Translate this page into:
Genome-wide and expression analysis to understand the DUF789 gene family during development of Arabidopsis thaliana
⁎Corresponding author. sfli@szu.edu.cn (Shuangfei Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Proteins with domains of unknown function (DUF) play an essential role in the growth of plants. However, we conducted a study on the genome-wide identification of DUF789 genes and the functional evolution of different members of the DUF789 gene family in the Arabidopsis thaliana genome. A total of 11 AtDUF789s were discovered in the A. thaliana genome, and a phylogenetic tree was constructed using sequences from A. thaliana, G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor. Gene structure analysis showed that the number of non-coding regions varied between 4 and 5, while the coding pattern ranged from 5 to 6. The promoter of AtDUF789s contains the cis-regulatory elements ABRE, MBS, and LTR, specifically. By analyzing the expression of the 11 AtDUF789s in tissues, we observed that these AtDUF789s were up-regulated in all observed tissues, which may indicate their involvement in plant growth. The study of the DUF789 gene family in A. thaliana provides new and valuable data for plant breeding and molecular studies.
Keywords
Domain
Development
Expression
Genes
Growth, Regulation
1 Introduction
Advancements in sequencing technology have led to an exponential increase in data related to genomics, transcriptomics, proteomics, and metabolomics. Despite the vast amount of data that has been generated, a significant portion of it remains unexplored (Chaudhari et al., 2024). There exists a group of conserved protein families that are composed of domains with unknown functions that have not yet been characterized. In 1998, researcher Chris Ponting was the first person to identify and designate these domains as DUF1 and DUF2 (Vishwakarma et al., 2024). After sequencing genomes in various species, additional DUF families were subsequently identified. Additionally, the Pfam database version 35.0 includes a total of 19,632 families, with 4,795 of them classified as DUF families. Certain proteins that possess DUFs play a crucial role in plant development. These proteins include DUF724, DUF1218, and DUF231 (Lv et al., 2023). Cellulose comprises the primary and secondary cell walls. Proteins containing the DUF266 domain are believed to play a role in cellulose biosynthesis, possibly functioning as glycosyltransferases. A mutation in the DUF266 gene significantly decreases cellulose synthesis in rice (Yang et al., 2017). The A. thaliana possesses six genes related to RUS, all of which code for proteins featuring the DUF647 domain. However, both RUS1 and RUS2 have been reported to have a significant role in regulating early seedling growth, vitamin B6 homeostasis and auxin transport (Tong et al., 2021). Furthermore, the expression of RUS6 was observed during various stages of plant growth, with a notably high level of expression in flowers. This indicates the significant role of RUS6 in the development of A.thaliana (Perry et al., 2021). Research conducted by Vergès et al. (2023) discovered that 23 genes were found to be expressed in the reproductive organs, of A. thaliana with the majority of them being expressed in the endosperm (Vergès et al., 2023). In A. thaliana DUF239 genes show a characteristic expression pattern that suggests a new role for plant neprosin-related proteins, particularly during seed maturation. Furthermore several studies have reported on the role of DUF gene families in A. thaliana and rice in responding to abiotic stress (Zhong et al., 2019). In A. thaliana, the TBL3 and TBR genes encode proteins that contain the DUF231 domain. This domain has significant function in the development of secondary cell walls in plants (Yuan et al., 2016). In A. thaliana the ESK1 gene (DUF231), acts as an inhibitor during cold acclimation (Yuan et al., 2013). Furthermore, the expression of the AtRDUF1 and AtRDUF2 genes in A. thaliana was found to be stimulated by abscisic acid (ABA) and drought stress. Conversely, when their expression is decreased, it leads to reduced drought stress tolerance. The salt-responsive gene TaSRHP (DUF581) was overexpressed in transgenic A. thaliana plants, resulting in increased resistance to stress (Hou et al., 2013). Furthermore, overexpression of the TaSRG (DUF662) transcription factor has been shown to enhance salt tolerance in both rice and A. thaliana. However, the gene SbSGL (DUF1645) has a significant function in sorghum by regulating the process of seed maturation (Zhang Bin et al., 2018). A mutation in the DUF1517 gene of A. thaliana renders it susceptible to cold stress (HAO et al., 2018). In this study, we analyzed the AtDUF789s at the genome-wide level. Our analysis included conducting a phylogenetic analysis, examining the gene structure, and assessing the chromosome location. Furthermore,we have identified members of the DUF789 family in several plant species including G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor in this study. Moreover, we conducted an analysis of the expression of AtDUF789s in various plant tissues, including leaves, flowers, fruits, and roots, utilizing RNA-seq data. This study provides a basis for future research on the role of AtDUF789 genes. Further investigation into AtDUF789 genes will also enhance our understanding of the regulatory mechanism of DUF789s in crops.
2 Materials and Methods
2.1 Identification of AtDUF789 genes
The Hidden Markov Model approach was used to search for DUF789 genes within the Arabidopsis thaliana genome. The genome of A. thaliana was obtained from the TAIR Arabidopsis database (http://www.arabidopsis.org/.). The HMM file containing the DUF789 domain (PF05623) was downloaded from the Pfam database (http://pfam.xfam.org). However, for further analysis we used HMMER 3.2.1 to search for amino acid sequences of DUF789 (PF05623) with an E-value less than 1e−5. The sequences of G. max, V. vinifera, S. tuberosum, M. truncatula, and S. bicolor were obtained from the JGI Phytozome 12.0 database (https://phytozome. jgi. doe. gov/pz/ portal. html). Additionally, the DUF789 sequences of A. thaliana, G. max, V. vinifera, S. tuberosum, M. truncatula, and S. bicolor were further validated through the CD-based search tool (https://www. ncbi. nlm. nih. gov/ Structure/ bwrpsb/ bwrpsb. cgi).
