Translate this page into:
First report on morphological and molecular characteristics of eight Agaricomycotina mushrooms in Saudi Arabia
⁎Corresponding author. gkhaled@ksu.edu.sa (Jamal M. Khaled)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Some fruiting body fungi, such as mushrooms, have valuable nutritional and medicinal properties. The present study aimed to report some mushroom species for the first time in several regions of Saudi Arabia: Abha, Al-Baha, Al-Taif, and Jeddah. Morphological and microscopic features, the internal transcribed spacer (ITS) region, and a Basic Local Alignment Search Tool (BLAST) analysis were studied to identify Crinipellis scabella, Parasola conopilea, Agrocybe pediades, Pyrofomes demidoffii, and Tulostoma fimbriatum in Al-Baha, and Conocybe macrospora, and Chlorophyllum palaeotropicum throughout Saudi Arabia. All mushrooms identified in this present work were Agaricomycotina mushrooms recorded for the first time in Saudi Arabia.
Keywords
Agaricomycotina
Mushrooms
Fruiting body fungi
Molecular phylogeny
1 Introduction
Recently, there has been a significant increase in research and food production of edible mushrooms worldwide, as they have a great medicinal and nutritional value (Adeniyi et al., 2018). It has been reported that some fruiting bodies of edible mushrooms are considered important natural ingredients in many medicinal agents such as anti-inflammatories, trehalose, oleic acid, gallic acid, melatonin, biotin, lycopene, and zinc can be extracted from Agaricus bisporus and Lentinus edodes (Muszyńska et al., 2018). Many scientific reports have confirmed that numerous fruiting bodies of mushrooms (such as Lentinus spp, Auricularia spp, Hericium spp, Grifola spp, Flammulina spp, Pleurotus spp, and Tremella spp) have immunomodulation, anti-obesity, hepato-prophylactic, antifungal, antibacterial, antiviral, antinociceptive, antidiabetic, antitumor, anticancer, and antioxidant agents properties (Ganesan and Xu, 2018).
The fruiting bodies of mushrooms are generally considered among the most biodiverse organisms on earth, for this reason, much research has been undertaken to investigate these microbes. The fungal kingdom includes more than 5 million species worldwide that play essential environmental roles and can colonize nearly all natural environments. They also have several nutritional and medicinal benefits (Martins, 2017). Global estimates indicate that there are more than 2300 and 2100 species of wild fruiting fungi that are edible and medicinal fungal strains, respectively (Martins, 2017).
Hibbett (2006) has reported that almost one-third of the identified fungal species belong to the Agaricomycotina, also called the hymenomycetes. Agaricomycotina is a subdivision belonging to the division Basidiomycota. The subdivision Agaricomycotina contains most of the described mushrooms which can produce fruiting bodies with hymenial layers. This subdivision consists of four classes “Agaricomycetes, Dacrymycetes, Walleomycetes and Tremellomycetes” (Lestari et al., 2019).
Biodiversity studies on mushrooms have been done in local environments in many countries, including India (Lakhanpal, 2014), China (Zhang et al., 2021), USA (Martins, 2017), Nigeria (Vyas et al., 2014), Indonesia (Siahaan et al., 2019), Egypt (Soliman and El-Sayed, 2021), Saudi Arabia (Abou-Zeid and Altalhi, 2006), and Iraq (Toma et al., 2013).
Obtaining maximum benefit from mushrooms depends on local survey studies to localize the cultivation of beneficial strains, then small- and large-scale production of bioactive compounds can be established. Hence, the present work aimed to study the biodiversity of wild mushrooms in four regions (Al-Taif, Al-Baha, Abha, and Jeddah) in Saudi Arabia.
2 Materials and methods
2.1 Study regions and design
The study regions included four governorates, Al-Taif, Al-Baha, Abha, and Jeddah, with GPS coordinates 21.2841°N, 40.4248°E; 20.0217°N, 41.4713°E; 18.2465°N, 42.5117°E; and 21.4858°N, 39.1925°E, respectively. Each region was divided into four large squares characterized by the presence of vegetation cover and humidity using a completely randomized design.
2.2 Collection of samples
The fruiting body fungi samples were collected from the above regions between November 1, 2020, and January 30, 2021. The dust and dirt were removed from the samples using a gentle air stream and soft brushes. The samples were kept in sterile plastic boxes then transferred to the Microbiology Laboratory, Botany and Microbiology Department, College of Science, King Saud University, Riyadh, Saudi Arabia.
