Translate this page into:
Heterologous WRKY and NAC transcription factors triggered resistance in Nicotiana benthamiana
⁎Corresponding author. zhongyujuan@gdaas.cn (Yu-Juan Zhong)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
SA (Salicylic acid) and JA (Jasmonic acid)/ET (Ethylene) are the defense and growth hormone regulators involved in alleviating the biotic and abiotic stresses. WRKY and NAC genes are contributors to plant resistance due to their active role in SA and JA/ET defense mechanism.
Methods
WRKY and NAC transcription factors (ptHR293 and ptHR759, respectively) were serially selected (by performing cDNA library functional screening, homology analysis, antioxidant enzymes, ROS burst, callose deposition and qRT-PCR analysis) from Pinellia ternata and transformed into Nicotiana benthamiana. Hybrids were generated to analyze the stability of disease resistance. UPLC-QTOF-MS was performed to study the bioactive compounds.
Results
Study demonstrated that ptHR293 and ptHR759 had potential to trigger ROS burst and callose deposition in N. benthamiana together with the activation of PR-genes and antioxidant enzymes. In transformed N. benthamiana, relative expression of PR-1a (Pathogenesis related-1a) and PDF1.2 (Plant defensin 1.2) was upregulated upto 21 folds and 4 folds for ptHR293 transformed N. benthamiana. While PR-1a and PDF1.2 exhibited 8 folds and 26 folds upregulation for ptHR759 transformed N. benthamiana. ptHR293 + ptHR759-F1 hybrids also exhibited a significant level of PR-gene expression. Significantly high resistance against Botrytis cinerea without influencing the standard seed germination, root and shoot length of transformed N. benthamiana was also observed. A significant induction of bioactive compounds was also observed in ptHR293 transformed N. benthamiana.
Conclusion
Conclusively, heterologous transcription factors, ptHR293 and ptHR759, perform their specific role in the activation of SA and JA/ET mediated defense mechanism.
Keywords
Hypersensitive response
Nicotiana benthamiana
Transcription factor
WRKY
3,4-Dichloromaleimide
1 Introduction
Plants combat with invading pathogens using bilayered innate immunity. The 1st layer is activated upon the recognition of microbe-associated molecular patterns (MAMPs) through pattern recognition receptors (PRRs). This kind of immunity is designated as PAMP/pattern triggered immunity (PTI). Arabidopsis thaliana AtFLS2 (FLAGELLIN- SENSING2) and AtEFR (EF-TU RECEPTOR) are examples for PRRs to spot the bacterial flagellin and elongation factor-Tu (EF-Tu), respectively (Hou et al., 2019). The pathogens adapted to the environment have an acquired number of effector proteins to inhibit the plant immunity including PTI. Plants also have evolved another layer of immunity to recognize these effectors. This recognition initiates the effector-triggered immunity (ETI). Leucine-rich repeat proteins are the examples for ETI receptors which comprises two sub-classes with distinct N-terminal domains (Stotz et al., 2014).
During PTI and ETI, plants activate hypersensitive response (HR), reactive oxygen species (ROS) burst, activation of pathogenesis-related (PR) genes, production of Phytoelaxins and transcriptional reprogramming (Wang et al., 2015). These immune responses collectively activate the local defenses and systemic defenses. Transcriptional reprogramming is directed by TFs (Transcription factors) and its co-regulatory proteins. After receptor stimulation and signal instigation, nominated TFs and their co-regulators in signaling pathways interpret the information and lead to required transcriptional changes (Meraj et al., 2020).
In Arabidopsis, a super family containing more than 70 members encodes unique class of transcription factors possessing WRKY (tryptophan [W], arginine [R], lysine [K], and tyrosine [Y]) zinc-finger motifs (Song et al., 2018). A number of WRKY genes are activated upon pathogen landing or application of defense molecules (Liu and Ekramoddoullah, 2009). Promotors of numerous plant pathogenesis-related genes including PR-genes and NPR1 possess W-box sequences that can be identified by WRKY proteins and work as a basic element for induced expression of these genes (Jiang et al., 2017). NAC (NAM, ATAF, CUC) genes construct the largest plant-specific TFs family present in different species (Nuruzzaman et al., 2013). NAC proteins have potential to regulate plant defense mechanism at infection site (Guo et al., 2017). Studies also suggested that NAC TFs are the contributors of JA/ET defense signaling (Nuruzzaman et al., 2013).
Nicotiana benthamiana genome size is 3 GB and it contains 19 pairs of chromosomes as a result of hybridization between two unknown ancestors. It is used as a model plant in plant–microbe interactions, RNA silencing and genetic engineering research (Baksa et al., 2015). Pinella ternata is an important medicinal Chinese herb and its aqueous extract has shown magical effects against cervical carcinoma. A number of studies revealed different compounds in P. ternata however, information regarding the presence of resistance genes is not sufficient (Li et al., 2013).
Herein, we proposed that genes from one plant species can elevate resistance level of another plant species. A research framework was designed with the purpose of identifying WRKY and NAC transcription factors named as ptHR293 and ptHR759, respectively, from P. ternata, with potential to activate the HR as well as SAR in N. benthamiana.
2 Materials and methods
2.1 Plant and pathogen
P. ternata was grown under controlled conditions at 22–26 °C and 14/10 h of light and dark intervals. N. benthamiana, Lycopersicon esculentum and Gossypium hirsutum were grown under 25–28 °C and 15/9 h of light and dark conditions. Pectobacterium carotovora and Botrytis cinerea (B05.10) were maintained on LB and PDA at 28 °C and 20 °C, respectively.