2.2 Analysis of physicochemical properties and prediction of subcellular location
To determine the isoelectric point, protein size, and molecular weight of the AtDUF789s protein, physicochemical analyses were performed using the ExPASy ProtParam program (http://us.expasy.org/tools/protparam.html). The subcellular localization of AtDUF789s was investigated using the CELLO online tool (http://cello. life. nctu. edu. tw/).
2.3 Chromosomal position and synteny analysis
The chromosomal position of DUF789 genes was obtained from the GFF file of A. thaliana. The TB-tools application was used to identify the chromosomal position of the AtDUF789 genes. The Circoletto tool was used to perform the synteny analysis of DUF789s. The sequences of A. thaliana, G. max, V. vinifera, S. tuberosum, M. truncatula, and S. bicolor species were used to analyze the synteny of DUF789.
2.4 Analysis of phylogenetic tree and chromosomal locations
We performed a multi-sequence alignment of the DUF789 genes of the following species:A. thaliana, G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor. We used Clustal W for the alignment. The alignment analysis used the protein sequences of A. thaliana, G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor. Redundant sequences were excluded prior to the alignment analysis. A phylogenetic tree was constructed using the Neighbor-Joining approach with a bootstrap value of 1000. The analysis included the genes of DUF789 from the following species A. thaliana, G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor. The software used to construct the phylogenetic tree was Mega and the tree was subsequently modified using iTOL.
2.5 Analysis of conserved motifs, domains and exon-intron arrangement
To analyze the conserved motif domains of AtDUF789s we used the Multiple Expectation Maximization for Motif Elicitation (MEME) online program with its default values. The amino acid sequences were also analyzed using the Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to identify domains. Moreover, the TB-tools application (Chen et al., 2020) was utilized to create a gene structure diagram for AtDUF789s.
2.6 Prediction of miRNA and gene expression analysis
The psRNATarget server (http://plantgrn. noble. org/psRNATarget/home) (Dai and Zhao, 2011) was utilized to predict miRNA targets for AtDUF789s using the CDS sequences of AtDUF789s. The standard settings were employed for this prediction. The expression datasets of DUF789 genes in different tissues of A. thaliana such as the leaves, flowers, roots, and fruits were obtained from NCBI (SRA: SRP128359: BioProject ID:2345). Gene expression levels were measured in fragments per kilobase of exon per million mapped reads (FPKM) (Ghosh and Chan, 2016).
3 Results
3.1 Identification of AtDUF789 and their localization
The Arabidopsis thaliana genome contains 11 DUF789s, with proteins ranging in size 186 to 409 amino acids and molecular weights between 21.15 and 45.81 kDa. The smallest identified protein is AtDUF789-9, while the longest identified protein is AtDUF789-11. A protein is deemed stable if its instability index is less than 40, and considered unstable if the instability index is greater than 40. Therefore, it was predicted that AtDUF789-1, AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-6, AtDUF789-7, AtDUF789-8, AtDUF789-9, AtDUF789-10, and AtDUF789-11 are unstable. The hydropathy grand average values suggest that the majority of AtDUF789s are mostly hydrophobic. The pI values range from 4.67 to 6.63, indicating that AtDUF789s do not have an overall electrical charge within this pH range. This study identified a total of seven genes in the forward direction and four genes in the reverse direction. The number of exons varied from 5 (AtDUF789-1, AtDUF789-6, AtDUF789-9) to 6 (AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-7, AtDUF789-8, AtDUF789-10, and AtDUF789-11). Interestingly, there are eight genes that have a maximum of 5 introns, while three genes have a minimal number of introns (4). These genes, namely AtDUF789-1, AtDUF789-6, and AtDUF789-9, are listed in Table 1. Additionally, the subcellular localization analysis has confirmed that AtDUF789s are present in both the chloroplast and nucleus. Notably, AtDUF789-10 is primarily located within the chloroplast, while AtDUF789-1, AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-6, AtDUF789-7, AtDUF789-8, AtDUF789-9, and AtDUF789-11 are located within the nucleus. Furthermore, 12 genes from Solanum tuberosum, 12 from Vitis vinifera, 13 from Medicago truncatula, 20 from Glycine max, and 11 from Sorghum bicolor have also been identified.
