Translate this page into:
Genetic diversity and antifungal activities of the genera Streptomyces and Nocardiopsis inhabiting agricultural fields of Tamil Nadu, India
⁎Corresponding authors. kumar.m@icar.gov.in (Murugan Kumar), saxena461@yahoo.com (Anil Kumar Saxena)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Actinomycetes receives much attention as plant-beneficial bacteria due to their distinct morpho-physiological, plant growth promoting and antifungal properties. Among the actinomycetes, members of the genus Streptomyces are of specific interest due to their metabolic versatility and ubiquity. The diversity of Streptomyces in different crop fields of Tamil Nadu has not been explored early. In this study, we explored phylogenetic diversity, morphological heterogeneity, and antifungal properties of cultivable actinomycetes isolated from the rhizosphere and bulk soils of different crops collected from 20 distinct districts of Tamil Nadu. A total of 65 actinomycetes were isolated from 40 soil samples including rhizosphere and bulk soils of different crops, and their diversity was analyzed using the 16S rRNA gene sequence. They were then characterized for cultural characteristics and antifungal activity against four fungal strains Fusarium udum, Fusarium oxysporum f.sp. ciceri, Macrophomina phaseolina and Sclerotium rolfsii. Out of the 65 isolates sequenced, 45 were found to be closely related to Streptomyces spp. while the remaining 20 showed similarities with Nocardiopsis spp. Cultural characterization on four different ISP media showed immense diversity among the members of the genera Streptomyces and Nocardiopsis. The strains of Streptomyces spp. and Nocardiopsis spp. showed varying levels of antifungal activity with all strains found antagonistic against both Fusarium udum and Fusarium oxysporum f.sp. ciceris. All the strains obtained in this study have been accessioned at the National Agriculturally Important Microbial Culture Collection (NAIMCC) to increase the database of characterized strains belonging to these two genera in the collection.
Keywords
Actinomycetes
Antifungal activity
Streptomyces spp.
Nocardiopsis spp.
1 Introduction
Actinomycetes are an important group of soil bacteria that are found to be more prevalent in dry soils than wet soils. Soil actinomycetes are known for their plant growth promotional activity as they exhibit various useful traits (Ma et al., 2020). They can also produce a variety of secondary metabolites, many of which are antibacterial or antifungal (Olanrewaju and Babalola, 2019). Most antibiotics used in human medicine are metabolites of actinomycetes, with many of them originating from Streptomyces spp. (Ma et al., 2020). The ability to synthesize a variety of chemical substances such as antibiotics, enzymes, and anti-tumor medicines, made Streptomyces spp. most common genus used in the pharma industry (Chaudhary et al., 2013). In soil, they grow as substrate mycelium and produce a range of enzymes including chitinase, xylanase and cellulase that breakdown complex organic polymers into constituent simple sugars (Seipke et al., 2012) thereby playing a direct role in biogeochemical cycles. They are significant candidates for natural fertilizers because of their ability to cycle nutrients. They are effective colonizers of rhizosphere and rhizoplane tissues with the potential to enhance plant growth, and development and boost yield (Dias et al., 2017). Streptomyces spp. promote plant growth through the production of phytohormones like auxins, cytokinins, and gibberellins, additionally they produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase whose activity is important in the suppression of plant stress (Sadeghi et al., 2012; Verma et al., 2011). They are implicated in phosphate solubilization (Jog et al., 2014) and siderophore production (Verma et al., 2011) as well. Yet the most important role of Streptomyces spp. in agriculture is their ability to control phytopathogens owing to their traits; siderophore production, antibiotics production and volatile compounds secretion (Olanrewaju and Babalola, 2019).
Isolation, morphological, biochemical, physiological, and molecular characterization of Streptomyces spp. are reported in many studies (Taddei et al., 2006). Taddei et al. (2006) studied the biochemical and morphological characterization of 71 Streptomyces spp. isolated from soil samples of Venezuela, of them 67 isolates were new strains. In a recent study, Kaur et al. (2019) tested a potent Streptomyces strain MR14 with antifungal and plant growth promoting properties. Cell free extracts of MR14 showed potent antagonistic activity against 13 different fungal phytopathogens. Despite their multifarious role, Streptomyces found beneficial to plants and agriculture overall, received lesser interest as compared to Streptomyces spp. studied for their role in pharmaceuticals (Rey and Dumas, 2017). It is only imperative, that the exploration of the phylogenetic and morphological diversity of this group of bacteria and enriching their cultural database could lead to identifying strains with unique potential. Agricultural fields especially plant rhizosphere is an important source of potent Streptomyces spp. with antagonistic activity against plant pathogens. In the present study, an attempt was made to isolate Streptomyces spp. from different crop rhizosphere and bulk soil samples collected from Tamil Nadu, India. The resultant isolates were characterized for morphological diversity, genetic diversity, and antagonistic potential against select phytopathogenic fungi. All the actinomycetes isolated in this study were submitted to the National Agriculturally Important Microbial Culture Collection (NAIMCC), a constituent unit of ICAR-National Bureau of Agriculturally Important Microorganisms, Mau India.
2 Materials and methods
2.1 Sample collection
A total of 40 soil samples including rhizosphere of different crops and bulk soils were collected in sterile bags from various locations in Tamil Nadu, India, details of the samples are depicted in Table 1. For rhizosphere samples, standing crops were uprooted at five random sites in a field and soils adhered to the roots were collected in a sterile poly bag. For bulk soil, the top 15 cm soil core was collected from five random sites in a sterile poly bag from each site. The samples once brought to the laboratory were shade dried aseptically and stored in a cold room (4 °C) until further processing.