2.2 Construction of cDNA library
For P. ternata cDNA library construction, tubers of P. ternata were infected with P. carotovora. Leaf samples collection was performed at 24, 36 and 48 h intervals directly in liquid nitrogen and preserved at −80 °C. cDNA library was constructed according to previous method (Wu et al., 2020). Colony PCR was performed with specific TF primers (Table S1). Clones were saved at −80 °C.
2.3 Analysis of cDNA library
Agrobacterium cells containing pTRV1, pTRV2 and pTRV2: target gene were grown on LB (50 mg/L kanamycin and rifampicin) at 28 °C for overnight. Agrobacterium cells were centrifuged for 10 min at 4000 rpm and re-suspended in infilteration solution (10 mmol/L MES, 20 g/L sucrose, 10 mmol/L MgCl2, 100 mmol/L acetosyringone, PH = 5.6) at OD600 = 0.8–1.0. For functional screening, 1 mL needless syringe was used to infilterate N. benthamiana, Lycopersicon esculentum and Gossypium hirsutum leaves with pTRV1 and pTRV2:target gene solutions (1:1 ratio). pTRV empty vector was used as control. HR symptoms were observed daily. Clones constantly showing HR symptoms were sequenced from Wuhan AuGCT (http://wh.augct.com/) with TF (reverse) primer. NCBI blast search was used for homology analysis and phylogenetic tree was constructed using MEGA X (Kumar et al., 2018).
2.4 Microscopy for ROS burst and callose deposition
N. benthamiana leaves injected with ptHR293, ptHR759 and pTRV empty vector were collected at the start of HR development. DAB kit (CWBIO) was used according to manufacturer instructions for ROS staining and amasses were detected under an optical microscope (Nikon Eclipse 55i), while callose deposits were stained according to previous protocol (Gómez-Gómez et al., 1999) and detected under ultraviolet epifluorescence (Nikon eclipse 80i).
2.5 Determination of antioxidant enzymes activity
N. benthamiana leaves injected with ptHR293, ptHR759 and pTRV empty vector were collected at 0, 24, 48, 72, 96, 120, 144 and 168 h intervals and preserved at −80 °C. Peroxidase (POD), superoxide dismutase (SOD) and polyphenol oxidase (PPO) were determined using previous protocols (Beauchamp and Fridovich, 1971; Mayer et al., 1966; Putter, 1974), respectively.
2.6 Determination of transcriptome level of PR-genes
Leaf samples were collected as mentioned above. Total RNA was extracted and cDNA was synthesized with HiFiScript Quick gDNA Removal cDNA kit (CWBIO). qRT-PCR was performed with SYBR® Premix Ex TaqTMII (TliRNaseH Plus) (TaKaRa Clontech). PR-genes specific primers are given in Table S1 (Muhammad et al., 2018). EF-1α served as an indigenous control. 2−ΔΔCT method (Livak and Schmittgen, 2001) was used for quantification.
2.7 Generation of ptHR293 and ptHR759 transformed N. benthamiana
Transgenic plants were obtained using previous method (Horsch et al., 1985). Positive seeds were screened on MS media (3 mg/L bialaphos), confirmed with PCR and sequence analysis. Hybridization was performed (Brito et al., 2015) to generate F1 hybrids.
2.8 Resistance assay
Leaves from 3 to 4 weeks old transformed and hybrid generation were used. Non-transformed leaves were used as a control. B. cinerea was inoculated according to previous method (Viaud et al., 2006) and data were recorded at 48 hpi. Following formula determined percent inhibition
2.9 Penalty-assay for ptHR293 and ptHR759 transformed N. Benthamiana
For the determination of ptHR293 and ptHR759 effect on transformed N. benthamiana, assay was conducted according to previous method (Muhammad et al., 2018).
2.10 UPLC-QTOF-MS/MS analysis and amino acid interaction
For the detection of induced bioactive compounds, samples were processed according to previous protocol (Hu et al., 2017). Non-transformed N. benthamiana was considered for comparison. Ribitol (50 µg/mL) was used as an internal standard and MZmine2 was used for raw data processing.
OmicsNet was used to process the genes and concomitant metabolites data. The reactant pairs module was discotinued and reaction classes were kept available (Wang et al., 2013). The number of active compounds was enlisted to draw structure transformation maps to show relationships between bioactive cmpounds and amino acids. Data were also compared with organism specific genome-based expansion data. Overall outcome was in the form of correlation of amino acids with target bioactive compounds in which each amino acid also carries representative gene numbers. Universally colored lines connecting circles (amino acids and compounds) were linked to BlastKOALA identifiers (Kanehisa et al., 2016) in pathway maps.
2.11 Statistical analysis
For significance analysis between control and treatment, student t-test was conducted by SPSS 25 at p ≤ 0.05 and p ≤ 0.01.
