 °C, except where otherwise specified. Escherichia coli DH5α competent cells were used for cloning. The above strains were presented by Professor William J. Steinbach of Duke University.
2.2 Construction of KpsF gene knockout strain of Aspergillus fumigatus and over-exp strain
Aspergillus fumigatus akuBKU80 pyrG-was cultured overnight in a basal medium supplemented with uridine and uracil, and hyphae were collected by filtration; genomic DNA was extracted. The primers were designed to amplify the DNA sequences of the 5′ and 3′ flanking sequences of the KpsF gene respectively, which were about 1.0 kb. First, the 3′ flanking sequence was amplified, cloned with restriction endonuclease EcoRI and SalI and ligated into the plasmid pJW24; next, the 5′ flanking sequence was amplified, and the 5′ flanking sequence was cloned with restriction endonucleases NotI and BamHI and ligated into the plasmid generated in the previous step to construct a knockout plasmid successfully; finally, knockout plasmid was digested with the restriction endonucleases NotI and SalI and then transformed with Aspergillus fumigatus protoplast transformation (Xia et al., 2018). The primers used in the test are shown in Table 1. The probe was designed, and the genomic DNA of Aspergillus fumigatus ΔKpsF transformant was digested with restriction endonuclease KpnI for verification of Southern blot. Construction of over-exp strain: the kpsF gene was cloned with a primer, digested with restriction endonuclease BamHI, ligated into pUCGH plasmid, and the constructed plasmid was directly transformed into Aspergillus fumigatus to obtain over-exp KpsF, which was verified by PCR.
| Primer | Sequence (5′ to 3′) | Fragment size |
|---|---|---|
| KpsF-promo-SalI-F | ACGTGTCGACCAATTTCACGGATAAGCAGG | 1042 bp |
| KpsF-promo-EcoRI-R | AGCTGAATTCGCTGTTCAATGGTCCATTATCG | |
| KpsF-term-BamHI-F | ACGTGGATCCAATGTTTCTCATTCCACTTACCC | 1000 bp |
| KpsF-term-NotI-R | AGTGCGGCCGCCAGTCATCCACTACATTGA | |
| pyrG-R-Screen | GGACATAGCGATAAGTCCAACC | 2300 bp |
| pyrG- KORT | TGGCGACCACACCCGTCCTGTG | |
| KpsF-probe-F | AAATCGACTCCTCAGCGACT | 500 bp |
| KpsF- probe-R | ATCGAAGGAGAAGACTGACG | |
| KpsF- BamHI -F | GACTGGATCCATGGGGCACCCCGAGCTG | |
| KpsF-BamHI-R | CATAGGATCCACCACAGTCACCCCAGA | 1400 bp |
KpsF-KpnI-F 5′GAC GGT ACC ATG GGG CAC CCC GAG CTG3′.
KpsF-BamHI-R:5′CAT AGG ATC CAC CAC AGT CAC CCC AGA 3′ 1400 bp.
KpsF-term-SbfI-F: 5′GCACCT GCA GGA ATG TTT CTC ATT CCA CTT ACC3′.
KpsF-term-HindIII-R: 5′GCA CAA GCT TCA GTC ATC CAC TAC ATT GA3′ 1000 bp.
2.3 Observation of colony diameter and morphological observation under microscope
On solid GMM medium, 1 × 106 conidia/ml of ΔKpsF, akuBKU80 and 10 μL of over-exp KpsF bacterial suspensions were incubated at 37 °C for 5 days the colony diameter was observed at 24 h, 48 h, 72 h, 96 h and 120 h respectively. The test was repeated 3 times. The radial growth of the strain was analyzed by repeated measurement of variance. Microscopy was performed with an Olympus DP71 microscope equipped with a digital camera after incubating the culture medium on Czapek’s medium (0.2% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, 0.001% FeSO4, 3% sucrose, 1.5% agar) for 1 week. The solid medium was cut into a size of about 1 × 1 cm2 and placed in a petri dish. Then, a total of 1 × 104 ∼ 2 × 104 spores were inoculated on the medium; covering by a piece of cover glass, the dish was placed in a wet box and cultured at 37 °C incubator for 48 h. Then, the cover glass was gently removed and dripped with a drop of lactic acid phenol neura staining solution. The arrangements of hyphae, branches, septum, and conidial head were observed, and the shapes of hyphae and spore head were observed and photographed. The specific steps were detailed in document (Ma et al., 2008).