2.3 Macroscopic and microscopic characteristics
The structure of the fruiting body and their spores (spore color, attachment, symmetry, size, ornamentation, shape) were recorded according to Largent et al., (1977).
2.4 Preparation of spore suspension
For each sample, a section of the gill of the mushroom was mixed well with sterile normal saline (0.089 % NaCl), and the spore suspension was collected by centrifugation at 3000×g for 15 min at 25 °C. The total count of spores was calculated using direct microscopic count by hemocytometer then the serial dilution was applied to obtain 1 mL of sterile normal saline solution containing approximately 5 ± 2 spores. The microscopic characteristics of the spores were studied using digital light microscopy (Motic Plus 2.0 ML, China); after that, the spore suspensions were stored in sterile glycerol solution (30 % v/v) at –80 °C till further analysis.
2.5 DNA extraction, amplification internal transcribed spacer (ITS) region, and sequencing PCR products
The DNA was extracted from the collected mushrooms using DNeasy Plant Mini Kit (Qiagen) following the manufacturer's protocol. The pure DNA product was preserved at –80 °C for future investigations. Amplification of ITS region was done using forward primer (5′-TCCGTAGGTGAACCTGCGG-3′) and reverse primer (5′-GCTGCGTTCTTCATCGATGC-3′) (Usyk et al, 2017). The program of PCR was designed using Optimase ProtocolWrtiter™ in a simple multiple-steps PCR protocol.
PCR product length was 600 bp. Purification of PCR products was done using a montage PCR clean-up kit (EMD Millipore, Burlington, MA, USA) to remove the PCR primers and deoxynucleotide triphosphates that were not included as part of the product. BigDye™ Terminator Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA) and Applied Biosystems model 3730XL automated DNA sequencing system were used to sequence the pure PCR products.
2.6 Identification and BLAST analysis
The sequenced PCR products were edited using Bioedit software (BioEdit Sequence Alignment Editor), then the nucleotide basic local alignment search tool (nucleotide – nucleotide BLAST) analysis was done in GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The identification was performed using the morphological and spore features, together with BLAS analysis. Evolutionary analysis of identified samples was done using the maximum likelihood test in MEGA-X v10.1.8 software according to Hasegawa et al., (1985) and Kumar et al. (2018).
3 Results
BLAST analysis, morphological and spore features of query-63801 sample collected from Al-Baha is a mushroom strain belonging to C. scabella (Figs. 1 and 2). For details of the analysis (the max score, total score, query cover, E value, percent identity, and accession length for all BLAST analysis), see tables in the supplementary section. Figs. 1 and 2 show the macroscopic and microscopic characteristics as well as the BLAST analysis of samples (query-35787) collected from Al-Baha, Saudi Arabia. The results confirmed that samples were P. conopilea (Table 2 in the supplementary section). The data obtained from Figs. 1 and 3 confirm that the mushroom sample (query-45217) collected from Al Baha were strains of A. pediades (Table 3 in the supplementary section).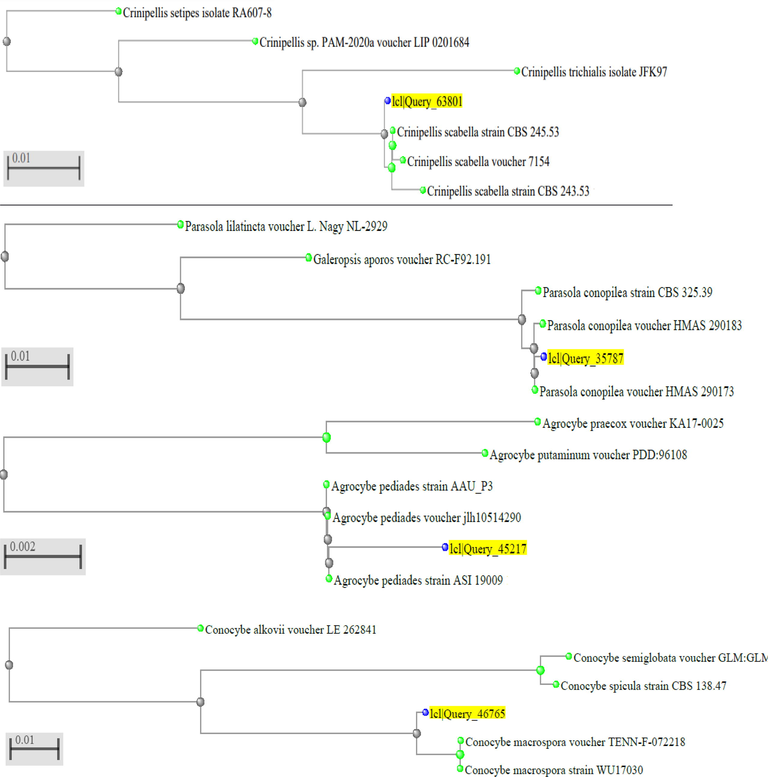
BLAST analysis of mushroom ITS sequence for query-63801 (C. scabella), query-35787 (P. conopilea), query-45217 (A. pediades) and query-46765 (C. macrospora).