GPS location
Address
Sample type
Strain no
9°53′22.0″N 78°09′22.0″E
Chinthamani, District Madurai, Tamil Nadu
Bulk soil coconut field
TN1, TN2
9°40′9.0″N 78°05′34.0″E
Kariapatti, District Virudhunagar, Tamil Nadu
Bulk soil after harvest of maize
TN3, TN4
9°29′37.0″N 78°06′25.0″E
Aruppukkottai, District Virudhunagar, Tamil Nadu
Bulk soil castor field
TN5, TN7
9°23′30.0N 78°07′40.0 E
Pandalgudi, District Virudhunagar, Tamil Nadu
Sunflower rhizosphere
TN6, TN8
9°08′50.0″N 78°00′23.0″E
Ettaiyapuram, District Thoothukkudi, Tamil Nadu
Bulk soil Jatropha field
TN9, TN10
8°51′04.0″N 78°07′15.0″E
Thoothukudi, Thoothukkudi District, Tamil Nadu
Sorghum rhizosphere
TN11, TN12
9°00′24.0″N 78°12′04.0″E
Vaippar, District Thoothukkudi, Tamil Nadu
Chilli rhizosphere
TN13, TN14
9°07′13.0″N 78°24′32.0″E
Sayalkudi, District Ramanathapuram, Tamil Nadu
Palm field bulk soil
TN15, TN16
9°13′17.0″N 78°36′37.0″E
Keerandai, District Ramanathapuram,Tamil Nadu
Rice rhizosphere
TN17, TN18
9°18′01.0″N 78°49′19.0″E
Pallamerkkulam, District Ramanathapuram, Tamil Nadu
Rice rhizosphere
TN19, TN20
9°22′57.0″N 78°50′08.0″E
Veethi, District Ramanathapuram, Tamil Nadu
Rice rhizosphere
TN21, TN22
9°37′21.0″N 78°55′50.0″E
Uppur, District Ramanathapuram, Tamil Nadu
Rice rhizosphere
TN23, TN24
10°03′58.0″N 79°13′48.0″E
Mumpalai, District Pudukkottai Tamil Nadu
Rice rhizosphere
TN25, TN26
10°18′10.0″N 79°20′27.0″E
Chinnaavudaiyarkoil, District Thanjavur, Tamil Nadu
Rice rhizosphere
TN27, TN28
10°25′32.0″N 79°32′52.0″E
Udayamarthandapuram, District Thiruvarur, Tamil Nadu
Rice rhizosphere
TN29, TN30
10°25′09.0″N 79°47′33.0″E
Kuravapulam, Nagapattinam District, Tamil Nadu
Rice rhizosphere
TN31, TN32
10°37′47.0″N 79°47′08.0″E
Meenamanallur, Nagapattinam District, Tamil Nadu
Rice rhizosphere
TN33, TN34
11°06′06.0″N 79°49′57.0″E
Chinnamedu, Mayiladuthurai District, Tamil Nadu
Rice rhizosphere
TN35, TN36
11°13′36.0″N 79°43′24.0″E
Sirkali, Mayiladuthurai District, Tamil Nadu
Rice rhizosphere
TN37, TN38
11°30′45.0″N 79°43′11.0″E
Chinnakomatti, District Cuddalore, Tamil Nadu
Rice rhizosphere
TN39, TN40
12°43′37.0″N 80°11′23.0″E
Thiruporu, District Chengalpattu, Tamil Nadu
Rice rhizosphere
TN41, TN42
12°53′48.0″N 80°14′50.0″E
Panaiyur, District Chennai, Tamil Nadu
Bulk soil coconut field
TN43, TN44
12°39′32.0″N 79°57′01.0″E
Grand Southern Trunk Road, Chengalpattu District Tamil Nadu
Rice rhizosphere
TN45, TN46
12°22′20.0″N 79°47′39.0″E
Kadamalaiputhur, District Kanchipuram, Tamil Nadu
Groundnut rhizosphere
TN47, TN48
12°11′02.0″N 79°37′25.0″E
Tindivanam, District Viluppuram, Tamil Nadu
Bhendi rhizosphere
TN49, TN50
11°40′51.0″N 79°17′18.0″E
Ulundurpet, District Kallakurichi, Tamil Nadu
Grassland
TN51
11°30′47.0″N 79°06′03.0″E
Kalpadi North, District Perambalur, Tamil Nadu
Brinjal rhizosphere
TN52
11°12′42.0″N 78°52′56.0″E
Pichandarkovil, District Tiruchirappalli, Tamil Nadu
Rice rhizosphere
TN53
10°54′11.0″N 78°43′30.0″E
Melapachchakudi, District Pudukkottai, Tamil Nadu
Rice rhizosphere
TN54
10°39′33.0″N 78°35′33.0″E
Kallupatti, District Tiruchirappalli, Tamil Nadu
Sorghum rhizosphere
TN55
10°25′40.0″N 78°26′07.0″E
Karungalakudi, District Madurai, Tamil Nadu
Brinjal rhizosphere
TN56
10°08′35.0″N 78°21′17.0″E
Kalikulam, District Tirunelveli, Tamil Nadu
Rice rhizosphere
TN57
9°59′50.0″N 78°15′23.0″E
Kalligudi, District Madurai, Tamil Nadu
Rice rhizosphere
TN58
9°43′33.0″N 77°58′46.0″E
Maniparaipatti, District Virudhunagar, Tamil Nadu
Redgram rhizosphere
TN59
9°25′23.0″N 77°55′08.0″E
Tenkasi, District Tenkasi, Tamil Nadu
Maize rhizosphere
TN60
9°12′41.0″N 77°47′45.0″E
Sankarankovil, District Tenkasi, Tamil Nadu
Rice rhizosphere
TN61
9°15′24.0″N 77°39′57.0″E
Alagiapandiapuram,District Tirunelveli, Tamil Nadu
Lemon rhizosphere
TN62
8°55′33.0″N 77°38′41.0″E
Palayamkottai, District Tirunelveli, Tamil Nadu
Rice rhizosphere
TN63
8°44′38.0″N 77°44′54.0″E
Amarapuram, District Thoothukudi, Tamil Nadu
Bulk soil banana field
TN64
8°32′31.0″N 78°06′43.0″E
Amarapuram, District Thoothukudi, Tamil Nadu
Bulk soil coconut field
TN66
2.2 Isolation of actinomycetes from soil
A selective media containing basal nutrients and antibiotics was used for the isolation of Streptomyces. Starch casein agar (SCA) containing soluble starch 10 gL-1, casein 0.3 gL-1, KNO3 2.0 gL-1, MgSO4·7H2O 0.05 gL-1, K2HPO4 2.0 gL-1, NaCl 2.0 gL-1, CaCO3 0.02 gL-1, FeSO4·7H2O 0.01 gL-1 and Agar 20.0 gL-1 supplemented with cycloheximide (50 mg/L), nystatin (50 mg/L) and rifampicin (50 mg/L) was used as the selective media (Nolan and Cross, 1988). The pH of the media was adjusted to 7. Standard serial dilution and plating method was followed to isolate Streptomyces. Aliquots (100 µL) of dilutions, 1/1000 and 1/10000 were plated on the selective media and incubated at 30 ± 1 °C for 7–10 days. Plates were observed regularly for the growth of the actinomycetes population. Different morphotypes from each of the samples were picked and purified in the selective media. The isolates were then stored as glycerol stocks (20 %) at −80 °C and slants at 4 °C until further study.
2.3 DNA isolation and 16S rRNA gene sequence amplification
Total DNA was extracted from fully grown actinomycetes cultures using Nucleo-pore gDNA Fungal Bacterial Mini Kit employing the manufacturer’s protocol. The quality of DNA was checked and ensured both by gel electrophoresis and nanodrop. Universal primers pA (AGAGTTTGATCCTGGCTCAG) and pH (AAGGAGGTGATCCAGCCGCA) were used for the amplification of the 16S rRNA gene. The PCR reactions were carried out in a peqSTAR thermal cycler by following the protocols of Kumar et al. (2014) with slight modifications. Briefly, the reactions were carried out in 25 μL volume comprising 12.5 μL Go Taq green Master Mix 2X (Promega, USA), 100 ng of primers (pA and pH), 50 ng DNA template and nuclease free water to adjust the volume. PCR conditions used were as follows; initial denaturation-95 °C for 5 min followed by 35 cycles of denaturation-95 °C for 30 s, annealing-52 °C for 40 s and extension-72 °C for 60 s; and a final extension at 72 °C for 10 min. The amplified PCR products were visualized in 1.5 % agarose gel electrophoresis (stained with ethidium bromide) using a gel documentation system (Bio Rad, USA).
2.4 16S rRNA gene sequencing and phylogenetic analysis
PCR products were purified using Gene JET PCR Purification Kit (Thermo Fisher, USA) and sequenced using the primer 1100R (GGGTTGCGCTCGTTG) (Engelbrektson et al., 2010) at GeneMatrix sequencing facility by following sanger dideoxy sequencing method in an ABI 3130xl Genetic Analyzer. Resultant sequences were then quality filtered in FinchTV and trimmed for better quality sequences (Stucky, 2012). Quality ensured sequences were then used to run a BLAST search against 16S rRNA gene sequences of type species available in EZ Biocloud to identify the isolates (https://www.ezbiocloud.net). Sequences were aligned and compared using the multiple sequence alignment tool, CLUSTAL W and a phylogenetic tree was constructed employing the neighbour joining method (Saitou and Nei, 1987). The Program used for phylogenetic tree construction is MEGA X (Kumar et al., 2018).