Gene ID
Chromosome
Strand
Start (bp)
End (bp)
Protein (AA)
M.W (kDa)
pI
GRAVY
Instability
Subcellular localization
AtDUF789-1
Chr1
1
900887
903003
308
35774.16
5.22
−0.556
51.58
Nucleus
AtDUF789-2
Chr1
1
5177529
5180049
360
40491.14
5.62
−0.466
49.17
Nucleus
AtDUF789-3
Chr1
−1
6135803
6138432
337
38413.95
4.87
−0.463
53.76
Nucleus
AtDUF789-4
Chr1
−1
27528052
27530952
314
36094.13
4.67
−0.638
60.33
Nucleus
AtDUF789-5
Chr2
−1
135244
137806
369
41643.29
5.57
−0.528
60.21
Nucleus
AtDUF789-6
Chr4
1
1511839
1514030
310
35808.13
5.25
−0.533
49.12
Nucleus
AtDUF789-7
Chr4
1
9105720
9108159
394
44658.91
5.69
−0.698
61.62
Nucleus
AtDUF789-8
Chr4
−1
13977303
13979006
285
33294.44
5.07
−0.392
56.25
Nucleus
AtDUF789-9
Chr5
1
2689713
2690974
186
21158.78
4.98
−0.384
43.95
Nucleus
AtDUF789-10
Chr5
1
7866742
7870302
301
33735.67
4.73
−0.406
48.72
Chloroplast
AtDUF789-11
Chr5
1
19956437
19958833
409
45812.54
6.63
−0.57
54.2
Nucleus
3.2 Cis-Regulatory element analysis
Specifically, this research examined eight cis-regulatory elements, four of which are related to phytohormones: gibberellin, methyl jasmonate, auxin, and abscisic acid. Furthermore, the components related to phytohormones include TGA, ABRE, TATC box, GARE motif, P-box, CGTCA motif, and TGACG motif. These components were found in various genes, emphasizing the crucial role of AtDUF789s in regulating phytohormones. Furthermore, we discovered components that respond to different types of stresses, namely anaerobic, low-temperature, drought, and salt. These components consist of the TCA-component, MBS, LTR, and ARE, indicating their involvement in stress response. Specifically, the MBS component, which is responsible for responding to drought, was predominantly discovered in AtDUF789-6, AtDUF789-7, and AtDUF789-8 (Supplementary Table S2).
The low-temperature responsive (LTR) element was found in six genes, namely AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-8, and AtDUF789-11. The anaerobic responsiveness (ARE) component was present in AtDUF789-1, AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-6, AtDUF789-7, AtDUF789-8, AtDUF789-9, AtDUF789-10, and AtDUF789-11 (Fig. 1).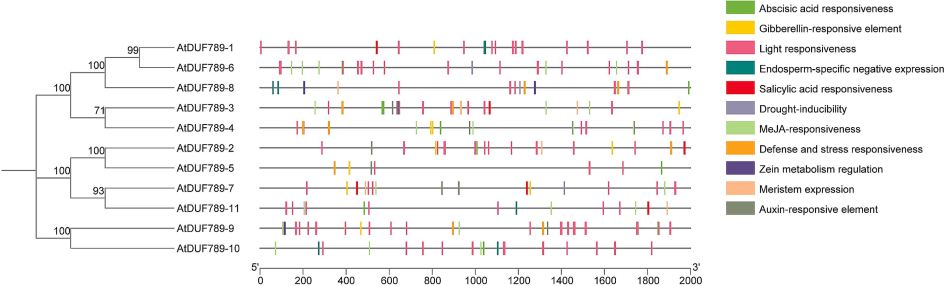
Cis-elements in the promoter regions of the AtDUF789 genes are linked with different hormone- and stress-responsive elements. Different color boxes show different identified elements.
3.3 Construction of phylogenetic tree for DUF789 genes
A phylogenetic tree for DUF789 was created using protein sequences from A. thaliana, G. max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, and Sorghum bicolor. The tree was divided into 5 groups, as shown in Fig. 2. Group I contained 15 proteins (4 S. bicolor, 4 M. truncatula, 2 S. tuberosum, 3 V. vinifera, and 2 G. max) (Fig. 2). Group II consisted of 15 proteins (2 M. truncatula, 1 S. tuberosum, 4 V. vinifera, 6 G. max, and 2 A. thaliana). Group III included 16 proteins (4 S. bicolor, 2 M. truncatula, 2 S. tuberosum, 2 V. vinifera, 4 G. max, and 2 A. thaliana). Group IV comprised 20 proteins (3 S. bicolor, 3 M. truncatula, 4 S. tuberosum, 4 A. thaliana, 4 G. max, and 2 Vitis vinifera). Group V consisted of 13 proteins (3 A. thaliana, 4 G. max, 1 V. vinifera, 3 S. tuberosum, and 2 M. truncatula). According to the constructed tree, Group IV had the highest number of proteins, while Group V had the lowest.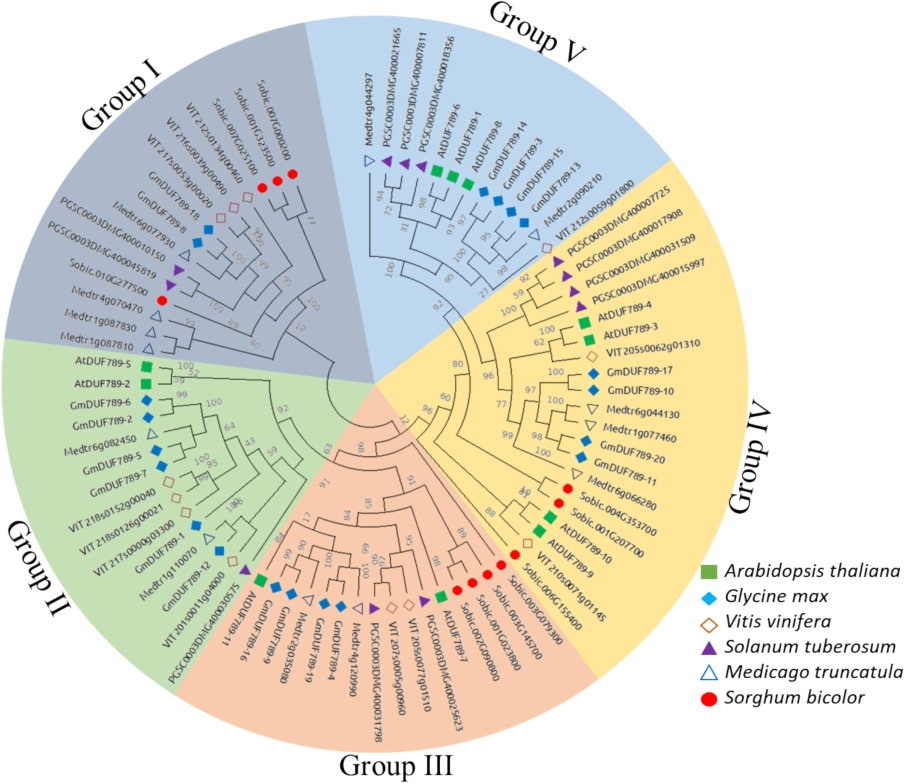
A neighbor-joining phylogenetic tree assessment of DUF789 genes from S. tuberosum, G. max, Sorghum bicolor, A. thaliana,Vitis vanifera and M. truncatula. Overall, AtDUF789 were clustered into five major classes.