3 Results
3.1 Induced hypersensitive response in N. benthamiana leaves
cDNA was ligated into expression vector and transformed into A. tumefaciens. Selected clones (1268) were saved at −80 °C for further investigation. Randomly 100 clones were selected for quality analysis, which revealed about 95% clones ranged from 100 to 1000 bp. Leaves of N. benthamiana, G. hirsutum and L. esculentum were infiltrated with each clone and typical HR reaction was observed around the infiltration site at 48 h of post infiltration (hpi), while no symptoms were observed for empty vector (Fig. 1A, B; Fig. S1A–D). Induction of typical HR reaction by 49/1268 clones, indicated that about 3.86% of P. ternata genes could be potential candidates for future studies. Further, P. ternata leaves were also infiltrated with 49 cDNA clones but no such HR reaction was observed (Fig. S1E and F).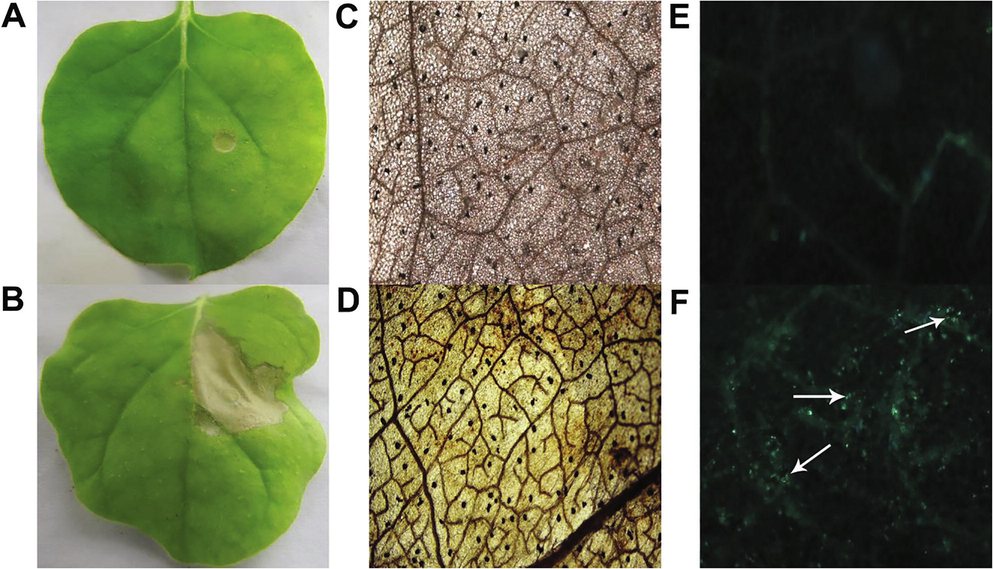
HR symptoms, ROS accumulation and callose deposits. (A–B) HR symptoms on leaves infiltrated by (A) control and (B) ptHR293. (C–D) ROS accumulates in leaves infiltrated by (C) control and (D) ptHR293. (E–F) Callose deposition in leaves infiltrated by (E) control and (F) ptHR293.
3.2 Sequence analysis of P. ternata cDNA clones
Homology analysis indicated that 25 clones shared homology with reported proteins, while 5 clones were the homolog of transcription factors and remaining 19 were not found to share homology. After homology analysis, sequences were named as ptHR (P. ternata hypersensitive response) genes. According to phylogenetic tree analysis, ptHR genes were divided into 3 clades (Fig. S2). Following initial screening and homology analysis (Table S2), ptHR293 (MN458409) and ptHR759 (MN458410) were selected for further experiments.
3.3 Activated ROS burst and callose deposition
Oxidative burst and callose deposition play a vital role in plant defense mechanism to avoid the spreading of pathogen. Infiltration of ptHR293 and ptHR759 in N. benthamiana leaves induced these two early mechanisms as revealed by microscopic images for ROS and callose (Fig. 1C–F).
3.4 Enhanced antioxidant enzymes activity in N. benthamiana
POD, PPO and SOD were measured from 0 to 168 hpi. The maximum activities of POD, PPO and SOD were recorded 72 hpi after ptHR293 and ptHR759 infiltration as compared to control, which started to decline with the progressive time period (Fig. 2A (a), B (a) and C (a)). These antioxidant enzymes were also considered in ptHR293 and ptHR759 transformed N. benthamiana. Significantly higher activity of these enzymes was recorded in transgenic N. bethamiana compared with non-transgenic N. bnethamiana (Fig. 2A (b), B (b) and C (b)).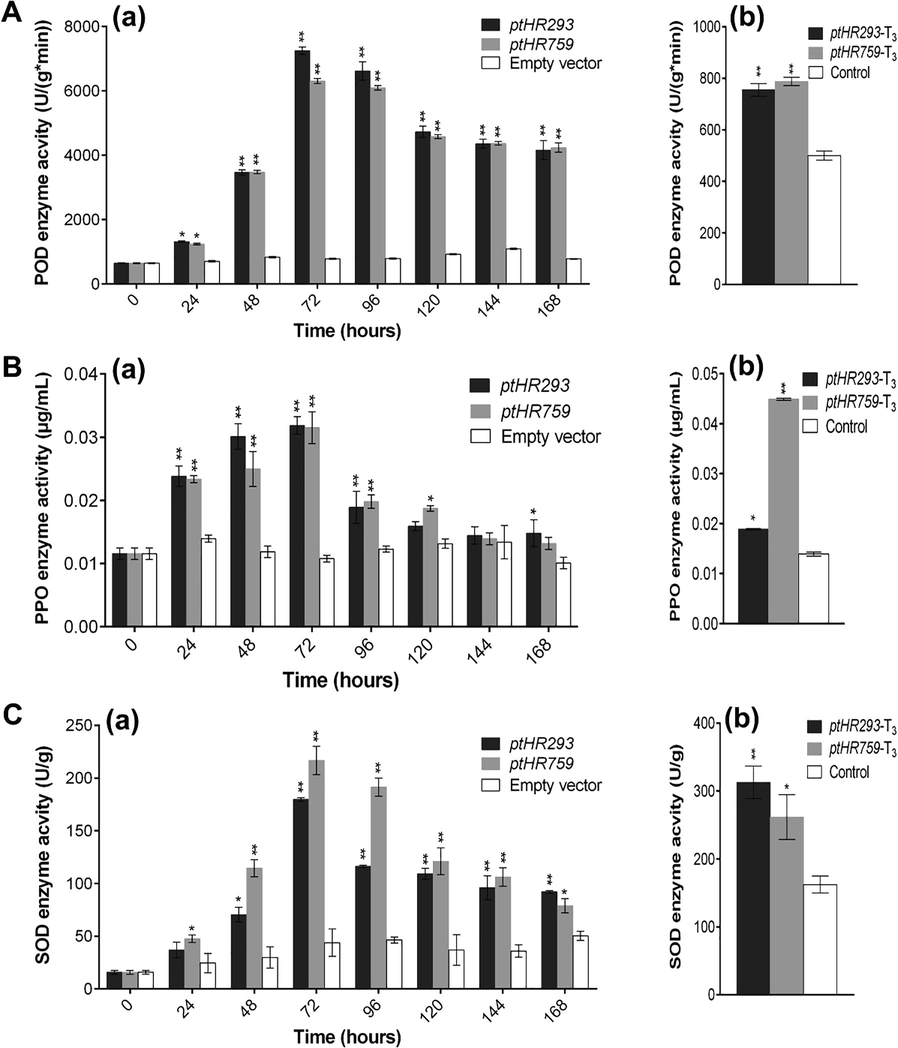
Kinetics of POD, PPO and SOD activities. (A) Kinetics of POD activity against (a) ptHR293 and ptHR759, and (b) in transformed N. benthamiana compared with non-transformed control, (B) Kinetics of PPO activity against (a) ptHR293 and ptHR759, and (b) in transformed N. benthamiana compared with non-transformed control, (C) Kinetics of SOD activity against (a) ptHR293 and ptHR759, and (b) in transformed N. benthamiana compared with non-transformed control. Data represent mean ± SD.