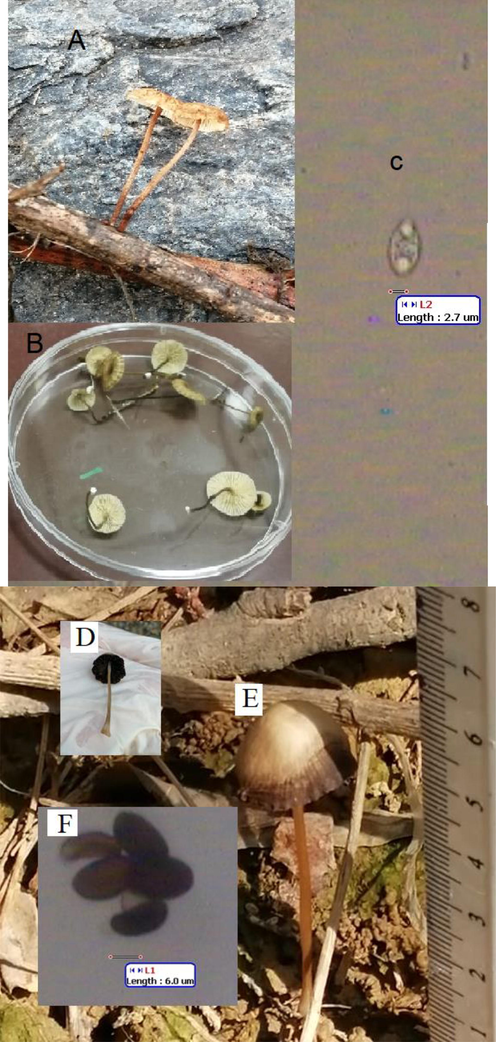
Morphologic and spore characteristics of C. scabella and P. conopilea collected from Al Baha, Saudi Arabia, in the field (A), in the laboratory (B) and image of single spore (C) for C. scabella, in the laboratory (D), in the field (E), and image of spores (F) for P. conopilea.

Morphologic and spore characteristics of A. pediades collected from Al Baha, Saudi Arabia in the laboratory (A), in the field (B) and image of spores (C).
The mushroom samples (query-46765) collected from Al-Taif were investigated using BLAST analysis and their morphological characteristics (Figs. 1 and 4). The data showed the samples belong to C. macrospora (Table 4 in the supplementary section). Figs. 5 and 6 show the BLAST analysis, macroscopic characteristics, and spores of the samples (query-57701) collected from Al-Baha. The data reported that the samples belong to Agrocybe pusiola strains (Table 5 in the supplementary section).
Morphologic and spore characteristics of C. macrospora collected from Al-Taif, Saudi Arabia. In the field (A and B), and image of spores (C).