3.4 Analysis of conserved motifs and domains
Ten unique motifs were identified and their locations were analyzed using the MEME tool for AtDUF789 proteins. Additionally, the full amino acid sequences were analyzed to identify conserved motifs. It was observed that all AtDUF789 proteins contain an AtDUF789 superfamily domain, indicating that this domain is conserved across all proteins.
During the motif analysis of AtDUF789 genes, it was found that genes within the same classes may contain different motifs, which could act as regulators for the various activities of the classes. For example, motif 1, with 50 amino acids (Supplementary Table S1), was found to be unique to group I, while motif 10 was also found in AtDUF789-3 and AtDUF789-4. The majority of genes have motifs 1, 4, and 6. It was also observed that motif 8 was unique to certain genes. For instance, motif 8 alone was present in GmDUF789-1, GmDUF789-6, GmDUF789-8, GmDUF789-7, and GmDUF789-11, while other motifs such as 1, 2, 3, 4, 5, 6, 7, 8, and 9 were identified in GmDUF789-1, GmDUF789-6, and GmDUF789-7 (Fig. 3).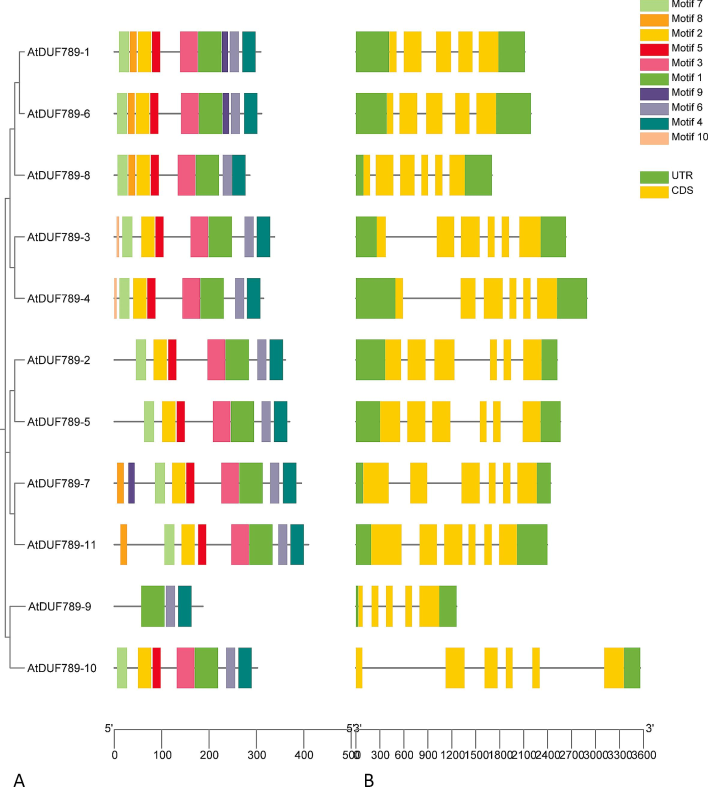
The gene structure and motif analysis of AtDUF789 genes. Based on phylogenetic relationships, the AtDUF789 were clustered into five major classes. (A) Conserved motif compositions were detected in AtDUF789. Different color boxes represent different motifs. (B) Gene structure of AtDUF789 genes. The light yellow color denotes exon and the black horizontal line symbolizes introns.
3.5 Gene structure analysis
Gene structure analysis showed that the number of non-coding regions varied between 4 and 5, while the coding pattern ranged from 5 to 6. As a result, we identified three genes with four introns and five exons, as well as eight genes with five introns and six exons (Fig. 3). Interestingly, members of AtDUF789s exhibited similar gene structures within their respective groups, indicating a potential shared evolutionary origin.