3.5 Activated PR-genes expression in N. benthamiana
ptHR293, ptHR759 and an empty vector as a control were infiltrated in N. benthamiana leaves to quantify the expression of PR-genes in SA and JA/ET mediated defense mechanism at different time intervals (Fig. 3; Fig. S3). Results revealed that PR-5 expression was maximum at 24 hpi, while PR-1a, PDF1.2, RBOHB and ERF1 were showing maximum expression at 48 hpi. The relative expression level of PAL was maximum at 72 hpi and then started to decline down. While, in case of NPR1, a continuous increase in expression was observed until 120 hpi. These results indicated that ptHR293 and ptHR759 have potential to enhance resistance in N. benthamiana.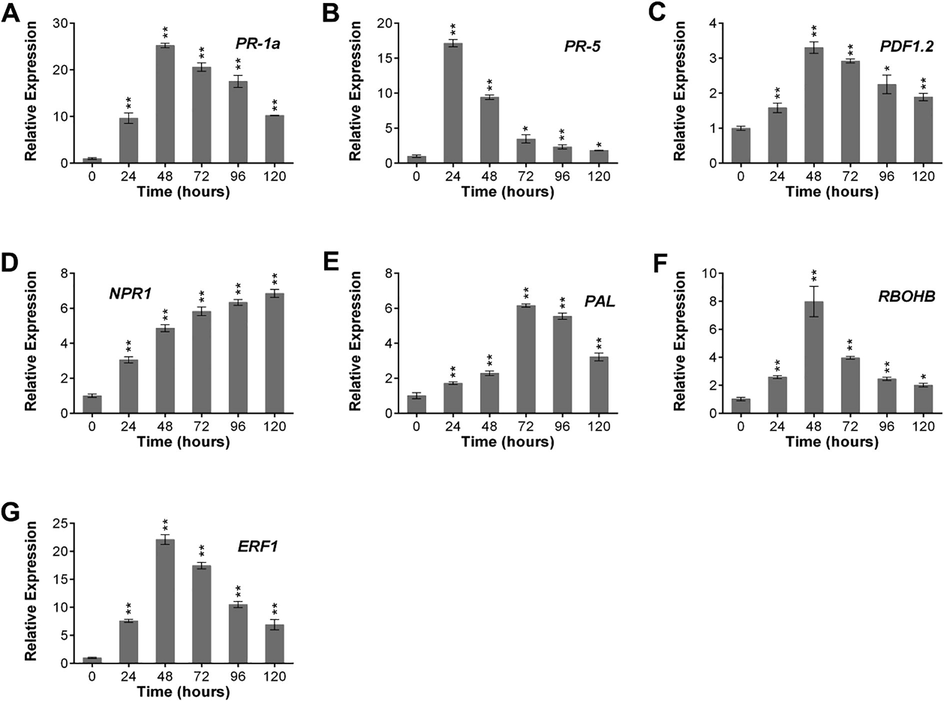
Transcription level of PR-genes against ptHR293. Relative expression of (A) PR-1a, (B) PR-5, (C) PDF1.2, (D) NPR1, (E) PAL, (F) RBOHB and (G) ERF1 compared with empty vector. Data represent mean ± SD.