2.4 Scanning electron microscopy observation of the effect of KpsF gene on sporulation of Aspergillus fumigatus
1 × 104 conidia/ml of ΔKpsF, akuBKU80 and over-exp KpsF Aspergillus fumigatus spores were added to 10 ml of GMM liquid medium and covered with sterile cover slips. Incubate it at 37 °C for 48 h, taken out the cover slips, place it in a PIPES buffer containing 3.5% formaldehyde for fixing, rinsing and dehydration, dry it in a dryer, spray it with gold and observe by scanning electron microscopy (scanning electron microscopy was Hitachi SU70 (Chiyoda, Tokyo, Japan). Digital images were obtained with the Axiocam MRm camera (Zeiss, Oberkochen, Germany). Axiovision 4.4 software (Zeiss, Oberkochen, Germany) was used for image acquisition and analysis. Micrographs were taken at 10,000× magnification)
2.5 Gene expression analysis by real-time reverse transcription-PCR (RT-PCR)
The CnaA and VcxA genes were searched from the Aspergillus fumigatus gene pool, and specific primers were designed, and β-tubulin was used as an internal reference (see Table 2 for experimental primers). Add 1 × 106 spores/ml 500 μL of KpsF, akuBKU80 and over-exp KpsF bacterial suspensions to 5 ml of liquid medium containing different concentrations of CaCl2, culture at 200 rpm in 37 °C constant temperature shaker for 18 h. The hyphae were collected by filtration, and stored in a refrigerator at −80 °C for standby use. The total RNA was extracted by the Trizol method, and cDNA was synthesized by reverse transcription (the mRNA required for the 20 μL system was not more than 1000 ng). RT-PCR amplification system: 1 μL of cDNA, 1 μL of positive and negative primers respectively, SYBR® Premix Ex Taq™ II 12.5 μL (TakaRa), and sterilized distilled water supplemented to 25 μL. The amplification conditions were as follows: predenaturation at 95 °C for 30 s, denaturation at 95 °C for 5 s, annealing extension at 56 °C for 30 s, 40 cycles repeated. The expressions of CnaA and VcxA were calculated by the ΔΔCt method, ΔΔCt = (target gene Ct value-internal reference gene Ct value) Experimental Group − (target gene Ct value-internal reference gene Ct value) Control Group. Data processing was performed through SPSS 17.0 software. The results were expressed as
| Primer | Sequence (5′ to 3′) | Fragment size |
|---|---|---|
| β-tubulin-(F) | TTCCCAACAACATCCAGACC | |
| β-tubulin-(R) | CGACGGAACATAGCAGTGAA | 139 |
| CnaA- (F) | GGAGGCAGACAATGATACCG | |
| CnaA-(R) | GGCGGACCTGTGCTTTATT | 109 |
| VcxA-(F) | TTCCGAGCAGACTTTCAACA | |
| VcxA-(R) | CCGTCGTGATTTTGGAGTGG | 131 |
2.6 Construction of KpsF-EGFP strain and observation of fluorescence localization
The KpsF localization observation was constructed by fusing the gene with the enhanced green fluorescent protein (EGFP). First, the KpsF gene was amplified by about 1.4 kb, and then cloned into two sites of the KpnI/BamHI of the plasmid pUCGH (Nabili et al., 2016) containing egfp. Approximately 1.0 kb of the KpsF flanking sequence was amplified and ligated into the plasmid SbfI/HindIII site constructed above, and the final plasmid was linearized by digestion with KpnI, and then transformed into Aspergillus fumigatus akuBKU80. Transformants were screened on medium containing hygromycin B to obtain KpsF-EGFP, which was verified by PCR. KpsF-EGFP localization was observed by laser scanning confocal microscopy. Take (1 × 106 cells/ml) 10 μL of KpsF-EGFP bacterial strain spore suspension constructed successfully, add to 10 ml of liquid GMM medium, mix and pour into the culture dish; Rinse the cover slip with 75% alcohol, and overheat with the alcohol lamp for sterilization, place it the culture dish after cooling slightly, make it sink to the bottom and remove air bubbles; protect from light, culture at 37 °C for 16 h; take out the cover slip, buckle it on the glass slide, and gently press the slide with toilet paper to remove excess the medium, and observe with confocal microscopy, which was as described in previous studies. (Model FV1000 for confocal microscope, Japan Olympus Corporation, software FV10-ASW 2.1) 10um ruler
3 Results
3.1 Bioinformatics analysis of KpsF gene of Aspergillus fumigatus
The KpsF gene in the Aspergillus fumigatus genome was identified by the bioinformatics method, numbered Afu6g08860. It was found from sequence analysis that the KpsF gene contained 1404 bases and encodes 443 amino acids to speculate that the gene may participate in the metabolism of carbohydrates. A homologous sequence comparative analysis of common model fungi using clustalw software revealed that the KpsF gene had similar PxIxIT motif-PVIAIT (see yellow part in Fig. 1). The homology of this gene and Neurospora crassa in Aspergillus fumigatus is 41.43%, and the homology of this gene in Cryptococcus neoformans is 20.18%. The homology of this gene in Candidaalbicans is 28.24%. It can be seen that there is high homology in the filamentous model fungus Neurospora crassa. In view of the importance of the PxIxIT motif, we constructed the Aspergillus fumigatus KpsF knockout strain (ΔKpsF). The schematic diagram of the ΔKpsF construction is shown in Fig. 2a. It can be seen from the figure that the Aspergillus fumigatus KpsF gene was replaced with the selection marker pyrG. Verification results of Southern blot can be seen in Fig. 2b to obtain two correct transformants, one of which was selected for subsequent experiments.