2.5 Characterization of soil actinomycetes based on pigment formation, growth pattern and mycelium formation
Cultural characteristics of all the actinomycete isolates were examined on four different ISP (International Streptomyces Project) media viz., ISP1, 2, 3 and 7, incubated at 28 ± 2 °C for 14 days. Morphological characteristics like pigment formation, growth pattern (recorded under three categories viz., poor, moderate and Good) and the colour of aerial and substrate mycelium were recorded.
2.6 Antifungal activity against phytopathogenic fungi
Four phytopathogenic fungi viz., Fusarium udum (NAIMCC-F-02017), Fusarium oxysporum f. sp. ciceris (NAIMCC-F-00889), Macrophomina phaseolina (NAIMCC-F-01260) and Sclerotium rolfsii (NAIMCC-F-01638) were collected from NAIMCC. They were grown on potato dextrose agar plates with an incubation at 28 ± 2 °C for 24–120 h and maintained on PDA slants and mineral oil for further use. Freshly grown cultures were used for antifungal assays. A dual culture assay was employed to test the antagonistic activity of Streptomyces spp. against four selected phytopathogens. SCA and PDA in the ratio of 1:1 was used as a medium to test the antagonistic activity. Fresh cultures of the bacterium were inoculated on two sides of the plate with the selected medium. The centre of the plate was inoculated with fresh mycelium of the test pathogens and the plates were incubated at 28 ± 2 °C for 96 h. Plates were observed twice a day during incubation for inhibition zones. Plates inoculated with test pathogen alone were also incubated simultaneously as a control. The zone of inhibition and percent inhibition of fungi were calculated using the following formula. where F = radius of fungal growth from the centre (cm) in the control plate.
A = the radius of radial fungal growth from the centre (cm) in the dual culture plate.
3 Results
3.1 Actinomycetes community composition
Isolation on the selective media resulted in a total of sixty-five isolates from 40 samples collected from Tamil Nadu (Table 1). Identification based on 16S rRNA gene sequencing and blast search against type species database revealed that these 65 isolates were affiliated to two genera belonging to Streptomyces and Nocardiopsis (Fig. 1).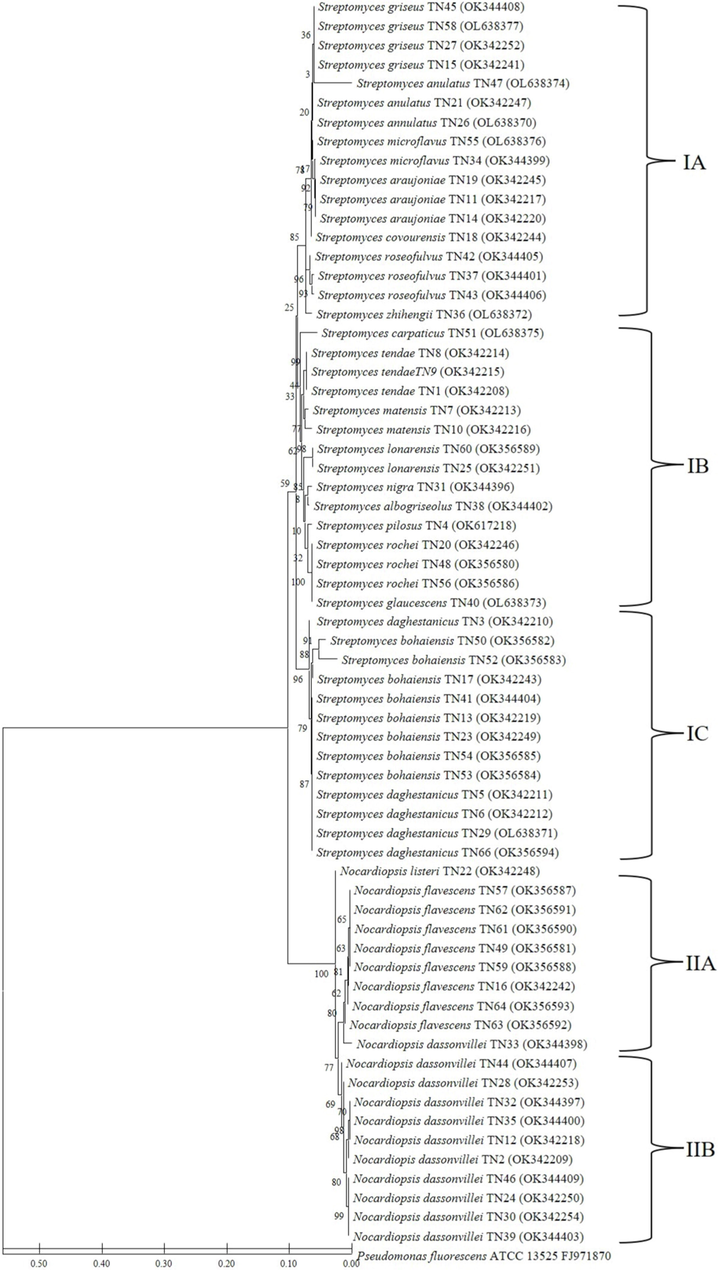
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationship between the 45 Streptomyces and 20 Nocardiopsis strains. Bootstrap value based on 1000 resampled datasets are shown at branch nodes.
3.2 Streptomyces
The genus Streptomyces represents 45 isolates accounting for 67.69 % of the total isolates. It was represented by 18 species viz., S griseus (4), S. anulatus (3), S. microflavus (2), S. covourensis (1), S.araujoniae (3), S. zhihengii (1), S. roseofulvus (3), S. daghestanicus (5), S. carpaticus (1), S. matensis (2), S. glaucescens (1), S. tendae (3), S. nigra (1), S. albogriseolus (1), S. pilosus (1), S. rochei (3), S. bohaiensis (8), S. lonarensis (2).
3.3 Nocardiopsis
Identified isolates include Nocardiopsis representing 32.31 % of the total isolates. Isolates belonging to Nocardiopsis are not as diverse as Streptomyces representing only three species viz., Nocardiopsis dassonvillei (11), Nocardiopsis flavescens (8) and Nocardiopsis listeria (1).
3.4 Phylogenetic diversity of actinomycetes based on 16S rRNA gene sequences
The relationship between the two genera obtained in the present study demonstrated through a phylogenetic tree is presented in Fig. 1. All the 65 strains were grouped into two genera Streptomyces and Nocardiopsis; the upper side of the dendrogram representing Streptomyces with 18 species, S. griseus, S. anulatus, S. microflavus, S. covourensis, S. araujoniae, S. zhihengii, S. roseofulvus, S. daghestanicus, S. carpaticus, S. matensis, S. glaucescens, S. tendae, S. nigra, S. albogriseolus, S. pilosus, S. rochei, S. bohaiensis and S. lonarensis. Streptomyces griseus, S. anulatus, S. microflavus and S. araujoniae separate in sub cluster 1A; Streptomyces griseus showed more close similarity with S. anulatus. Streptomyces covourensis, S. zhihengii, S. carpaticus, S. matensis, S. glaucescens, S. nigra, S. albogriseolus, and S. pilosus, were represented by single nodes which separate in different sub cluster. The larger cluster was represented by Streptomyces bohaiensis which separates in subcluster 1C, which is also represented by S. daghestanicus. Major cluster 2 is represented by the genus Nocardiopsis and it has been divided into two sub clusters, 2A and 2B. Sub cluster 2A presents all Nocardiopsis flavescens and single node Nocardiopsis listeria while sub cluster 2B represents all Nocardiopsis dassonvillei.