3.6 Enhanced PR-genes expression in ptHR293 and ptHR759 transformed plants
ptHR293 and ptHR759 transformed plants were attained using Agrobacterium-mediated transformation technique. Seeds from transformed N. benthamiana were collected to grow the next generation for the confirmation of transformation stability. From the analysis of the transcriptome level of PR-genes, it was found that PDF1.2, RBOHB, ERF1, NPR1, PR-1a and PAL were showing the high relative expression level in ptHR293 transformed N. benthamiana (Fig. 4A), while in case of ptHR759 transformed N. benthamiana, NPR1, ERF1, PR-1a, PAL and PDF1.2 were expressing high relative to non-transformed control (Fig. 4B). Analysis of ptHR293 + ptHR759-F1 hybrid revealed the high relative expression of RBOHB, PR-5, PDF1.2, PR-1a, PAL, NPR1, and ERF1 (Fig. 4C). Relative expression level of ptHR293 and ptHR759 was down regulated in T3 generation, while significantly upregulated in F1 generation (Fig. 4D). From results, it was speculated that ptHR293 and ptHR759 are the contributors of SA and JA/ET signaling pathways.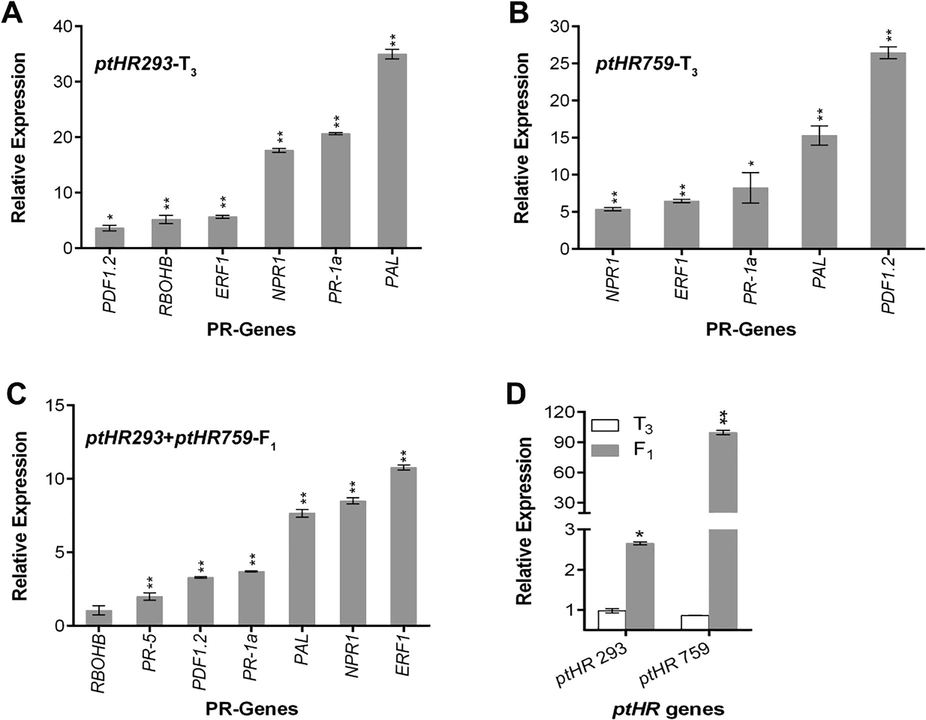
Influence of ptHR293 and ptHR759 on transcriptome level of PR-genes. (A) Relative expression of PR-genes in ptHR293 transformed plants as compared to control, (B) Relative expression of PR-genes in ptHR759 transformed plants as compared to control, (C) Relative expression of PR-genes in ptHR293 + ptHR759-F1 plants as compared to control, (D) Relative expression of ptHR293 and ptHR759 in transformed T3 and F1 N. benthamiana compared with respective control. Data represent mean ± SD.
3.7 Enhanced resistance response of transformed N. benthamiana
The uniformed size leaves were detached from T3 transformed, F1 hybrid and non-transformed N. benthamiana and inoculated with B. cinerea. Percent inhibition was determined and found that ptHR293 and ptHR759 transformed N. benthamiana, and ptHR293 + ptHR759-F1 hybrid confers significant resistance against B. cinerea compared to control (Fig. 5A–E).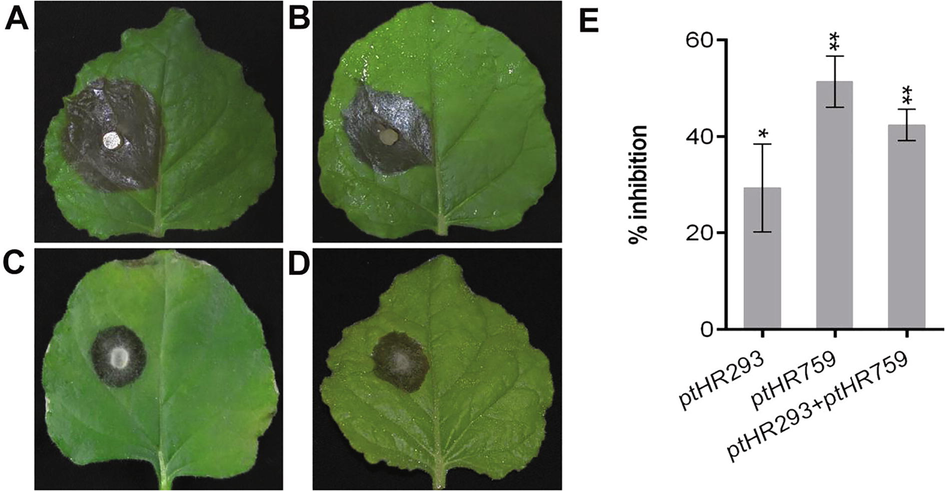
Resistance analysis against B. cinerea. (A) Non-transformed control leaves (lesion diameter 24.3 ± 0.130 mm), (B) ptHR293 transformed leaves, (C) ptHR759 transformed leaves, (D) ptHR293 + ptHR759-F1 leaves, and (E) percent inhibition against B. cinerea as compare to control.