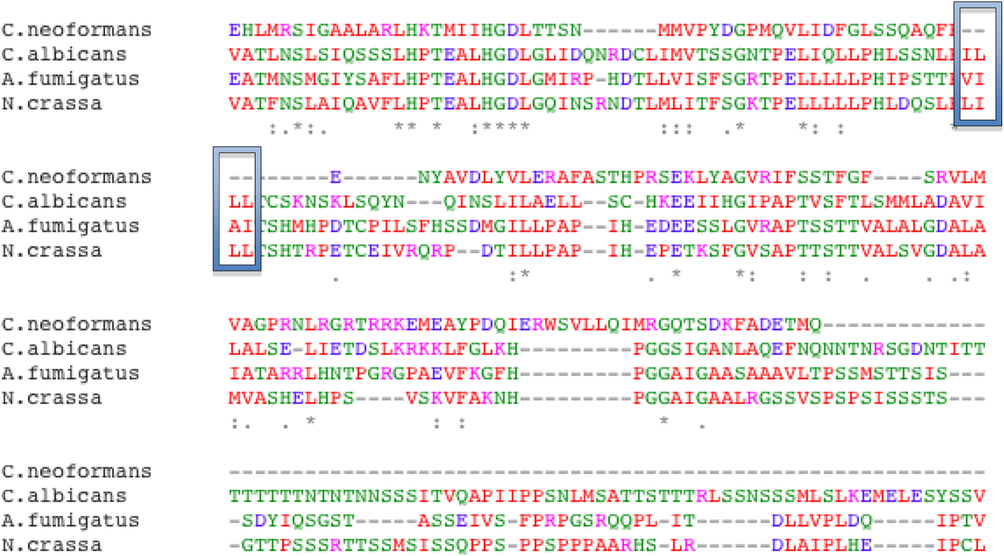
- The homologous sequence alignment results of KpsF gene in common fungi. It presents the homologous sequence alignment results. We performed sequence homology alignment of the kpsF gene in common model fungi. As shown in the figure, the kpsF gene in Aspergillus fumigatus contains PVIAIT motif. It has high homology with the gene sequence in N. crassa and C. albicans (yellow mark in the picture).
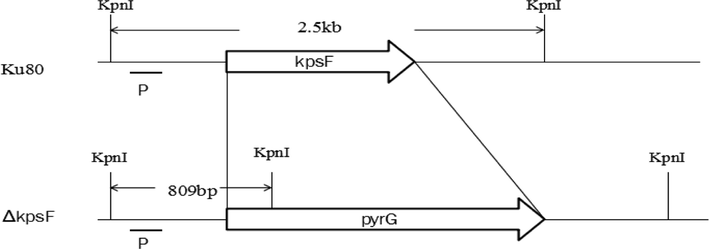
- Sketch map of KpsF gene deletion, KpsF gene was replaced by selection marker pyrG from A. parasiticus. KpnI sites are present in the KpsF promoter gene and the KpsF terminator in wild type. KpnI sites are present in the KpsF promoter gene and the pyrG in the mutant. Conclusion: the number 1 and 4 are true transformations.