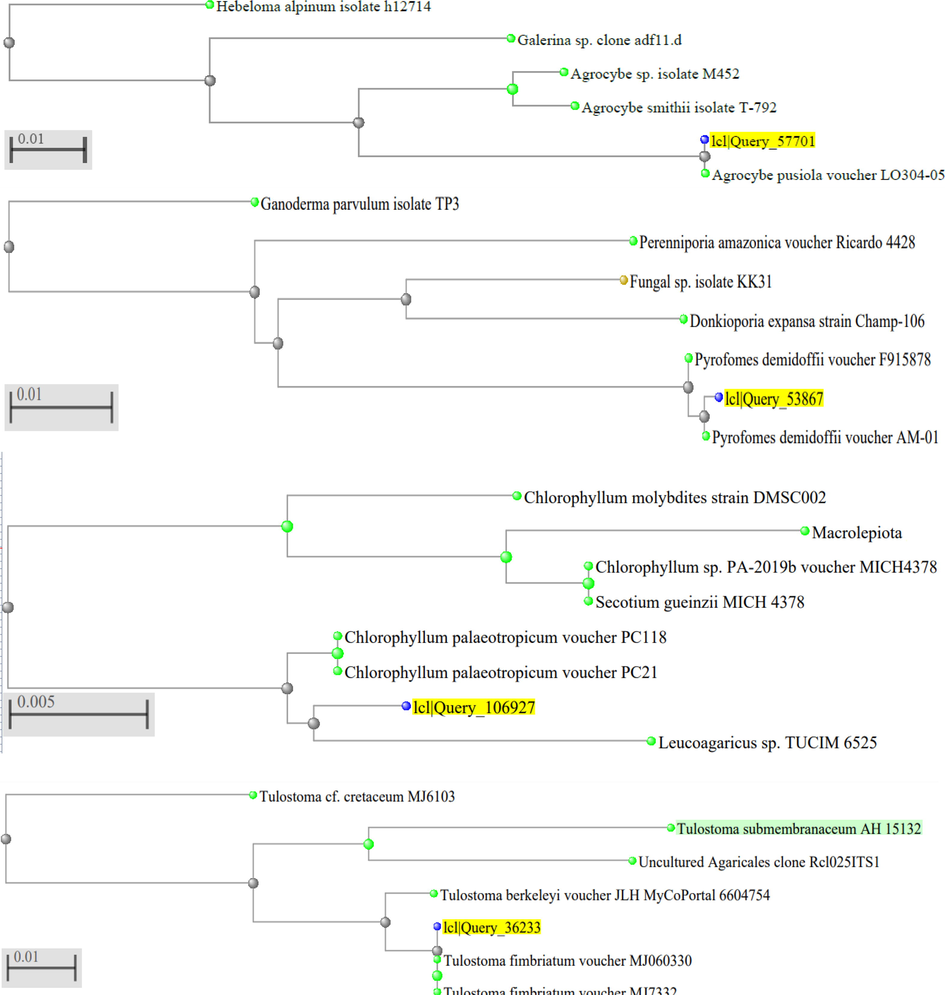
BLAST analysis of mushroom ITS sequence for query-57701 (A. pusiola), query-53867 (P. demidofii), query-106927 (C. palaeotropicum), and query-36233 (T. fimbriatum).
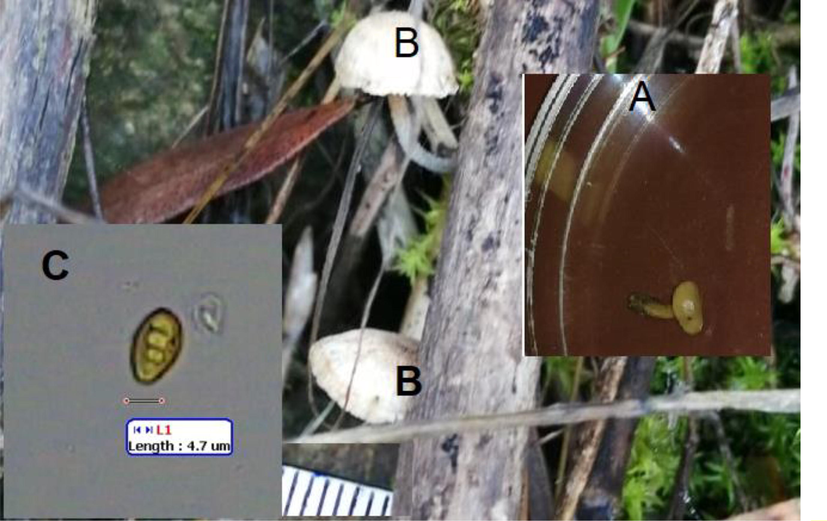
Morphologic and spore characteristics of A. pusiola collected from Al Baha, Saudi Arabia. In the laboratory (A), in the field (B) and image of single spore.
The morphological features and BLAST analysis of the samples (query-53867) collected from Al-Baha indicated the mushroom samples were Pyroformes demidofii strains (Figs. 5 and 7). The identification parameters of BLAST were listed in Table 6 in the supplementary section.
Morphologic and spore characteristics of P. demidoffii collected from Al-Baha, Saudi Arabia. In the field (A) and image of single spore (B).