3.6 Chromosomal positions and synteny analysis of AtDUF789s
The chromosomal positions of all 11 AtDUF789 genes indicate that the genes were unequally distributed across chromosomes. Table 1 provides the specific chromosome positions for the AtDUF789 genes. The results show that four chromosomes contain the AtDUF789 genes, with chromosomes 1, 2, 4, and 5 all having AtDUF789 genes. In total, there were eleven AtDUF789 genes across these four chromosomes, with three genes located in chromosomes 4 and 5 (Fig. 4).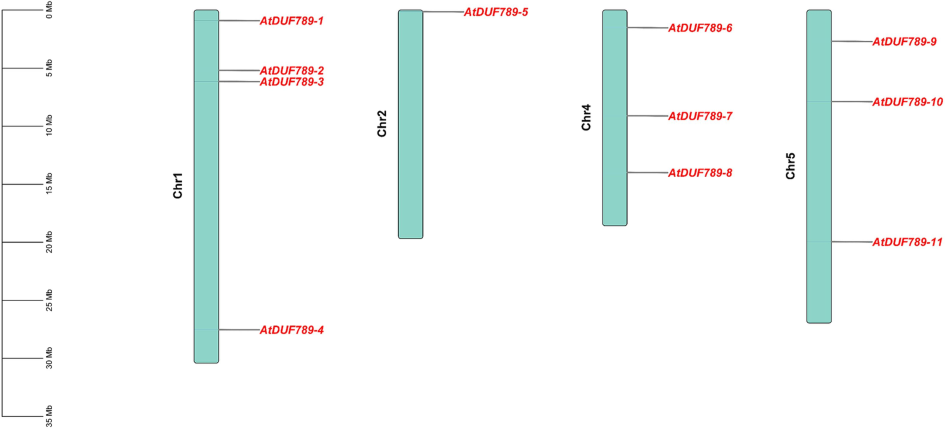
Chromosomal distribution of AtDUF789 genes. The green color showed Chromosomes. Red color genes.
Additionally, this investigation reveals the presence of synteny among Vitis vinifera, S. tuberosum, G. max, A. thaliana, S. bicolor, and M. truncatula plants. The purpose of this investigation was to determine the role, evolution, expression, and duplications of DUF789 genes. Our results indicate a synteny relationship between the sequences VIT_205s0077g01510, AtDUF789-11, and GmDUF789-7. Furthermore, AtDUF789-2 and AtDUF789-5 of A. thaliana and the gene VIT_201s0011g04000 of M. truncatula also exhibited synteny. Notably, AtDUF789-6 of A. thaliana showed synteny with the S. tuberosum gene PGSC0003DMG400021665 sequence (Fig. 5).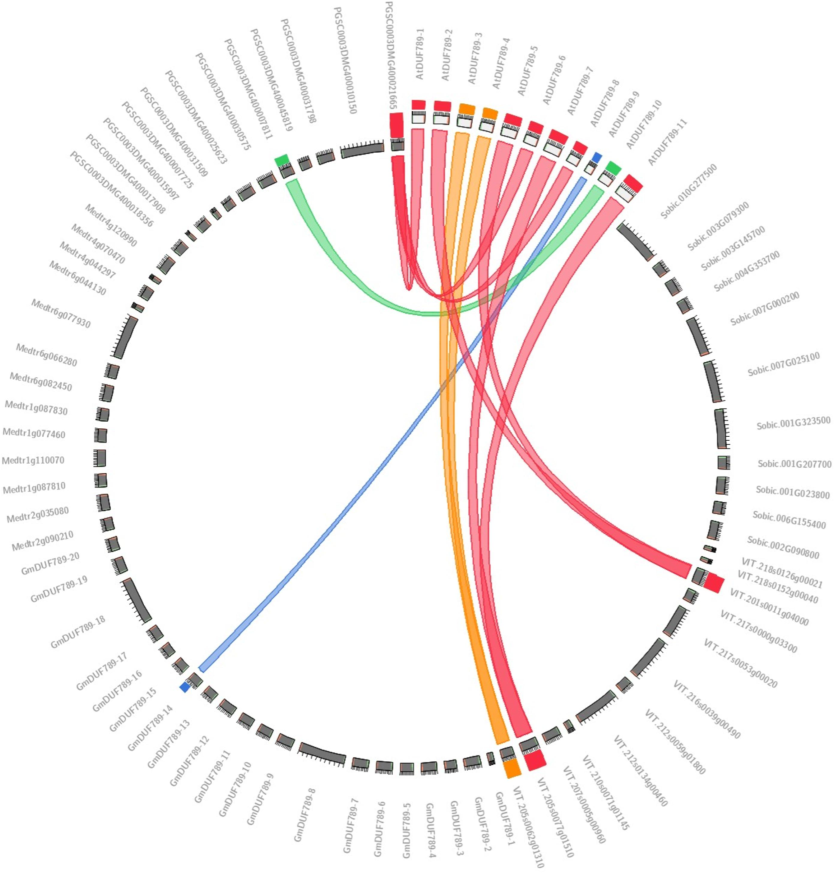
Synteny map among all identified DUF789 sequences of S. tuberosum, M. truncatula, A. thaliana, and G. max.
3.7 miRNA prediction
Several studies have emphasized the significance of miRNA in plant stress response. In summary, the studies revealed that ath-miR156 targets three genes (AtDUF789-1, AtDUF789-2, and AtDUF789-8), ath-miR159 targets two genes (AtDUF789-2 and AtDUF789-8) (Supplementary Table S3), ath-miR172 targets AtDUF789-2, ath-miR8167 targets AtDUF789-8, ath-miR845 targets AtDUF789-8, ath-miR415 targets AtDUF789-2, ath-miR854 targets AtDUF789-2, and ath-miR1888 targets AtDUF789-11. Additionally, ath-miR5021 targets AtDUF789-1, AtDUF789-4, AtDUF789-5, and AtDUF789-11, while ath-miR5656 targets AtDUF789-4, AtDUF789-1, AtDUF789-6, and AtDUF789-11. Moreover, two members of the ath-miR3932 family target AtDUF789-4 and AtDUF789-5.