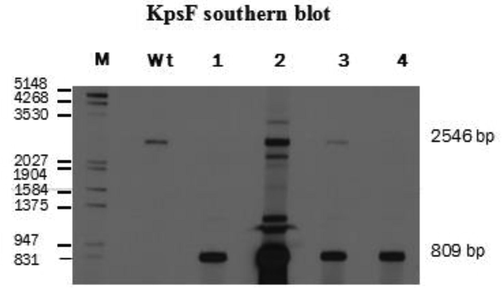
- The verification of ΔKpsF strains by Southern blot, wt as the control strains akuBKU80, M: DNA marker (bp) Genomic DNAs from the WT and four transformants #1, #2, #3 and #4 were digested with KpnI for 15 h and probed with DIG-labeled KpsF probe (500 bp).
3.2 Overexpression of KpsF gene of Aspergillus fumigatus affects the morphology of Aspergillus fumigatus (growth rate and phenotypic observation of KpsF, akuBKU80 and over-exp KpsF strain)
By constructing kpsF knockout strains and over-exp strains, it was found that there was no significant difference in the growth rate of diameter of ΔKpsF and akuBKU80 colonies, and there was no obvious abnormality in colony morphology. Lactic phenol medan staining showed that, under low magnification, ΔKpsF hypha was slender and arranged neatly, and the branches were mostly at an angle of 45°. The high-power microscope showed that the membrane of the hyphae was clear and distinct, the conidial head was short columnar, and the sporophore wall was smooth, and there was no significant difference compared with the control strain akuBKU80. In contrast, radial growth of the over-exp KpsF colony was significantly reduced compared with ΔKpsF and akuBKU80, which decreased approximately 45% compared with akuBKU80, and over-exp KpsF colonies showed deeper wrinkles (Figs. 3a, 3b). There were many branches of hyphae, so that the membrane of over-exp strain is relatively small; the conidial head was relatively short, the top capsule was poorly developed, and the single layer small stem on the top capsule was not obvious. These spores poorly developed were significantly increased compared with KpsF and akuBKU80 (Fig. 3c), indicating that the KpsF gene defect had no effect on the radial growth and morphology of Aspergillus fumigatus strains, while the KpsF gene overexpression made the colony radial growth limited, and the branching angle of the hypha was irregular with much hyphae branching. In order to further verify the phenomena that we have seen, we carried out microscopic observation by scanning electron microscopy and found that the morphology of akuBKU80 spores was relatively full and smooth, and its spines were evenly distributed. Although there were no spores on the individual stems of KpsF, the surface of overall spores was smooth and the spine was evenly distributed, which was not significantly different from the control strain. However, the surface spines on the over-exp KpsF strain increased significantly, and the surface spines of the spores of the gene knockout strain and the control strain were not evenly distributed (Fig. 3d). This was also consistent with our general observation.
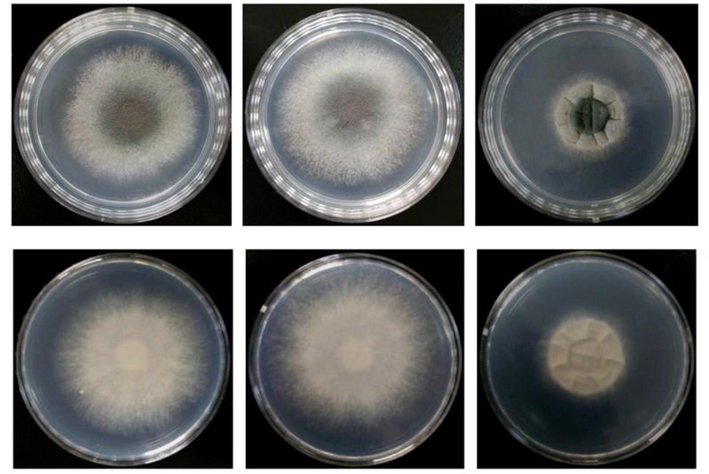
- From the figure through spot-assay, colonial morphology of three strains akuBKU80, ΔKpsF and over-exp KpsF cultured for 72 h can be seen. The diameters of ΔKpsF and akuBKU80 colonies are not differed much, the colonies were villiform, and the middle was smoke green. The surrounding and the back were milky white; for KpsF over-exp strain, the colony diameter was smaller than akuBKU80 and ΔKpsF, the texture was villiform, the center was dark green, and the colonies had deep folds, the surrounding and the back were white, and the cracks were everywhere in radial distribution.