3.8 ptHR293 and ptHR759 transformed N. benthamiana exhibited normal morphology
To determine the effect of ptHR293 and ptHR759 on transformed plants, seed germination, root length and shoot length were observed. Results showed that growth of transformed plants was as normal as non-transformed. There was no significant difference for seed germination, and root and shoot length, between transformed and non-transformed plants (Fig. 6A and B). The findings revealed that ptHR293 and ptHR759 enhanced the plant basic resistance without influencing its normal growth parameters.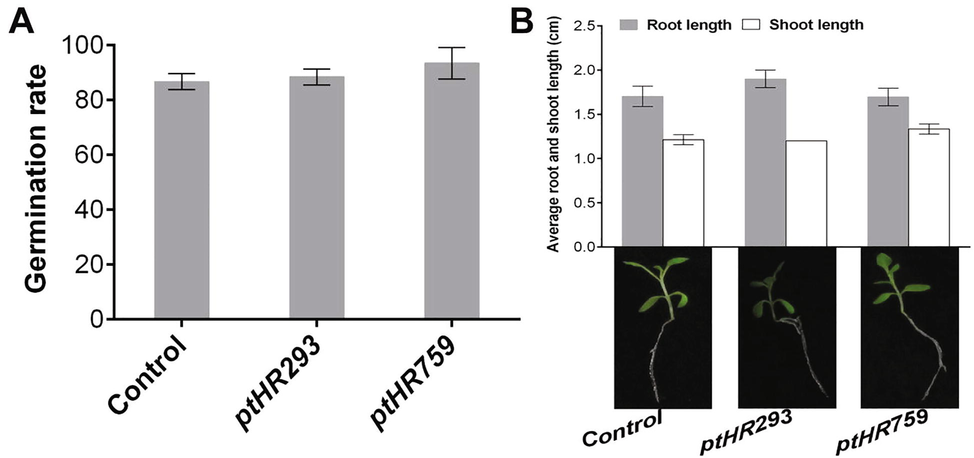
Penalty analysis of ptHR293 and ptHR759 transformed N. benthamiana. (A) Germination rate of transformed seeds compared with non-transformed. (B) Average root and shoot lengths of transformed plants in comparison with non-transformed. Data represent mean ± SD.
3.9 Detected bioactive compounds in transformed N. benthamiana and their interaction
In order to observe the perturbations in plant metabolomics inside transformed N. benthamiana, UPLC-QTOF-MS was conducted and compounds were identified on the basis of mass spectra using NIST compound library. Results indicated the production of several compounds elicited by ptHR293 transformation. These compounds were not found in non-transformed control (Table 1; Fig. S4; Table S3). Among these compounds, 3,4-Dichloromaleimide was considered as biomarker for ptHR293 because of its consistent presence and detection.
Compounds
Content (µg ribitol equivalent/g of dry weight)
Functions
References
Ethoxzolamide
56.14 ± 4.34
1) Carbonic anhydrase inhibitor
2) M. tuberculosis PhoPR regulon inhibitorYasin et al. (2018)
Pyrrolidine
16.85 ± 1.30
1) Antioxidative, antifungal and antibacterial.
Ahmad et al. (2020)
Lignocerane
112.82 ± 8.73
1) Anti-parasitic and antibacterial.
2) Cytotoxic against AGS, MDA-MB-231, HT-29 and NIH 3 T3 cells.Hafeez et al. (2019)
3,4-Dichloromaleimide
2650.60 ± 213.17
1) Chitin synthase inhibitor.
Ahmed et al. (2017)
Sulfanilamide
12.91 ± 0.99
1) β-class carbonic anhydrase inhibitor.
Maresca et al. (2014)
2-Undecanone
10.40 ± 0.79
1) Antibacterial, antifungal and insecticidal.
Akram et al. (2014)
ptHR293 showed positive interaction with different amino acids viz; L-Serine, Cholin, Glycine, Glycerol and several other amino acids (Fig. 7A), while in case of ptHR759, negative interaction was observed with these amino acids (Fig. 7B). From results, we can conclude that amino acids showing positive interaction with ptHR293 were responsible for the generation of bioactive compounds (Table 1) detected in ptHR293 transformed plants. Negative interaction of ptHR759 with amino acids can be considered as main reason for the generation of no bioactive compounds in ptHR759 transformed plants.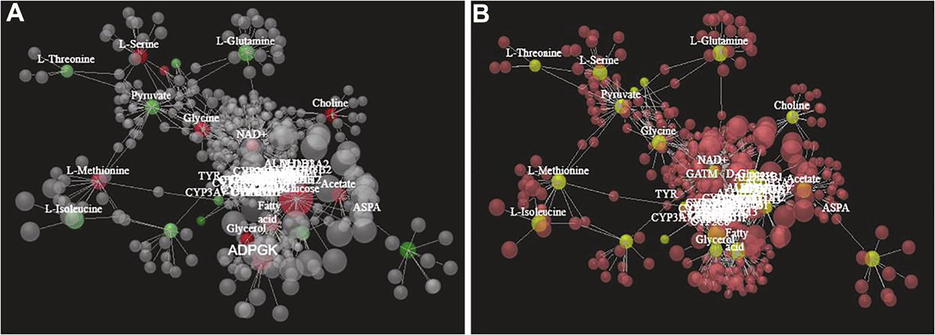
Interaction of ptHR genes with amino acids. Red circle represent positive interaction and green circle represent negative interaction. Tagged red and green circles are surrounded with already reported genes. (A) Interaction of ptHR293 with amino acids and, (B) Interaction of ptHR759 with amino acids.
4 Discussion
We attempted to improve the basal resistance level of N. benthamiana using ptHR293 and ptHR759 genes from P. ternata. To achieve the objectives, a cDNA library was created and 49 different unique sequences were found with potential to activate the hypersensitive response. Out of these 49 unique sequences, ptHR293 and ptHR759, showed homology with WRKY and NAC transcription factors, respectively, were selected for further study.