The BLAST analysis, morphologic characteristics, and spore features of C. palaeotropicum collected from Jeddah (query-106927) are arranged in Figs. 5 and 8 and the details of BLAST analysis was arranged in Table 7 in the supplementary section.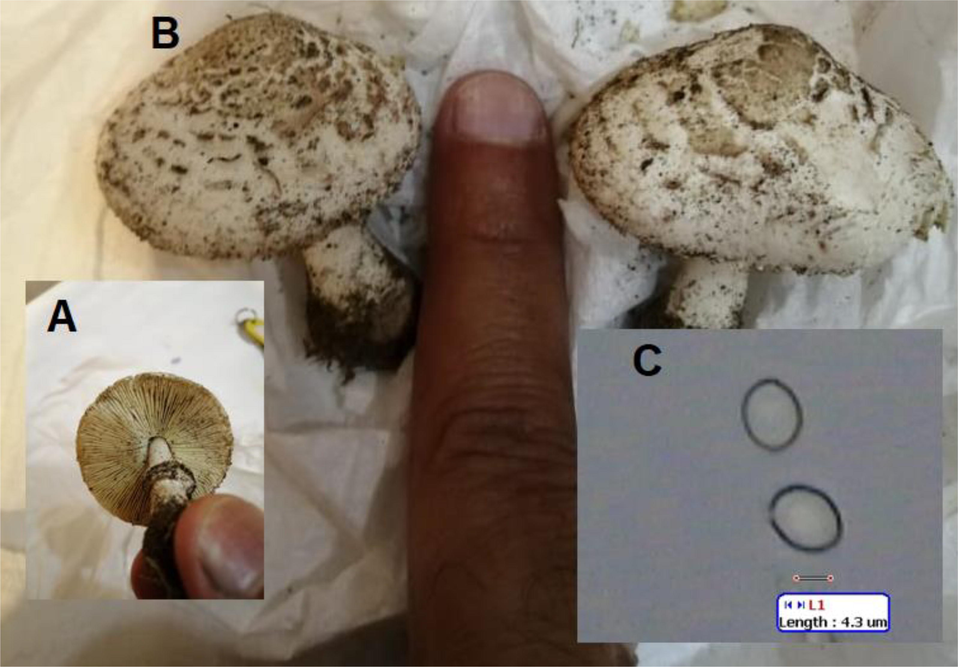
Morphologic and spore characteristics of C. palaeotropicum collected from Jeddah, Saudi Arabia. In the laboratory (A and B) and image of spores (C).
Figs. 5 and 9 show the BLAST analysis, morphologic characteristics, and spores features of T. fimbriatum collected from Al-Baha (query-36233). The identification parameters of BLAST analysis was arranged in Table 8 in the supplementary section.
Morphologic and spore characteristics of T. fimbriatum collected from Al-Baha, Saudi Arabia. In the laboratory (A), in the field (B) and image of spores (C).
Phylogenetic analyses and models of DNA sequence evolution (substitution model) of the identified mushroom samples were done using the maximum likelihood method and Hasegawa-Kishino-Yano model. The relative rates of various changes among all samples are shown in Fig. 10. The results showed that C. palaeotropicum and P. demidofii originated (derived) from ancestral strains of the tested mushroom samples with the 56 certainty degree (it represents the value of certainty the fungal strains originated (derived) from ancestral strains). C. macrospora, T. fimbriatum and C. scabella originated from ancestral fungal strains with 82 certainty degree. The certainty degree of A. pediades, A. pusiola and P. conopilea was 56. The taxonomy analysis showed that the mushroom samples comprised eight species belonging to five families (Maramiaceae, Psathyrellaceae, Bolbitiaceae, Agaricacea, and Polyporaceae) in two orders (Agaricales and Polyporales) and in one class (Agaricomycotina) (Figure 1 in the supplementary section).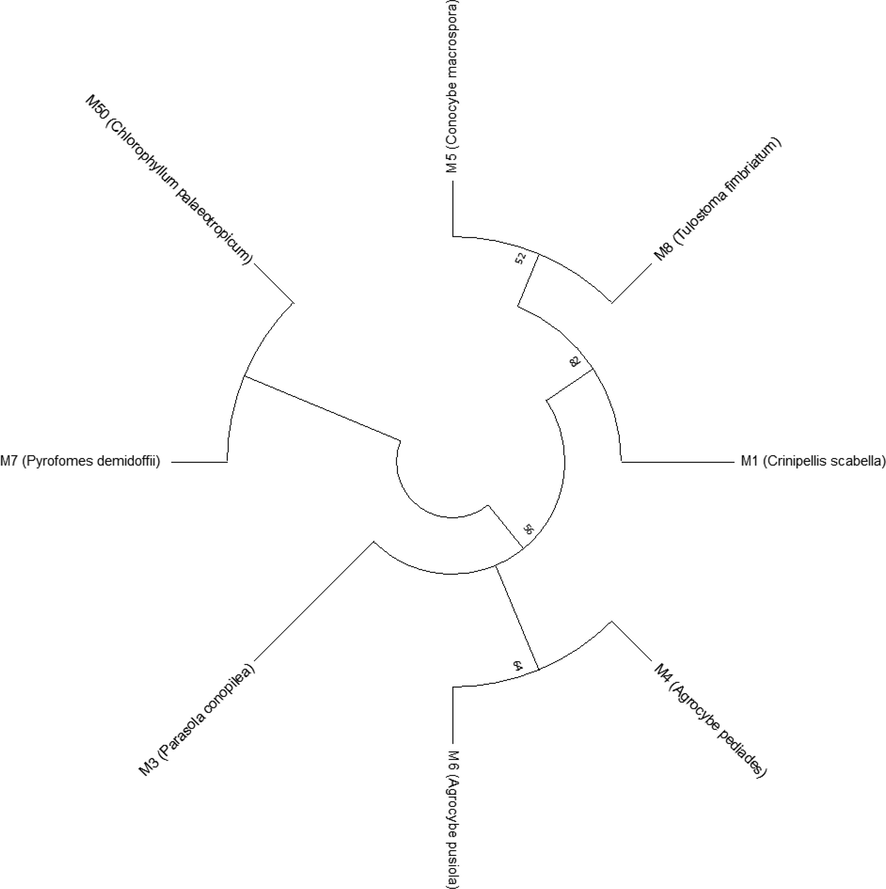
The evolutionary history using the maximum likelihood method and Hasegawa-Kishino-Yano model.
4 Discussion
In the present study, as far as we know, we were able to collect and identify-eight mushroom species for the first time in Saudi Arabia. On the other hand, we could not collect (and include in our work) some mushroom species identified by previous studies of the same regions. In 2019, Manzelat published results reporting that Morchella spp., Agaricus spp., Agrocybe spp., Amanita spp., Boletus spp., Coprinus spp., Lepiota spp, Pleurotus