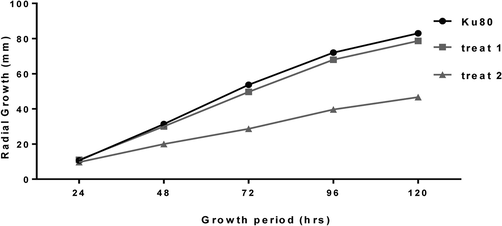
- The radial growth results of ΔKpsF, akuBKU80 and Over-exp KpsF on the GMM medium at 37 °C for 24 h, 48 h, 72 h, 96 h, 120 h.
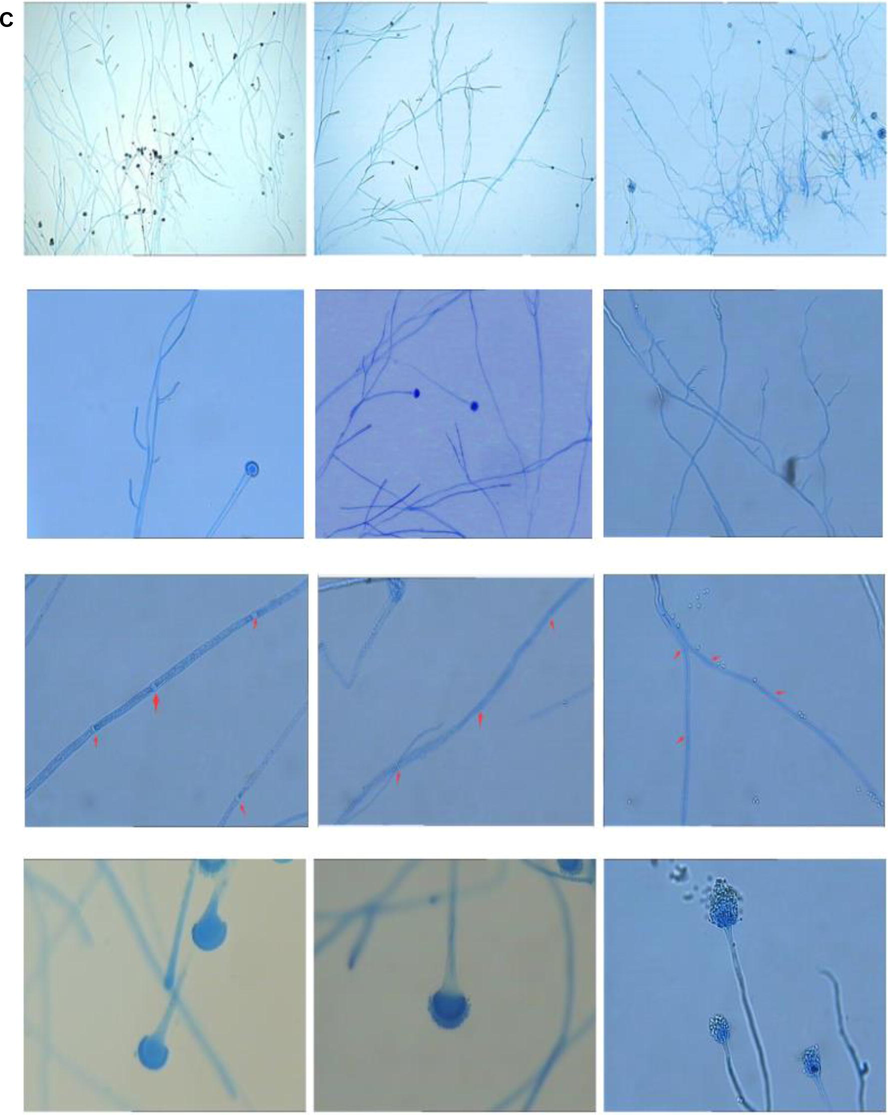
- Under low magnification, mycelial growth of over-exp KpsF was denser than that of KpsF and wild strain Ku80, and the arrangement was disordered; the branches were more and shorter, and the branching angle was relatively larger. There was no significant difference between hyphae of the ΔKpsF and Ku80. The hyphae were slender and arranged neatly, and the branches were mostly 45° (Medan staining, ×200) (see Fig. 3–3 for details). Under high magnification, ΔKpsF and Ku80 were not much different through observation. The separations of the hyphae were clear and distinct, the conidial head was short columnar, and the sporophore wall was smooth. KpsF over-exp strains had relatively few hyphae branches, relatively few separations; conidial heads were relatively short, top capsules were poorly developed, and monolayer small sterigma on the top capsule was not obvious. These spores poorly developed significantly increased compared with KpsF and Ku80 (Medan staining, ×400) (see Fig. 3–3 for details).