The phylogenetic tree presenting the relationship among the 45 strains of the genus Streptomyces is presented in Fig. 2. It showed the grouping of sequences into three distinct groups I, II, and III. Group I present a maximum of 17 Streptomyces strains; all three strains of S.araujoniae isolated from three different crop fields which showed 99 % sequence similarity formed a distinct branch in this group. Similarly, in this group S. griseus, S. anulatus, S. microflavus and S. roseofulvus forms separate cluster; these were isolated from different crop fields and showed more than 99 % sequence similarities. One strain of S. covourensis and S. zhihengii in cluster I were collected from the rice rhizosphere. Two strains of S. lonarensis separated in cluster II which showed 99–100 % similarity with respective species and were collected from rice and maize rhizosphere. Interestingly major cluster II comprises 15 strains separated into three subclusters, with a maximum of eight strains that belong to S. bohaiensis, followed by S. daghestanicus. Cluster III presents five species with single isolates of each which includes S. glaucescens, S. pilosus, S. nigra, S. albogriseolus and S. carpaticus; these strains were collected from rice, bulk soil, and grass field as presented in Table 1. Additionally, this cluster also presents 3 strains each of S. rochei and S. tendae; all these six strains were isolated from six different crop fields. Fig. 3 presents the relationship among the members of Nocardiopsis which exhibits the grouping of sequences in two major groups; cluster I comprise a maximum of 10 strains which showed 99–100 % sequence similarities with Nocardiopsis dassonvillei. Out of these 10 strains, seven were isolated from rice rhizosphere, two from coconut fields and one from sorghum rhizosphere. Further cluster II comprises eight strains that showed 99 % sequence similarities with Nocardiopsis flavescens these isolates were from six different crop fields which includes palm, okra, rice, redgram, lemon, and banana. Additionally in this cluster, 1 strain showed 99 % sequence similarities with Nocardiopsis listeri isolated from rice rhizosphere grouped as a separate subgroup.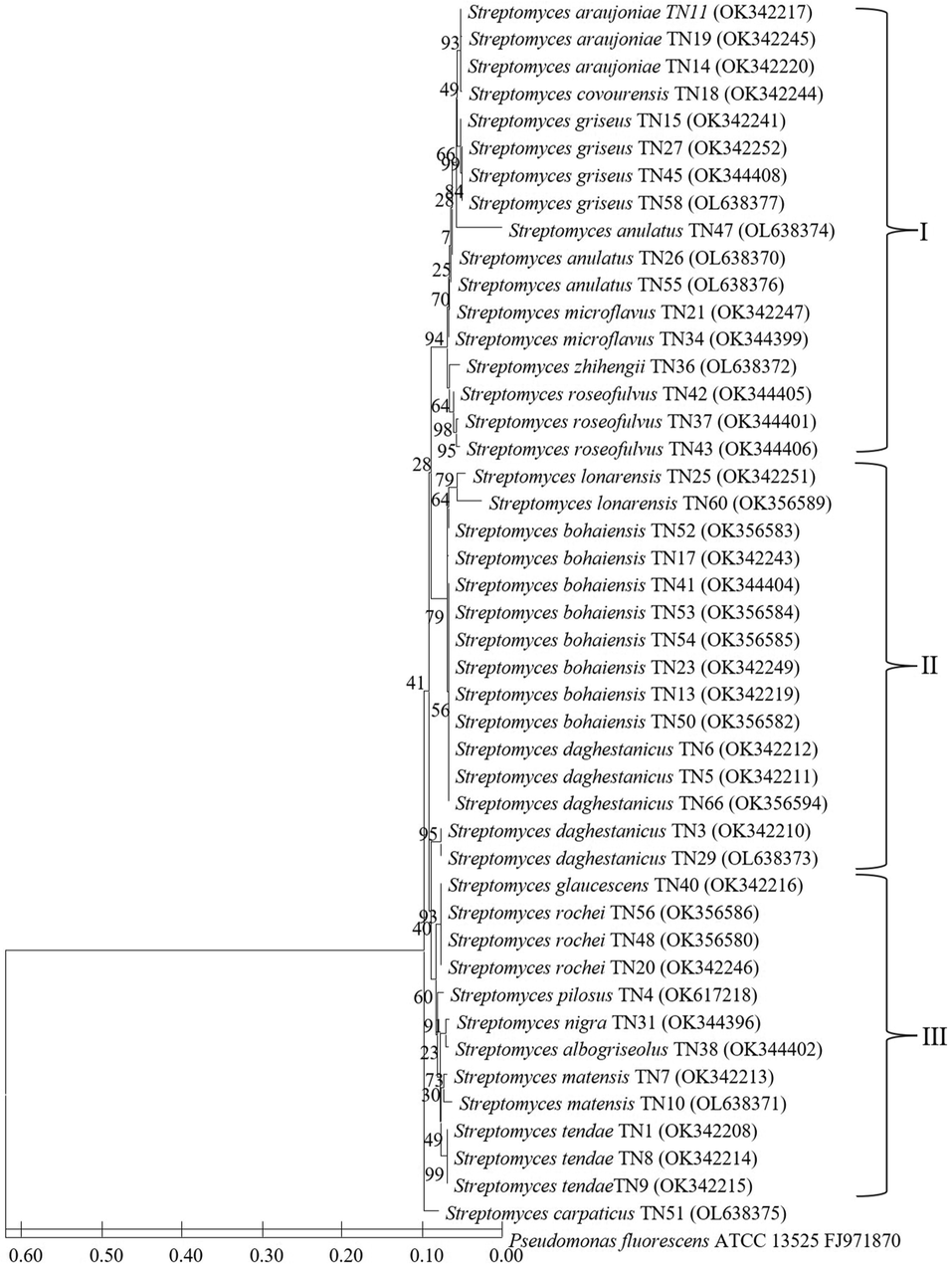
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationship among the 45 Streptomyces strains. Bootstrap value based on 1000 resampled datasets are shown at branch nodes.
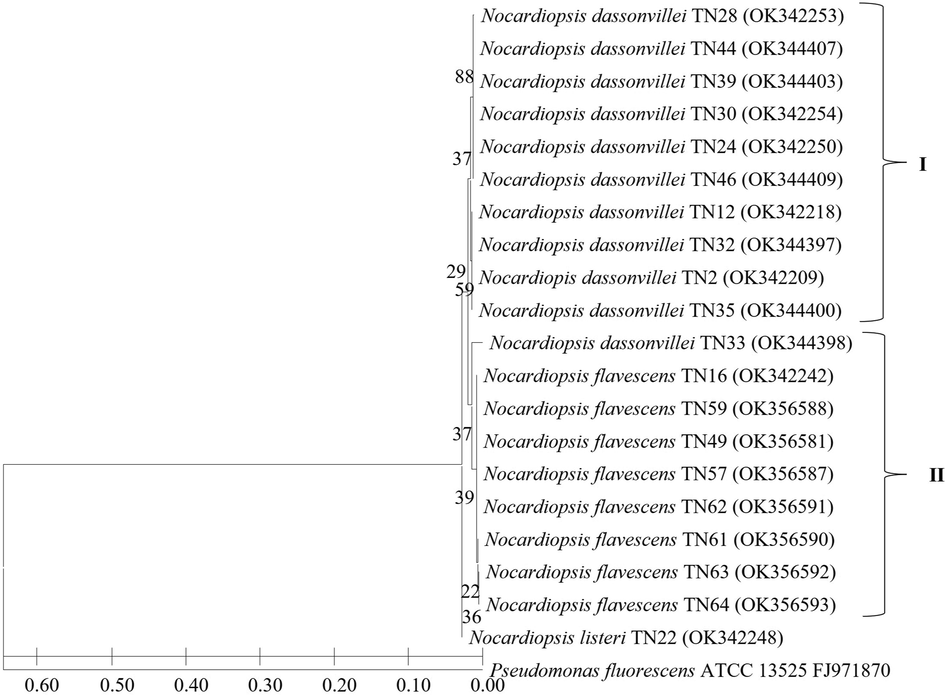
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationship among the 20 Nocardiopsis strains. Bootstrap value based on 1000 resampled datasets are shown at branch nodes.