P. ternata is a famous Chinese traditional medicinal herb which is still being used to cure a number of diseases. Several studies have reported this herb for the identification of its unique compounds (Zhu et al., 2013). According to the best of our knowledge, there is no single study performed to unveil its potential against the inhibition of plant pathogens. Functional screening of P. ternata cDNA library has revealed that certain P. ternata genes have potential to enhance the resistance level of N. benthamiana.
On the plant cell wall, at the site of interaction with a pathogen, papillae are formed and considered to be the initial defense signal at cellular level. Previously callose has been revealed as main component of papillae (Voigt, 2014). ROS are reported to be involved in plant defense response and developmental mechanisms. Oxidative burst as an early event, generates localized ROS to cause cell death for the inhibition of pathogen spread (Keshavarz-Tohid et al., 2016). We confirmed from microscopic images that ptHR293 and ptHR759 have induced the ROS burst and callose deposition in N. benthamiana leaves.
Defense related antioxidant enzymes are the basic proteinaceous compounds produced in plants to remediate the damages induced by biotic and abiotic stresses. Plants with certain level of resistance must have some activity of these enzymes. POD reported to be involved in polymerization of lignins from monolignols and convert plant tissue into the hard structure against pathogen (Yang et al., 2017). For the oxidation of phenolic compounds into toxic quinones, PPO plays an important role and contribute resistance towards pathogen. Lu et al. (2015) reported SOD as an important antioxidant enzyme to protect plants from oxidative damages. In our study, a significant increase in these enzymes was observed to support the vigor use of ptHR293 and ptHR759 in plant resistance mechanism.
Different PR-proteins are associated with different biochemical pathways, such as PR-1a and PR-5 represent the SA pathway, PDF1.2 represents the JA/ET pathway and ERF1 as ET pathway marker gene (Leonetti et al., 2017). NPR1 has been reported to be involved in the activation of PR-genes to trigger the SAR (Wang et al., 2015). PAL is reported for the synthesis of antimicrobial compounds (phytoalexins, lignins and other phenolic compounds) in phenylpropanoid pathway (Wang et al., 2015). RBOHB stimulates the ROS burst, especially H2O2 to combat with biotic and abiotic stresses (Deng et al., 2016). In present investigation, an interesting phenomenon was observed when 2 different ptHR genes were expressed together in N. benthamiana, they can induce PR-5 (SA marker).
Many biochemical interactions among pathogen enzymes and host inhibitors occur in apoplastic space. Previous studies reported that localized infection and NPR1 gene activates the defense signals in whole plant (Wang et al., 2015). For resistance evaluation, a necrotrophic fungal pathogen B. cinerea (Viaud et al., 2006) was used, and a significant level of resistance was observed. Findings revealed the potential of ptHR293 and ptHR759 to be used for the development of future resistant cultivars of economically important crops.
Secondary metabolites from ptHR293 transformed N. benthamiana are reported for their biological activities as Ethoxzolamide is an essential bioactive compound with antibacterial and carbonic anhydrase inhibitor properties (Yasin et al., 2018). Pyrrolidine, accompanied by pyridine, synthesizes nicotine and plays role in plant defense. It contains DNA binding affinity and cytotoxicity as its action mechanism (Ahmad et al., 2020). Lignocerane has been reported in Chrysanthemum and Stevia species as a part of plant defenses against herbivory (Hafeez et al., 2019). 3,4-Dichloromaleimide is a cyclic imide and well-known for its pain-relieving pharmacological activities (Ahmed et al., 2017). Sulfanilamide reduced the activity of Gly decarboxylase complex and serine hydroxymethyltransferase. Structural analogs of this compound also enhance the mitochondrial efficacy under stress (Maresca et al., 2014). 2-Undecanone determines the ultimate defensive properties of plants (Akram et al., 2014). Induction of these compounds in ptHR293 transformed plants can avoid the agricultural losses without affecting the normal growth.
Transcriptomic data showed a clear difference in the expression pattern of WRKY genes. Gene-specific motif networks identified the WRKY gene at the center and positively interacting with NAC domain making the plant tolerant against abiotic stresses. NAC transcription factors had mono, bi, and multipartite nuclear localization signals and encode multiple protein chimera (Bashir et al., 2016). In this way, NAC alone could regulate the interactome network of protein–protein interaction. On the other side, WRKY was associated with W-box promoter elements, thus imperative to understand WRKY transcription (Chi et al., 2013). WRKY domain, along with NAC domain and their conserved motifs modulates the physiological processes.
5 Conclusion
Transcription factors, WRKY and NAC, from P. ternata have potential to enhance plant resistance. Here, it is also proved that when heterologous transcription factors were transformed into N. bentamiana, its basal resistance was significantly improved up to several folds. This idea still needs further investigations, and also can be helpful to open new directions of plant–microbe interaction research.
Funding information
Work supported by the National Major Project for Transgenic Organism Breeding (2016ZX08003-001), Research and Development Plan Project in key areas of Guangdong Province (2018B020202007) and Guangdong Academy of Agricultural Sciences Foundation of the Dean Project (BZ201905).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Metabolic and proteomic perspectives of augmentation of nutritional contents and plant defense in Vigna unguiculata. Biomolecules. 2020;10:224.
- [CrossRef] [Google Scholar]
- Characterization of anti-bacterial compounds from the seed coat of chinese windmill palm tree (Trachycarpus fortunei) Front. Microbiol.. 2017;8
- [CrossRef] [Google Scholar]
- Basal susceptibility of tomato varieties against different isolates of fusarium oxysporum f. sp. lycopersici. Int. J. Agric. Biol.. 2014;16
- [Google Scholar]
- Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genomics. 2015;16:1025.