- The spores of the control strain were relatively full and smooth, and the spines distributed on the spores were relatively uniform. In the gene-deficient strain, the relative spore growth was not full, and there were no spores on individual phialide, the overall surface of the spores was smooth and the spines were evenly distributed, which was not significantly different from the control strains. The surface of the over-exp strain increased significantly, and the spores of the surface were less evenly distributed compared with the gene knockout strain and the control strain.
3.3 Aspergillus fumigatus KpsF gene has negative regulating effect on calcium stress response gene CnaA
To prove whether the KpsF gene is involved in the transcriptional regulation of calcium ions, we used real-time quantitative PCR to observe the effect of KpsF gene deletion on CnaA and VcxA expression. Studies have shown that the CnaA expression of ΔKpsF in GMM liquid medium was 2.81 times than akuBKU80, and the CnaA expression of ΔKpsF was slightly higher than akuBKU80; at 10 mM CaCl2 concentration, the CnaA gene expression of ΔKpsF was 29.94 times than akuBKU80, which was significantly increased (see Table 3a, Fig. 4a). At 100 mM CaCl2 concentration, there was no significant difference in the relative expression of CnaA between ΔKpsF and akuBKU80. One-way analysis of variance showed that ΔKpsF had a statistically significant difference in CnaA expression relative to akuBKU80 at different Ca2+ concentrations (P < 0.05), and the difference was significant at low concentration of Ca2+ (P < 0.01). This suggested that this gene may be involved in the negative feedback regulation of CnaA. The expression of VcxA gene in over-exp KpsF at 10 mM CaCl2 concentration was significantly higher than that in the control group akuBKU80 and ΔKpsF, the difference was statistically significant (P < 0.01). There was no significant difference in the expression of VcxA gene compared with the KpsF-deficient strain and the control group akuBKU80 (Table 3b, Fig. 4b). There was no significant difference in the relative expression of VcxA of KpsF and akuBKU80 at 100 mM CaCl2 concentration. Overexpression of the KpsF gene increased VcxA gene expression in a low concentration calcium chloride pressure response.
| Strains | Relative expressions (
|
||
|---|---|---|---|
| 0 mM | 10 mM | 100 mM | |
| akuBKU80 | 1.003 ± 0.054 | 1.006 ± 0.027 | 1.035 ± 0.202 |
| ΔKpsF | 2.819 ± 0.269 | 29.94 ± 1.001 | 1.417 ± 0.059 |
| Over-exp KpsF | 1.085 ± 0.169 | 1.571 ± 0.076 | 1.745 ± 0.208 |
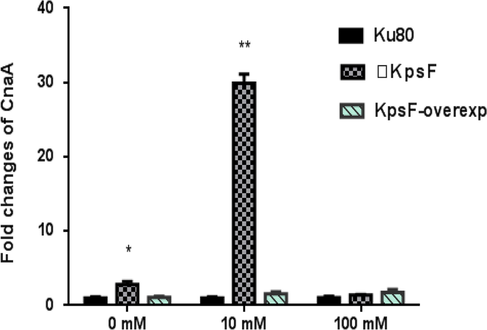
- Relative expression of CnaA genes under treatment conditions of different concentrations CaCl2, ** expresses (P < 0.01). * expresses (P < 0.05), □KpsF in the figure refers to △KpsF, over-exp KpsF refers to over-exp strain.
| Strain types | Relative expression (
|
||
|---|---|---|---|
| 0 mM | 10 mM | 100 mM | |
| AkuBKU80 | 1.058 ± 0.217 | 1.029 ± 0.156 | 1.030 ± 0.142 |
| ΔKpsF | 1.113 ± 0.192 | 1.354 ± 0.017 | 1.54 ± 0.004 |
| KpsF over-exp strain | 1.304 ± 0.052 | 2.726 ± 0.361** | 1.090 ± 0.114 |
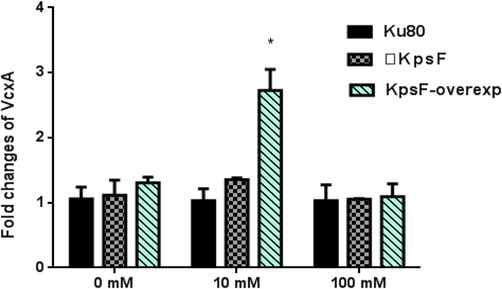
- Relative expression under treatment conditions at different concentration of CaCl2, ** expresses (P < 0.01), * expresses (P < 0.05).