3.5 Morphological characteristics of actinomycetes
The morphological properties of all 65 actinomycetes strains are presented in Supplementary Table 1. The strains showed differential growth patterns on different ISP media. Six strains viz., S. daghestanicus TN6, S. araujoniae TN14, N. flavescens TN14, S.bohaiensis TN17, S. griseus TN27 and N. dassonvillei TN33 were characterized as showing good growth in all the four ISP media. Twenty-one strains were characterized as showing good growth in atleast three ISP media while seventeen strains showed good growth in atleast two ISP media. Pigment production was observed in twenty strains in atleast one media. S. zhihengii TN 36 produced pigment on all four ISP media and S. araujoniae TN19 produced pigments on three ISP media while the strains S. araujoniae TN11, S. araunoniae TN14 and S. anulatus TN55 produced pigments on atleast two ISP media. The Media ISP-3 facilitated the production of pigment by 18 strains TN1, TN3, TN6, TN8, TN9, T11, TN13, TN14, TN16, TN17, TN19, TN28, TN31, TN35, TN38, TN55, TN63, and TN66; Six strains which include TN1, TN8, TN9, TN11, TN19, and TN28, exhibit pigment formation on ISP-1 media. Five strains showed pigmentation on ISP-4 media which includes TN4, TN5, TN9, TN14, and TN19 while ISP-7 facilitated pigment formation by only one strain (TN9). The colour of aerial and substrate mycelium of all the 65 actinomycetes strains was also recorded which includes white, brown, pale green, pale yellow, yellow, grey, greenish-yellow, light green, green, light yellow and dark green.
3.6 Antifungal activity of actinomycetes strains
Antifungal activities of all the 65 actinomycetes strains against four fungal strains, F. udum, F. oxysporum f.sp. ciceris, M. phaseolina and S. rolfsii. are represented in Table 3. All 65 strains showed antifungal activity against F. udum with inhibition ranging from 1.96 to 63.99 %. S. daghestanicus TN3, N. dassonvillei TN12 and S. araujoniae TN19 showed maximum inhibition with respective inhibition percentages of 63.99, 63.96 and 63.96. Twenty strains showed more than 50 % inhibition against F. udum. Similarly, all the strains exhibited antifungal activity against F. oxysporum f.sp. ciceris with inhibition ranging from 1.79 to 63.81 %. Nine strains showed more than 50 % inhibition against F. oxysporum while strains S. araujoniae TN11 and S. daghestanicus TN3 showed maximum inhibition with a percent inhibition of 63.81 and 61.33, respectively. Out of 65 strains, 48 showed the ability to inhibit Macrophomina phaseolina on plates with inhibitions ranging from 3.75 to 82.50 %. Twenty strains in the study have shown inhibition percent of 70 and above against M. phaseolina while strains S. rochei TN56, N. flavescens TN49 and S. daghestanicus TN 66 showed maximum inhibitions with percent inhibitions of 82.50, 81.67 and 79.17 respectively. Against S. rolfsii 39 strains showed inhibition and inhibition percent was recorded between 3.33 and 52.50. Only three strains viz., S. araujoniae TN11, S. araujoniae TN19 and S. griseus TN27 showed more than 40 % inhibition. A total of 27 strains showed antifungal activity against all four fungal strains, with six strains viz., S. pilosus TN4 (Fig. 4), S. tendae TN8, S. araujoniae TN11, N. dassonvillei TN12 S. araujoniae TN19 and S. griseus TN27 showing considerable percent inhibition against all the four pathogens tested, while 33 strain showed antifungal activity against atleast three fungal strains.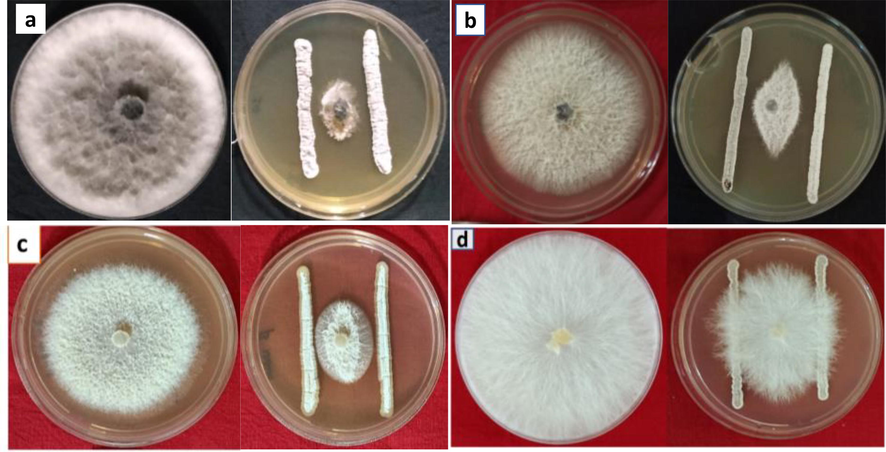
Antifungal activity of Streptomyces pilosus TN4 against different fungal pathogens; Macrophomina phaseolina (a), Fusarium oxysporum (b), Fusarium udum (c) and Sclerotium rolfsii (d).
3.7 Nucleotide sequence accession numbers
The 16S rRNA gene sequences of 65 strains identified in the current study have been submitted to the GenBank nucleotide sequence database (https://www.ncbi.nlm.nih.gov/nucleotide). All the strains isolated and characterized in this study were submitted to NAIMCC (National Agriculturally Important Microbial Culture Collection, ICAR-NBAIM, India (Table 2).