- [CrossRef] [Google Scholar]
- Tomato plant proteins actively responding to fungal applications and their role in cell physiology. Front. Physiol.. 2016;7
- [CrossRef] [Google Scholar]
- Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem.. 1971;44:276-287.
- [CrossRef] [Google Scholar]
- Pollination triggers female gametophyte development in immature Nicotiana tabacum flowers. Front. Plant Sci.. 2015;6:561.
- [CrossRef] [Google Scholar]
- Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant. 2013;6:287-300.
- [CrossRef] [Google Scholar]
- Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana. Sci. Rep.. 2016;6:20579.
- [CrossRef] [Google Scholar]
- A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J.. 1999;18:277-284.
- [CrossRef] [Google Scholar]
- An NAM domain gene, GhNAC79, improves resistance to drought stress in upland cotton. Front. Plant Sci.. 2017;8
- [CrossRef] [Google Scholar]
- Hafeez, M., Liu, S., Jan, S., Ali, B., Shahid, M., Fernández-Grandon, G.M., Nawaz, M., Ahmad, A., Wang, M., 2019. Gossypol-induced fitness gain and increased resistance to deltamethrin in beet armyworm, Spodoptera exigua (Hübner) [WWW Document]. Pest Manag. Sci. https://doi.org/10.1002/ps.5165
- A simple and general method for transferring genes into plants. Science (80-.). 1985;227:1229-1231.
- [CrossRef] [Google Scholar]
- Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci.. 2019;10
- [CrossRef] [Google Scholar]
- Hu, W., Pan, X., Muhammad, H., Abbas, K., Li, F., Dong, W., 2017. Metabolites contributing to Rhizoctonia solani AG-1-IA maturation and sclerotial differentiation revealed by UPLC-QTOF-MS metabolomics 0–16. https://doi.org/10.1371/journal.pone.0177464
- WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol.. 2017;59:86-101.
- [CrossRef] [Google Scholar]
- BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol.. 2016;428:726-731.
- [CrossRef] [Google Scholar]
- The role of nitric oxide in basal and induced resistance in relation with hydrogen peroxide and antioxidant enzymes. J. Plant Physiol.. 2016;199:29-38.
- [CrossRef] [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [CrossRef] [Google Scholar]
- Induction of SA-signaling pathway and ethylene biosynthesis in Trichoderma harzianum-treated tomato plants after infection of the root-knot nematode Meloidogyne incognita. Plant Cell Rep.. 2017;36:621-631.
- [CrossRef] [Google Scholar]
- Study on anti-ehrlich ascites tumor effect of Pinellia ternata polysaccharide in vivo. Afr. J. Trad. Complem. Altern. Med.. 2013;10:380-385.
- [Google Scholar]
- Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome. 2009;52:77-88.
- [CrossRef] [Google Scholar]
- Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402-408.
- [CrossRef] [Google Scholar]
- The role of Cu/Zn-SOD and Mn-SOD in the immune response to oxidative stress and pathogen challenge in the clam Meretrix meretrix. Fish Shellfish Immunol.. 2015;42:58-65.
- [CrossRef] [Google Scholar]
- Dihalogenated sulfanilamides and benzolamides are effective inhibitors of the three β-class carbonic anhydrases from Mycobacterium tuberculosis. J. Enzyme Inhib. Med. Chem.. 2014;28:384-387.
- [CrossRef] [Google Scholar]
- Assay of catechol oxidase—a critical comparison of methods. Phytochemistry. 1966;5:783-789.
- [CrossRef] [Google Scholar]
- Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes (Basel). 2020;11:346.
- [CrossRef] [Google Scholar]
- Muhammad, H., Abbas, K., Xiang, J., Ahmad, Z., Wang, L., Dong, W., 2018. Enhanced Nicotiana benthamiana immune responses caused by heterologous plant genes from Pinellia ternate, 1–14.
- Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol.. 2013;4:1-16.
- [CrossRef] [Google Scholar]
- Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci.. 2014;19:491-500.
- [CrossRef] [Google Scholar]
- Viaud, M., Fillinger, S., Liu, W., Polepalli, J.S., Le Pêcheur, P., Reddy Kunduru, A., Leroux, P., Legendre, L., 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant-Microbe Interact. MPMI 19, 1042–1050. https://doi.org/10.1094
- Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci.. 2014;5:168.
- [CrossRef] [Google Scholar]
- PeBL1, a novel protein elicitor from Brevibacillus laterosporus strain A60, activates defense responses and systemic resistance in Nicotiana benthamiana. Appl. Environ. Microbiol.. 2015;81:2706-2716.
- [CrossRef] [Google Scholar]
- Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc.. 2013;8:2502-2515.
- [CrossRef] [Google Scholar]
- Cell membrane-interrupting antimicrobial peptides from isatis indigotica fortune isolated by a bacillus subtilis expression system. Biomolecules. 2020;10:1-17.
- [CrossRef] [Google Scholar]
- Yang, Q., He, Y., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., Bian, Y., El Kasmi, F., Yang, L., Teixeira, P., Kolkman, J., Nelson, R., Kolomiets, M., L Dangl, J., Wisser, R., Caplan, J., Li, X., Lauter, N., Balint-Kurti, P., 2017. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–1372. https://doi.org/10.1038/ng.3919
- Halotolerant plant-growth promoting rhizobacteria modulate gene expression and osmolyte production to improve salinity tolerance and growth in Capsicum annum L. Environ. Sci. Pollut. Res.. 2018;25:23236-23250.
- [CrossRef] [Google Scholar]
- A comparative proteomic analysis of Pinellia ternata leaves exposed to heat stress. Int. J. Mol. Sci.. 2013;14:20614-20634.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.08.005.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