3.8 Expression of AtDUF789s in different tissues
RNA-seq data were used to examine the expression of AtDUF789s in leaf, flower, fruit, and root tissues. The analysis revealed that several AtDUF789 genes, namely AtDUF789-1, AtDUF789-2, AtDUF789-3, AtDUF789-4, AtDUF789-5, AtDUF789-6, AtDUF789-7, AtDUF789-8, AtDUF789-9, AtDUF789-10, and AtDUF789-11, showed high expression levels in leaf, flower, fruit, and root tissues (Fig. 6). For instance, AtDUF789-1 had high expression in both leaves and roots, while AtDUF789-6 showed high expression in flower and root tissues (Supplementary Table S5). These genes were found to be highly expressed throughout plant development, along with other genes.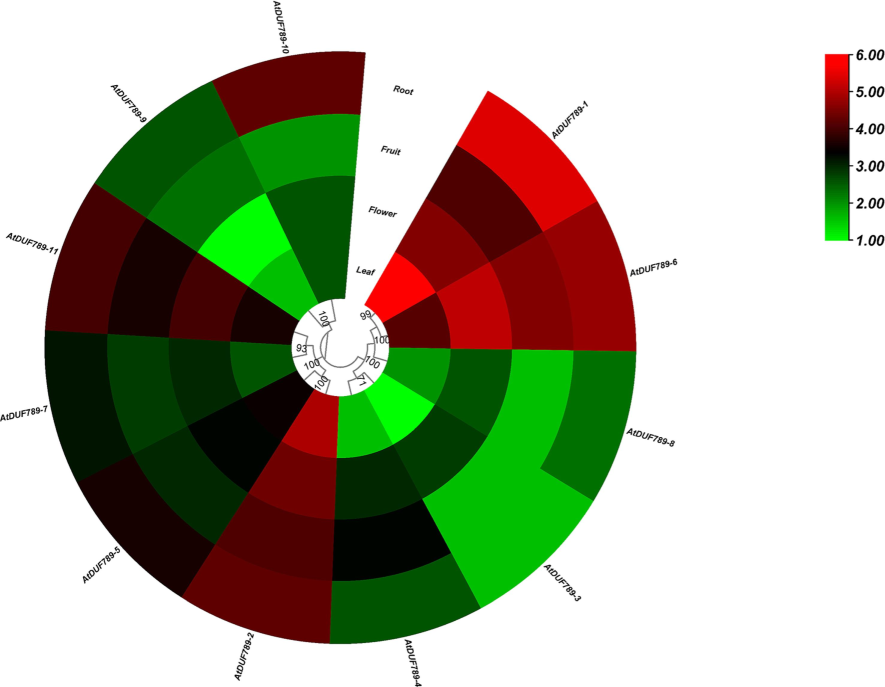
Expression profiling of AtDUF789 genes in various developmental tissues. The red,white, and green colors display high to low expression. levels.
4 Discussion
The DUF gene family is known to regulate various biological functions in plants (Zaynab et al., 2022). However, there has not been a thorough investigation of the AtDUF789 gene family in A. thaliana. In this study, we aim to fill this gap by conducting a genome-wide identification of DUF789 genes. Additionally, we examined the functional evolution of different members of the DUF789 gene family in the Arabidopsis thaliana genome. Furthermore, the current study has discovered a total of 11 AtDUF789s within the genome of Arabidopsis thaliana. In order to identify the distinctive qualities of the AtDUF789s within the family, we also analyzed the physicochemical features of the eleven members. This analysis revealed that each AtDUF789 possesses its own unique characteristics. Zaynab et al. (2023) reported that each GmDUF668 protein exhibited unique characteristics (Zaynab et al., 2023b). We conducted subcellular localization prediction to determine the specific location of AtDUF789 proteins within intracellular organelles. Upon examination of subcellular localization, it was found that the majority of AtDUF789 proteins were primarily located within the nucleus (Paulo et al., 2013). This suggests that these proteins play a crucial role in this organelle.
Exons play a crucial role in the protein synthesis process by containing essential information. On the other hand, introns have a protective function, preventing specific random mutations that could potentially harm the coding proteins (Lesk, 2010). The analysis has significant implications for researchers who study the evolutionary perspective, structure, and function of the DUF789 gene family. Furthermore, the examination of the gene structure has unveiled the quantity of exons and introns in the AtDUF789 genes. This analysis of gene structure yields crucial insights into the evolutionary patterns of exon–intron distribution and the factors that influence the diverse range of biochemical functions (Gelfman et al., 2012). Furthermore, it was observed that various AtDUF789s shared the same structure. Furthermore the analysis of the gene structure of AtDUF789 revealed variations in the number of exons and introns. Moreover, the differences in the number of exons observed among the AtDUF789 genes imply functional variations within this gene family. Similarly, the analysis of conserved gene motifs suggests that some AtDUF789 genes share the same set of motifs. The fact that the AtDUF789 genes have similar motifs suggests that they are evolutionarily related and serve the same biological purpose.