As shown in Table 3a and Fig. 4a: The relative expression of CnaA gene in the ΔKpsF-deficient strain (9.94 ± 1.001) was significantly higher than that in the Ku80 (1.006 ± 0.027) of Control Group and over-exp KpsF (1.571 ± 0.076) at 10 mM CaCl2 concentration. The expression of ΔKpsF was 29.76 times (P < 0.01) higher than that of Ku80 of Control Group, and the KpsF over-exp strain was 1.56 times higher than tKu80 (P < 0.05) of Control Group. And at a concentration of 100 mM CaCl2.
As shown in the experimental results of VcxA gene at 10 mM CaCl2 concentration in Table 3b and Fig. 4b: KpsF over-exp strain (2.726 ± 0.017) was significantly higher than Ku80 (1.029 ± 0.156) in Control Group and ΔKpsF defective strain (1.354 ± 0.017), and the difference had statistical significance (P < 0.01). ΔKpsF defective strain had no obvious change compared with Ku80 in Control Group. KpsF over-exp strain was 2.65 times larger than expression of Ku80 in Control Group.
3.4 KpsF-EGFP localization indicates that it may be located at the membrane under normal growth conditions
We initially observed the KpsF gene localization by constructing a KpsF fluorescent localization strain. It could be found from the study that under normal growth conditions (basal medium GMM culture conditions), the gene was diffusely distributed in the cytoplasm, but there was a clear accumulation in the separation, but not located in the nucleus, hyphae top or cell wall and other special structures. We speculate that it may be related to the separation of Aspergillus fumigatus.
4 Disscussion
Calcineurin (CaN) is a calmodulin-dependent protein phosphatase that regulates various processes in fungi, including morphogenesis, ion homeostasis, toxicity and stress response (Bader et al., 2006). In Cryptococcus, it reduces the antifungal sensitivity of Candida and affects the virulence of Candida (Xu et al., 2019; Seyedmousavi et al., 2015). Calcineurin includes two subunits: CnaA and CnaB, which recognize a variety of substrates and affect the development and physiological functions of eukaryotic cells. It is a target protein of immunosuppressive agent cyclosporine A (CsA) and tacrolimus (FK506) commonly used clinically. However, due to the systemic side effects (such as nephrotoxicity, hypertension, neurotoxicity, diabetes, gastrointestinal disorders, etc.) caused by FK506 and CsA, it is the main obstacle in clinical application (Bruneau et al., 2001), while its use in the treatment of patients with invasive Aspergillus infection is greatly limited. Therefore, the study of Calcineurin-related substrates or target proteins of Aspergillus fumigatus and recognition of endogenous Calcineurin inhibitors is an important strategy for the study of new antifungal drugs (Juvvadi et al., 2011).
Previous studies showed that CaN in Aspergillus fumigatus localized at the apex and separation of hyphae and had a significant effect on the pathogenicity of Aspergillus fumigatus. Studies showed that the role of CaN was closely related to the presence of PxIxIT motif, and similar motifs appeared in the downstream substrates Crz1 (Damasio et al., 2017), Slm1, Slm2 (Leonardelli et al., 2016), Hph1 and other genes. We have found a similar motif (PVIAIT) in the KpsF gene of the genome of Aspergillus fumigatus by bioinformatics method.
We used pyrG as a selection marker to construct a knockout strain of this gene, an over-exp strain and a green fluorescent protein-labeled localization strain, and expected to clarify some characteristics of the gene. With the deletion of Calcineurin, CbpA, CrzA (Cramer et al., 2008) and other genes, the germination of spores of Aspergillus fumigatus would be affected and the growth of hyphae were different. We constructed the green fluorescent protein-labeled KpsF gene and observed its localization in Aspergillus fumigatus under the condition without any stress. Our study showed that the gene was diffusely distributed in the hypha, but there was a clear accumulation of green fluorescence at the separation. We speculate that the gene may be associated with separation of Aspergillus fumigatus. We will further observe its localization changes in response to different pressure conditions.
Regulation of calcium ion balance plays an important role in fungal pathogenesis (Kanhayuwa and Coutts, 2016). Different from previous studies, this study showed that under the condition of low concentration Ca2+, the expression of ΔKpsF was significantly higher than that of akuBKU80, while under the condition of high concentration \Ca2+, there was no difference in the expression of CnaA and the control strain ΔKpsF. The role of VCX1 in Ca2+ tolerance is lower in strains with functional Calcineurin, and higher in the strains in absence of activity of Calcineurinis (Kidd et al., 2015). The results showed that the expression of VcxA gene in KpsF over-exp strain was significantly higher than that in Control Group and ΔKpsF at 10 mM Ca2+ concentration, while ΔKpsF has no significant change compared with Control Group, indicating that under the condition of low concentration Ca2+, overexpression of KpsF gene may stimulate the expression of VcxA gene by promoting the activity of Calcineurin. Although CbpA, CrzA and KpsF have similar PxIxIT motifs, they have distinct mechanisms of interaction with Calcineurin.