Strain No
Identity
NAIMCC accession number
NCBI accession number
TN1
Streptomyces tendae
NAIMCC-B-02880
OK342208
TN2
Nocardiopis dassonvillei
NAIMCC-B-02865
OK342209
TN3
Streptomyces daghestanicus
NAIMCC-B-02874
OK342210
TN4
Streptomyces pilosus
NAIMCC-B-03007
OK617218
TN5
Streptomyces daghestanicus
NAIMCC-B-02875
OK342211
TN6
Streptomyces daghestanicus
NAIMCC-B-02876
OK342212
TN7
Streptomyces matensis
NAIMCC-B-02878
OK342213
TN8
Streptomyces tendae
NAIMCC-B-02881
OK342214
TN9
Streptomyces tendae
NAIMCC-B-02882
OK342215
TN10
Streptomyces matensis
NAIMCC-B-02879
OK342216
TN11
Streptomyces araujoniae
NAIMCC-B-02868
OK342217
TN12
Nocardiopsis dassonvillei
NAIMCC-B-02866
OK342218
TN13
“Streptomyces bohaiensis
NAIMCC-B-02871
OK342219
TN14
Streptomyces araujoniae
NAIMCC-B-02869
OK342220
TN15
Streptomyces griseus
NAIMCC-B-02877
OK342241
TN16
Nocardiopsis flavescens
NAIMCC-B-02867
OK342242
TN17
Streptomyces bohaiensis
NAIMCC-B-02872
OK342243
TN18
Streptomyces covourensis
NAIMCC-B-02873
OK342244
TN19
Streptomyces araujoniae
NAIMCC-B-02870
OK342245
TN20
Streptomyces rochei
NAIMCC-B-02904
OK342246
TN21
Streptomyces microflavus
NAIMCC-B-02901
OK342247
TN22
Nocardiopsis listeri
NAIMCC-B-02908
OK342248
TN23
Streptomyces bohaiensis
NAIMCC-B-02897
OK342249
TN24
Nocardiopsis dassonvillei
NAIMCC-B-02889
OK342250
TN25
Streptomyces lonarensis
NAIMCC-B-02900
OK342251
TN26
Streptomyces anulatus
NAIMCC-B-02894
OL638370
TN27
Streptomyces griseus
NAIMCC-B-02899
OK342252
TN28
Nocardiopsis dassonvillei
NAIMCC-B-02890
OK342253
TN29
Streptomyces daghestanicus
NAIMCC-B-03002
OL638371
TN30
Nocardiopsis dassonvillei
NAIMCC-B-02891
OK342254
TN31
Steptomyces nigra
NAIMCC-B-02903
OK344396
TN32
Nocardiopsis dassonvillei
NAIMCC-B-02892
OK344397
TN33
Nocardiopsis dassonvillei
NAIMCC-B-02893
OK344398
TN34
Streptomyces microflavus
NAIMCC-B-02902
OK344399
TN35
Nocardiopsis dassonvillei
NAIMCC-B-02894
OK344400
TN36
Streptomyces zhihengii
NAIMCC-B-03010
OL638372
TN37
Streptomyces roseofulvus
NAIMCC-B-02905
OK344401
TN38
Streptomyces albogriseolus
NAIMCC-B-02896
OK344402
TN39
Nocardiopsis dassonvillei
NAIMCC-B-02895
OK344403
TN40
Streptomyces glaucescens
NAIMCC-B-03003
OL638373
TN41
Streptomyces bohaiensis
NAIMCC-B-02898
OK344404
TN42
Streptomyces roseofulvus
NAIMCC-B-02906
OK344405
TN43
Streptomyces roseofulvus
NAIMCC-B-02907
OK344406
TN44
Nocardiopsis dassonvillei
NAIMCC-B-02985
OK344407
TN45
Streptomyces griseus
NAIMCC-B-03004
OK344408
TN46
Nocardiopsis dassonvillei
NAIMCC-B-02986
OK344409
TN47
Streptomyces anulatus
NAIMCC-B-02995
OL638374
TN48
Streptomyces rochei
NAIMCC-B-03008
OK356580
TN49
Nocardiopsis flavescens
NAIMCC-B-02992
OK356581
TN50
Streptomyces bohaiensis
NAIMCC-B-02997
OK356582
TN51
Streptomyces carpaticus
NAIMCC-B-03001
OL638375
TN52
Streptomyces bohaiensis
NAIMCC-B-02998
OK356583
TN53
Streptomyces bohaiensis
NAIMCC-B-02999
OK356584
TN54
Streptomyces bohaiensis
NAIMCC-B-03000
OK356585
TN55
Streptomyces anulatus
NAIMCC-B-02996
OL638376
TN56
Streptomyces rochei
NAIMCC-B-03009
OK356586
TN57
Nocardiopsis flavescens
NAIMCC-B-02987
OK356587
TN58
Streptomyces griseus
NAIMCC-B-03005
OL638377
TN59
Nocardiopsis flavescens
NAIMCC-B-02988
OK356588
TN60
Streptomyces lonarensis
NAIMCC-B-03006
OK356589
TN61
Nocardiopsis flavescens
NAIMCC-B-02989
OK356590
TN62
Nocardiopsis flavescens
NAIMCC-B-02990
OK356591
TN63
Nocardiopsis flavescens
NAIMCC-B-02991
OK356592
TN64
Nocardiopsis flavescens
NAIMCC-B-02993
OK356593
TN66
Streptomyces daghestanicus
*Submitted to NAIMCC
OK356594
Strain
F. udum
F. oxysporum
Macrophomina phaseolina
S. rolfsii
TN1
51.04 ± 9.11
12.50 ± 0.634
40.00 ± 2.04
14.17 ± 0.393
TN2
52.02 ± 1.89
42.38 ± 2.36
71.67 ± 3.12
20.83 ± 1.42
TN3
63.99 ± 2.53
61.33 ± 2.20
63.33 ± 3.12
0.00
TN4
49.05 ± 4.28
51.79 ± 3.78
75.00 ± 4.08
38.33 ± 2.47
TN5
29.03 ± 3.06
5.36 ± 0.186
0.00
21.67 ± 1.57
TN6
1.96 ± 0.080
8.93 ± 0.493
16.25 ± 2.04
0.00
TN7
42.01 ± 2.54
41.71 ± 6.88
45.00 ± 3.06
4.17 ± 0.393
TN8
47.98 ± 3.98
42.05 ± 4.37
47.50 ± 2.04
38.33 ± 5.96
TN9
57.99 ± 4.98
12.50 ± 0.892
23.75 ± 1.02
15.83 ± 1.44
TN10
37.05 ± 4.14
56.95 ± 3.15
71.67 ± 3.12
0.00
TN11
62.03 ± 3.36
63.81 ± 4.38
75.00 ± 4.25
52.50 ± 6.29
TN12
63.96 ± 2.88
49.81 ± 2.69
76.67 ± 4.38
36.67 ± 2.33
TN13
10.01 ± 1.02
5.90 ± 0.224
0.00
0.00
TN14
59.00 ± 3.71
53.57 ± 2.92
74.17 ± 2.36
0.00
TN15
6.98 ± 0.322
5.36 ± 0.148
0.00
0.00
TN16
29.03 ± 3.06
32.62 ± 2.36
32.50 ± 2.04
0.00
TN17
57.96 ± 2.96
46.14 ± 3.27
77.50 ± 4.78
15.00 ± 1.02
TN18
13.01 ± 1.09
37.14 ± 5.31
75.00 ± 3.88
0.00
TN19
63.96 ± 2.88
53.57 ± 3.79
78.75 ± 8.66
44.17 ± 2.85
TN20
50.98 ± 3.96
16.07 ± 0.835
0.00
0.00
TN21
5.98 ± 0.072
4.52 ± 0.124
67.50 ± 5.10
7.50 ± 0.282
TN22
16.04 ± 1.03
23.21 ± 1.86
31.25 ± 2.04
11.67 ± 0.632
TN23
10.01 ± 0.672
20.21 ± 2.77
60.83 ± 5.62
0.00
TN24
51.99 ± 2.56
22.43 ± 1.59
65.42 ± 2.36
12.50 ± 0.552
TN25
36.99 ± 2.63
21.43 ± 1.08
10.00 ± 1.32
0.00
TN26
7.01 ± 0.470
43.10 ± 2.63
0.00
10.83 ± 0.447
TN27
35.23 ± 2.71
45.81 ± 3.70
72.50 ± 6.42
44.17 ± 3.88
TN28
48.93 ± 2.98
46.14 ± 3.27
0.00
25.00 ± 1.65
TN29
1.99 ± 0.090
42.38 ± 2.36
3.75 ± 0.18
10.00 ± 0.435
TN30
3.95 ± 0.230
42.43 ± 3.45
70.00 ± 4.08
10.00 ± 0.242
TN31
46.94 ± 5.64
37.50 ± 2.69
45.00 ± 2.04
0.83 ± 0
TN32
6.98 ± 0.19
46.43 ± 2.92
75.00 ± 6.62
0.00
TN33
5.02 ± 0.185
42.38 ± 2.36
0.00
12.50 ± 0.752
TN34
37.82 ± 2.88
47.10 ± 3.76
65.83 ± 3.12
0.00
TN35
6.98 ± 0.183
50.81 ± 3.95
76.67 ± 5.32
11.67 ± 0.668
TN36
7.01 ± 0.292
8.93 ± 0.672
0.00
8.33 ± 0.244
TN37
7.01 ± 0.578
32.62 ± 2.36
0.00
8.33 ± 0.435
TN38
53.00 ± 4.24
46.14 ± 3.27
66.67 ± 5.65
24.17 ± 1.18
TN39
56.00 ± 3.71
47.00 ± 4.24
0.00
11.67 ± 0.662
TN40
6.98 ± 0.468
32.14 ± 2.92
0.00
10.83 ± 0.543
TN41
4.99 ± 0.178
11.67 ± 0.578
70.00 ± 2.88
0.00
TN42
8.02 ± 0.541
12.50 ± 0.449
0.00
12.50 ± 0.652