The promoters of AtDUF789s contain cis-acting components that demonstrate their responsiveness to different hormones and stresses. This suggests that AtDUF789s can be regulated by various stresses (salinity, drought, and low temperature) as well as phytohormones (auxin, ABA, MeJA, and GA). The current study findings are supported by multiple studies that have reported the role of cis-components in a plant's response to stress (Raza et al., 2020). In addition, TaGAPC1 also responds to various stresses and its expression pattern is controlled in response to salinity and osmotic stress. During stress, the LTR, GT1, DRE, and MBS elements of the TaGAPC1 promoter undergo alterations due to methylation (Wen et al., 2021). This modification serves to counterbalance the effects of stress and regulate the gene's expression levels. MicroRNAs, also known as miRNAs, are a group of small RNAs that are non-coding and endogenous in nature. They typically measure 20–22 nucleotides in size (Ling et al., 2013). These miRNAs play a significant role in various cellular and physiological processes, including plant development, growth, and response to environmental stress conditions (Kumar, 2014). Several studies have shown that miRNAs play a potential role in various biological processes. In this study, we identified several miRNAs that target 11 AtDUF789 genes. Importantly, our findings are in line with previous research and serve to confirm our results. Wang et al. (2020) examined the role of miR156 in A. thaliana (Ye et al., 2020), while Wang et al. (2020) focused on investigating the developmental role of miR156 in rice (Wang et al., 2023). Additionally, Zhao et al. 2024 reported that miR172s play a positive regulatory role in development (Zhao et al., 2024). Previous studies have also confirmed the significance of miR172 and its target expression in plant developmental processes. Spanudakis et al and Jackson reported the essential role of miR159 at different stages of plant growth (Spanudakis and Jackson, 2014). Therefore, these findings suggest that miRNAs play a vital role in plant development, growth, and stress tolerance.
To investigate the involvement of AtDUF789 genes in plant development, we examined the expression of 11 specific genes in leaves, flowers, fruits, and roots. This analysis was carried out using an RNA-seq dataset from the BioProject PRJNA168212. Furthermore, previous studies have demonstrated that the expression patterns of certain DUF genes undergo significant variations across different tissue types throughout the process of development. A study conducted by Zhang et al. (2024) revealed that OsDUF247 genes were expressed in rice (Zhang et al., 2024). Their findings revealed that OsDUF247 was expressed in seedlings, roots, stems, and leaves. Zaynab et al. (2023) conducted a study on the expression of DUF668 genes in soybean (Zaynab et al., 2023b). Their findings revealed that GmDUF668 was expressed in flowers, nodules, roots, and leaves. The expression analysis demonstrated higher levels of gene expression in these tissues. Specifically, genes such as GmDUF668-30, GmDUF668-28, GmDUF668-27, GmDUF668-26, GmDUF668-22, GmDUF668-19, GmDUF668-17, GmDUF668-16, GmDUF668-15, GmDUF668-13, GmDUF668-10, GmDUF668-9, GmDUF668-8, GmDUF668-7, GmDUF668-4, and GmDUF668-3, showed upregulation across roots, nodules, leaves, and flowers (Zaynab et al., 2023b). The expression of GhDUF4228 genes in various tissues of cotton, including stem, root, leaf, bract, pistil, sepal, petal, anther, and filament was also investigated. The gene expression profiles at different developmental stages provided information about various genes (Rochette et al., 2008). It was reported that certain genes exhibited upregulation in different parts of the plant, including the stem, root, leaf, bract, pistil, sepal, petal, anther, and filament (Rochette et al., 2008). Furthermore, Aulakh et al. (2014) conducted a tissue-specific expression analysis for sweet potato using RNA-seq data (Aulakh et al., 2014). The findings revealed that several IbDUF668 genes had increased expression patterns across different tissues during development. Multiple IbDUF668 genes were found to exhibit increased expression in tissues during development. Upon examining the transcriptome data for the GmDUF4228 genes, it was found that these genes were expressed in leaves, root hairs, nodules, and pods. It is worth noting that GmDUF4228-56, GmDUF4228-70, and GmDUF4228-73 exhibit particularly strong expression in roots (Leng et al., 2021). The study found that several DUF599 genes were expressed in potato tissues. To assess the importance of StDUF599-6 and StDUF599-9 in development, their expression in tissues was examined (Zaynab et al., 2023a). These findings support that AtDUF789s play a crucial role in plant development.
5 Conclusions
In this investigation, we identified a total of 11 AtDUF789s. These AtDUF789s were classified into five clusters based on a phylogenetic tree. To create this tree, we used protein sequences from G.max, Vitis vinifera, Solanum tuberosum, Medicago truncatula, Sorghum bicolor, and A.thaliana. Furthermore, the proteins ranged from 186 to 409, and with molecular weights between 21.15 and 45.81 kDa. Analysis of cis-elements suggests that AtDUF789 genes may be involved in stress responses. Most of the identified AtDUF789 genes have shown significant upregulation during the expression stages. This suggests that AtDUF789s play a crucial role in plant development. Further analysis of DUF789 genes will yield valuable data for the molecular breeding of A. thaliana, aiming to improve plant growth.
CRediT authorship contribution statement
Madiha Zaynab: Conceptualization, Data curation, Formal analysis. Yasir Sharif: Investigation, Methodology. Rashid Al-Yahyai: Software, Supervision, Validation. Athar Hussain: Writing – original draft. Monther Sadder: Writing – review & editing. Kahkashan Perveen: Data curation, Formal analysis. Najat A. Bukhari: Methodology. Shuangfei Li: Writing – review & editing.