In conclusion, this study shown that overexpression of KpsF gene affected the growth rate of Aspergillus fumigatus; fluorescence localization showed that KpsF gene may be related to hyphal separation, and KpsF gene involved in the balance regulation of Ca2+ concentration of Aspergillus fumigatus. As the gene newly discovered, specific functions of KpsF is still in its preliminary exploration. We will construct a ΔKpsFΔCnaA to further explore the relationship between KpsF and CaN, and conduct a more comprehensive study on the KpsF gene.
Acknowledgments
The project was funded by the following fund items. “Project support by the Natural Science Foundation of China (Grant No 81101233)”; “Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (Grant No2018-123)”; “Research Project Supported by Shanxi Scholarship Council of China (Grant No 2016-052)”; “Key Research and Development Projects of Shanxi Province (Grant No201803D3111).”
I would like to express my heartfelt gratitude to Professor William J. Steinbach and Professor Praveen R. Juvvadi of Medical Center of Duke University for their great help in experimental design, methods and related materials for experiments!
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Role of Calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild-type strain. Infect. Immun.. 2006;74(7):4366-4369.
- [Google Scholar]
- Proteome analysis of Aspergillus fumigatus identifies glycosylphosphatidylinositol-anchored proteins associated to the cell wall biosynthesis. Electrophoresis. 2001;22(13):2812-2823.
- [Google Scholar]
- Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot. Cell. 2008;7(7):1085-1097.
- [Google Scholar]
- Xyloglucan breakdown by endo-xyloglucanase family 74 from Aspergillus fumigatus. Appl. Microbiol. Biotechnol.. 2017;101(7):2893-2903.
- [Google Scholar]
- Characterization of the rax1 gene encoding a putative regulator of G protein signaling in Aspergillus fumigatus. Biochem. Biophys. Res. Commun.. 2017;487(2):426-432.
- [Google Scholar]
- Localization and activity of the Calcineurin catalytic and regulatory subunit complex at the septum is essential for hyphal elongation and proper septation in Aspergillus fumigatus. Mol. Microbiol.. 2011;82(5):1235-1259.
- [Google Scholar]
- Short interspersed nuclear element (SINE) sequences in the genome of the human pathogenic fungus Aspergillus fumigatus Af293. PLoS One. 2016;11(10):e0163215
- [Google Scholar]
- Multi-triazole-resistant Aspergillus fumigatus infections in Australia. Mycoses. 2015;58(6):350-355.
- [Google Scholar]
- Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob. Agents Chemother.. 2016;60(9):5420-5426.
- [Google Scholar]
- The sho1 sensor regulates growth, morphology, and oxidant adaptation in Aspergillus fumigatus but is not essential for development of invasive pulmonary aspergillosis. Infect. Immun.. 2008;76(4):1695-1701.
- [Google Scholar]
- High prevalence of clinical and environmental triazole resistant Aspergillus fumigatus in Iran: is it a challenging issue? J. Med. Microbiol.. 2016;65(6):468-475.
- [Google Scholar]
- Investigation an antifungal activity of diclofenac sodium against hyphae formation in Aspergillus fumigatus with attention to the expression of Ef-1 gene. Iran J. Public Health. 2018;47(5):770-772.
- [Google Scholar]
- Aspergillus fumigatus responds to natural killer (NK) cells with upregulation of stress related genes and inhibits the immunoregulatory function of NK cells. Oncotarget. 2016;7(44):71062-71071.
- [Google Scholar]
- Pharmacodynamics of isavuconazole in an Aspergillus fumigatus mouse infection model. Antimicrob. Agents. Chemother.. 2015;59(5):2855-2866.
- [Google Scholar]
- Relationship between methylation status of RASSF2A gene promoter and endometriosis-associated ovarian cancer. J. Biol. Regul. Homeost. Agents.. 2018;32(1):21-28.
- [Google Scholar]
- Age-associated changes of cytochrome P450 and related phase-2 gene/proteins in livers of rats. PeerJ.. 2019;7:e7429
- [Google Scholar]