TN43
11.02 ± 0.672
9.29 ± 0.326
0.00
5.83 ± 0.432
TN44
6.98 ± 0.135
8.93 ± 0.552
0.00
20.83 ± 1.88
TN45
6.00 ± 0.224
10.71 ± 0.637
77.50 ± 7.72
14.17 ± 0.542
TN46
57.99 ± 2.54
41.07 ± 2.89
73.33 ± 5.36
0.00
TN47
8.02 ± 0.376
8.93 ± 0.472
38.75 ± 2.76
0.00
TN48
57.99 ± 2.54
42.71 ± 2.74
70.83 ± 7.32
17.50 ± 1.12
TN49
59.98 ± 3.92
50.19 ± 2.69
81.67 ± 9.12
0.00
TN50
10.99 ± 1.08
5.36 ± 0.154
67.50 ± 1.98
0.00
TN51
1.96 ± 0.073
8.93 ± 0.438
0.00
0.00
TN52
6.00 ± 0.080
1.79 ± 0.052
0.00
0.00
TN53
2.97 ± 0.240
12.50 ± 0.472
40.00 ± 2.66
10.83 ± 0.242
TN54
44.98 ± 1.92
46.76 ± 3.33
74.17 ± 4.65
0.00
TN55
60.01 ± 2.84
48.67 ± 5.79
75.83 ± 8.44
5.00 ± 0.680
TN56
57.96 ± 2.96
47.14 ± 2.10
82.50 ± 6.35
0.00
TN57
1.96 ± 0.177
8.93 ± 0.558
64.17 ± 5.42
3.33 ± 0.182
TN58
3.98 ± 0.134
12.50 ± 0.738
6.25 ± 0.252
0.00
TN59
4.96 ± 0.273
16.07 ± 1.19
67.50 ± 3.44
5.00 ± 0.152
TN60
6.00 ± 0.082
8.93 ± 0.662
65.00 ± 5.88
12.50 ± 0.822
TN61
7.93 ± 0.495
5.36 ± 0.293
40.00 ± 3.77
10.00 ± 0.680
TN62
56.16 ± 3.52
8.93 ± 0.548
47.50 ± 4.29
0.00
TN63
7.96 ± 0.269
8.93 ± 0.437
0.00
9.17 ± 0.393
TN64
9.95 ± 0.358
23.21 ± 1.79
67.50 ± 5.32
0.00
TN66
63.09 ± 3.96
55.76 ± 3.51
79.17 ± 7.98
14.17 ± 1.35
4 Discussion
Actinomycetes are endowed with metabolic potential to survive and prosper in varied environments including agricultural fields where their presence impacts the survival and growth of other bacteria, as well as plants (Verma et al., 2011; Sadeghi et al., 2012; Bennur et al., 2016; Olanrewaju and Babalola, 2019; Ma et al., 2020). Among the actinomycetes, the genus Streptomyces is endowed with a plethora of metabolic capabilities and hence considered as bacteria of immense importance both in natural environments and Industry. To explore the potential of this group of bacteria it is necessary to understand their diversity, physiology, and ecology (Ma et al., 2020). The aim of this study was to specifically isolate Streptomyces and study their genetic and cultural diversity and explore their antifungal potential against various phytopathogens. A selective media was used to isolate Streptomyces spp. from crops’ rhizosphere and bulk soil collected from different fields and different regions of Tamil Nadu, India. A total of 65 putative Streptomyces isolates were collected, characterized, and identified. Identification based on 16S rRNA gene sequence similarity revealed the isolates fall under two genera viz., Streptomyces and Nocardiopsis. It's worth noting that the 16S rRNA gene sequence's utility as a phylogenetic and taxonomy identifier is restricted. The core genome divergence between Streptomyces strains with 97 percent 16S rRNA gene sequence identity can be as high as 30 %, with an ANI of 100–78.3 % (van Bergeijk et al., 2020). 16S rRNA gene sequence identity of 98.6 % is widely used for bacterial species identification and delineation (Chun et al., 2018); however, for actinomycetes, this threshold is raised to 99 % based on comparative studies between 16S rRNA gene sequences (Guo et al., 2015). As a result, we used a 99–100 % threshold to assign actinomycetes isolates to distinct clusters. Since more than 30 % of the isolates obtained in the study belonged to Nocordiopsis we have taken those isolates too in further analysis. Like Streptomyces spp. members of the genus Nocardiopsis too can survive under different environmental conditions and produce a range of bioactive compounds (Bennur et al., 2016). In the current study, 45 isolates were assigned to Streptomyces spp., and 20 isolates to Nocardiopsis spp. based on 16S rRNA gene sequences. Streptomyces spp. is reported to have advanced adaptability to exist in different extreme environments and habitant of diverse conditions like frozen soils, deserts, and oceans (Okoro et al., 2009; Passari et al., 2018). The findings of this study have shown that there is a high level of diversity within Streptomyces spp. present in the agriculture fields of Tamil Nadu. The isolates collected from different crop rhizosphere and bulk soils of agriculture fields fell under 18 species forming three clades. A similar study on the diversity of streptomycetes in prairie soils resulted in 34 different OTUs, albeit the 16S rRNA gene sequence used for diversity analysis is less than 300 bp in the study (Davelos et al., 2004). In our earlier study with soils of Meghalaya, India the diversity of Streptomyces spp. is higher representing 26 different species (Singh et al., 2022).
Streptomyces bohaiensis is the most abundant (8 isolates) among the 45 Streptomyces spp. isolates found in the rhizosphere of rice, okra, brinjal and chilli and in bulk soil, while S. daghestanicus is the second most abundant isolate (5 isolates). A study earlier has shown the presence of Streptomyces spp. in 20 different plant communities (Adil et al., 2017). In the present study, we have isolated Streptomyces spp. from 34 samples among the 40, representing rhizosphere of different crops like rice, maize, sorghum, groundnut, sunflower, castor, brinjal, okra and chilli, bulk soils of coconut, jatropha and palm fields, thus confirming their ubiquity in soils. Cultural characterization on different ISP media is a routine study involved in the characterization of actinomycetes. In our study, cultural characterization on four different ISP media has shown immense diversity among the soil actinomycetes. Isolates belonging to the same species gave different characterizations among themselves reiterating the fact that there are strain level differences. More than twenty strains showed pigment production in various ISP media used. Earlier many researchers reported similar pigmentation by different actinomycetes strains (Fernandes et al., 2021; Amsaveni et al., 2015).