Acknowledgement
The authors would like to acknowledge the support provided by Researchers Supporting Project Number RSP2024R229, King Saud University, Riyadh, Saudi Arabia. We thank Shenzhen Science and Technology Program KCXFZ20230731094059009 and KCXST20221021111206015.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Characterization and RNA-seq analysis of underperformer, an activation-tagged potato mutant. Plant Mol. Biol.. 2014;84:635-658.
- [Google Scholar]
- Biological big-data sources, problems of storage, computational issues, and applications a comprehensive review. Knowl. Inf. Syst. 2024:1-51.
- [Google Scholar]
- TBtools an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194-1202.
- [Google Scholar]
- psRNATarget a plant small RNA target analysis server. Nucleic Acids Res.. 2011;39:W155-W159.
- [Google Scholar]
- Changes in exon–intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res.. 2012;22:35-50.
- [Google Scholar]
- Analysis of RNA-Seq data using TopHat and Cufflinks. Plant Bioinform.: Methods Protocols 2016:339-361.
- [Google Scholar]
- Overexpression of AmDUF1517 enhanced tolerance to salinity, drought, and cold stress in transgenic cotton. J. Integr. Agric.. 2018;17:2204-2214.
- [Google Scholar]
- A novel ABA-responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in Arabidopsis thaliana. Plant Mol. Biol. Report.. 2013;31:791-801.
- [Google Scholar]
- Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol.. 2014;174:93-115.
- [Google Scholar]
- Genome-wide analysis of the DUF4228 family in soybean and functional identification of GmDUF4228–70 in response to drought and salt stresses. Front. Plant Sci.. 2021;12:628299
- [Google Scholar]
- Introduction to protein science architecture, function, and genomics. USA: Oxford University Press; 2010.
- MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov.. 2013;12:847-865.
- [Google Scholar]
- Unraveling the diverse roles of neglected genes containing domains of unknown function (DUFs) progress and perspective. Int. J. Mol. Sci.. 2023;24:4187.
- [Google Scholar]
- Subcellular fractionation enhances proteome coverage of pancreatic duct cells. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2013;1834:791-797.
- [Google Scholar]
- RUS6, a DUF647-containing protein, is essential for early embryonic development in Arabidopsis thaliana. BMC Plant Biol.. 2021;21:232.
- [Google Scholar]
- Arabidopsis thaliana model plant for the study of abiotic stress responses. the plant family brassicaceae. Bio. Physiol. Resp. Environ. Stresses 2020:129-180.
- [Google Scholar]
- Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:1-26.
- [Google Scholar]
- The role of microRNAs in the control of flowering time. J. Exp. Bot.. 2014;65:365-380.
- [Google Scholar]
- Arabidopsis ROOT UV-B SENSITIVE 1 and 2 interact with aminotransferases to regulate vitamin B6 homeostasis. bioRxiv 2021:2021-2103.
- [Google Scholar]
- The Arabidopsis DUF239 gene family encodes Neprosin-like proteins that are widely expressed in seed endosperm. The Plant Genome. 2023;16:e20290.
- [Google Scholar]
- AnnoDUF A Web-based tool for annotating functions of proteins having domains of unknown function (DUFs) bioRxiv 2024:2024-2106.
- [Google Scholar]
- Superstar microRNA, miR156, involved in plant biological processes and stress response. a review. Sci. Hortic.. 2023;316:112010
- [Google Scholar]
- Comprehensive in silico characterization and expression profiling of TCP gene family in rapeseed. Front. Genet.. 2021;12:794297
- [Google Scholar]
- Overexpression of a domain of unknown function 266-containing protein results in high cellulose content, reduced recalcitrance, and enhanced plant growth in the bioenergy crop Populus. Biotechnol. Biofuels. 2017;10:1-13.
- [Google Scholar]
- The role of miR156 in rejuvenation in Arabidopsis thaliana. J. Integr. Plant Biol.. 2020;62:550-555.
- [Google Scholar]
- The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O-and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol.. 2013;54:1186-1199.
- [Google Scholar]
- TBL3 and TBL31, two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol.. 2016;57:35-45.
- [Google Scholar]
- Genome-wide identification and expression profiling of DUF221 gene family provides new insights into abiotic stress responses in potato. Front. Plant Sci.. 2022;12:804600
- [Google Scholar]
- Expression profiling of DUF599 genes revealed their role in regulating abiotic stress response in solanum tuberosum. J. King Saud University-Science. 2023;35:102368
- [Google Scholar]
- Genome-wide analysis and expression profiling of DUF668 genes in Glycine max under salt stress. Plants. 2023;12:2923.
- [Google Scholar]
- Zhang Bin, Z.B., Zhang Xin, Z.X., Xu GuoYun, X.G., Li MingJuan, L.M., Cui YanChun, C.Y., Yin XuMing, Y.X., Yu Yan, Y.Y., Xia XinJie, X.X., Wang ManLing, W.M., 2018. Expression of sorghum gene SbSGL enhances grain length and weight in rice.
- Zhang, F., Yang, J., Sohail, A., Lu, C., Xu, P., 2024. Genome-wide characterization and analysis of rice DUF247 gene family.
- Exploring MicroRNAs associated with pomegranate pistil development. an identification and analysis study. Horticulturae. 2024;10:85.
- [Google Scholar]
- Characterization and functional divergence of a novel DUF668 gene family in rice based on comprehensive expression patterns. Genes. 2019;10:980.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103478.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