Antifungal activity of actinomycetes especially, Streptomyces spp. are reported in various studies (Rey and Dumas, 2017; Tamreihao et al., 2018; Marimuthu et al., 2020; Sholkamy et al., 2020; Gebily et al., 2021). They have been found to protect a variety of plants from soil-borne fungal diseases to varying degrees. Similarly, the members of the genus Nocardiopsis are known for their antifungal activity against different phytopathogens (Bennur et al., 2016). The inherent problems of resistance development by the target fungi and residue in the environment, associated with disease control measures by chemical fungicides led to the search for alternative plant protection measures. Due to the lack of novel antimicrobial metabolites, more and more researchers are focusing their efforts on different ecosystems. In the present study, antifungal activity assay against four fungal pathogens has shown that all 65 actinomycetes strains showed antifungal activity against both F. udum and F. oxysporum f. sp. ciceris. This is consistent with the results of Amini et al. 2016 who reported all 112 actinobacteria isolated as antifungal against F. oxysporum f.sp. ciceris. In the current study, 74.25 % strain showed antifungal activity against M. phaseolina and 60.60 % against S. rolfsii. Singh et al. (2016) tested the antifungal properties of 80 actinomycetes strains against Rhizoctonia solani, Fusarium solani, Macrophomina phaseolina, Sclerotium rolfsii, and Colletotrichum truncatum, and found various level of antifungal properties of different strains. Kamara and Gangwar (2015) isolated 100 actinomycetes strains from 30 rhizospheric soil samples of Catharanthus roseus and Withania somnifera from different locations of Ludhiana, India, and tested their antifungal activity against viz: Sclerotium rolfsii, Rhizoctonia solani, Helminthosporium oryzae, Macrophomina phaseolina, Penicillium sp., Fusarium oxysporum and Alternaria alternata. These findings along with our results suggest that rhizospheric soils have the prospective to identify actinomycetes with potent antifungal activity. To the best of our understanding, the present study offers for the first time a prelude about the unexplored streptomycetes and Nocardiposis diversity associated with different crop root rhizosphere and bulk soil from different locations in Tamil Nadu, India.
5 Conclusion
Our findings suggest that there exists a higher level of diversity among the members of the genera Streptomyces and Nocardiopsis in agricultural fields of Tamil Nadu although the latter is less diverse as compared to Streptomyces spp. The study also demonstrates that members of these genera inhabiting the rhizosphere and bulk soils of the study area possess great potential as antifungal agents. Their utilities as biocontrol agents are to be explored in further studies.
Authors contributions
PT, MTZ and SCK performed isolation, morphological characterization, and antifungal activity assay. WAA performed the phylogenetic analysis and compiled the data. MK planned and supervised the experiments and prepared the first draft. HC and AKS collected samples, planned experiments, and improved the manuscript.
Acknowledgments
Authors are grateful for Indian Soil Microbiome Project, ICAR-NBAIM, Mau, India, for providing funds required for sample collection and experimentation. Authors also express sincere gratitude to the Director ICAR-NBAIM, for providing infrastructure facility.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Plant community richness mediates inhibitory interactions and resource competition between Streptomyces and Fusarium populations in the rhizosphere. Microbiol. Ecol.. 2017;74:157-167.
- [Google Scholar]
- Evaluation of Streptomyces spp. against Fusarium oxysporum f. sp. ciceris for the management of chickpea wilt. J. Plant. Prot. Res.. 2016;56(3):1-8.
- [Google Scholar]
- Screening and isolation of pigment producing Actinomycetes from soil samples. J. Biosci. Nanosci. 2015;2(2):24-28.
- [Google Scholar]
- Nocardiopsis species: a potential source of bioactive compounds. J. Appl. Microbiol.. 2016;120(1):1-16.
- [Google Scholar]
- Diversity and versatility of actinomycetes and its role in antibiotic production. J. Appl. Pharm. Sci.. 2013;3(8):S83-S94.
- [Google Scholar]
- Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol.. 2018;68(1):461-466.
- [Google Scholar]
- Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microbial Ecol.. 2004;48(4):601-612.
- [Google Scholar]
- Plant growth and resistance promoted by Streptomyces spp. in tomato. Plant Physiol. Biochem.. 2017;118:479-493.
- [Google Scholar]
- Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J.. 2010;4(5):642-647.
- [Google Scholar]
- Isolation and identification of pigment producing actinomycete Saccharomonospora azurea SJCJABS01. Biomed. Pharmacol. J.. 2021;14(4):2261-2269.
- [Google Scholar]
- Characterization and potential antifungal activities of three Streptomyces spp. as biocontrol agents against Sclerotinia sclerotiorum (Lib.) de Bary infecting green bean. Egyptian J. Biol. Pest Control. 2021;31(1):1-15.
- [Google Scholar]
- Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol.. 2015;81(9):3086-3103.
- [Google Scholar]
- Mechanism of phosphate solubilization and antifungal activity of Streptomyces spp. isolated from wheat roots and rhizosphere and their application in improving plant growth. Microbiol.. 2014;160(4):778-788.
- [Google Scholar]
- Antifungal Activity of Actinomycetes from Rhizospheric Soil of Medicinal plants against phytopathogenic fungi. Int. J. Curr. Microbiol. Appl. Sci.. 2015;4(3):182-187.
- [Google Scholar]
- Biocontrol and plant growth promoting potential of phylogenetically new Streptomyces sp. MR14 of rhizospheric origin. AMB Express. 2019;9(1):1-14.
- [Google Scholar]
- MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [Google Scholar]
- Deciphering the diversity of culturable thermotolerant bacteria from Manikaran hot springs. Annals Microbiol.. 2014;64(2):741-751.
- [Google Scholar]
- Phylogenetic and physiological diversity of cultivable actinomycetes isolated from alpine habitats on the Qinghai-Tibetan Plateau. Front. Microbiol. 2020:2218.
- [Google Scholar]
- Antifungal activity of Streptomyces sp. SLR03 against tea fungal plant pathogen Pestalotiopsis theae. J. King Saud Univ. Sci.. 2020;32(8):3258-3264.
- [Google Scholar]
- Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 2009;95(2):121-133.
- [Google Scholar]
- Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol.. 2019;103(3):1179-1188.
- [Google Scholar]
- Bioprospection of actinobacteria derived from freshwater sediments for their potential to produce antimicrobial compounds. Microb. Cell Factories. 2018;17(1):1-14.
- [Google Scholar]
- Plenty is no plague: Streptomyces symbiosis with crops. Trends Plant Sci.. 2017;22(1):30-37.
- [Google Scholar]
- Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol.. 2012;28(4):1503-1509.
- [Google Scholar]
- The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.. 1987;4(4):406-425.
- [Google Scholar]
- Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev.. 2012;36(4):862-876.
- [Google Scholar]
- Antimicrobial quercetin 3-O-glucoside derivative isolated from Streptomyces antibioticus strain ess_amA8. J. King Saud Univ. Sci.. 2020;32(3):1838-1844.
- [Google Scholar]
- Genetic Diversity and Anti-Oxidative Potential of Streptomyces spp. Isolated from Unexplored Niches of Meghalaya, India. Curr. Microbiol.. 2022;79(12):379.
- [Google Scholar]
- Characterization of actinomycetes against phytopathogenic fungi of Glycine max. Asian J. Pharm. Clin. Res.. 2016;9(Suppl 1):216-219.
- [Google Scholar]
- SeqTrace: a graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech.. 2012;23(3):90.
- [Google Scholar]
- Isolation and identification of Streptomyces spp. from Venezuelan soils: morphological and biochemical studies. Microbiol. Res.. 2006;161(3):222-231.
- [Google Scholar]
- Acidotolerant Streptomyces sp. MBRL 10 from limestone quarry site showing antagonism against fungal pathogens and growth promotion in rice plants. J. King Saud Univ. Sci.. 2018;30(2):143-152.
- [Google Scholar]
- Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat. Rev. Microbiol.. 2020;18(10):546-558.
- [Google Scholar]
- Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol.. 2011;51(5):550-556.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102619.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