spp, and Podaxis spp could be collected from the Abha and Jizan regions (Manzelat, 2019). Before that, in 2006, Abou-Zeid and Altalhi collected mushrooms samples from some regions of Al-Taif; these were identified according to their morphological characteristics as A. arvensis, A. bisporus, A. augustus, Lepiota procera, Lepiota rhacodes, Lepiota cristata, Pleurotus ostreatus, Pleurotus cornucopiae, Coprinus comatus, A. cylindracea, Pleurotus pistillaris, Inocybe splendens, Phaelepiota aurea, and Boletus edulis (Abou-Zeid and Altalhi, 2006).
We could not collect many mushrooms species like collected previously in the above studies such as Morchella spp., Amanita spp., Boletus spp., Coprinus spp., Lepiota spp, Pleurotus spp, Podaxis spp, Inocybe spp., and Phaelepiota spp. In fact, in our present study, all mushroom species (C. scabella, P. conopilea, A. pediades, C. macrospora, P. demidofii, C. palaeotropicum, and T. fimbriatum) that were collected and identified were new mushroom species.
In 2010, Antonín and Noordeloos described C. scabella as having a small fruiting body (basidiocarp) with orange-brown to red-brown cap (pileus) hairs on a pale cream white background, and its basidiospores were wide with differently fashioned cheilocystidia (large cells on the edge of the gill) (Antonín and Noordeloos, 2010). Adamczyk reported that C. scabella grows on grasses that are considered special substrates for this mushroom (Adamczyk, 2016). In our present study, we collected C. scabella from one of the deep valleys of Al-Baha containing perennial trees, hardwood, shade, water, and grass. This valley is characterized by little human activity. Adamczyk has reported that more than 40 mushroom species (including C. scabella) showed biodiversity related to abandoned lands and the development stage of those lands (Adamczyk, 2016). According to https://www.gbif.org/species/2538113 (8/20/2021), C. scabella has recorded 2369 occurrences (not including Saudi Arabia) worldwide, and there are 4 infraspecies of this fungus. There are only 17 nucleotides of C. scabella strains that have been identified and their data have been deposited in NCBI, and according to BLAST tree analysis, there were only 7 strains of C. scabella (data not shown).
P. conopilea (formerly named Psathyrella conopilea) was collected from the same valley as C. scabella. P. conopilea features a breakable main body, dark basidiospores, and a hygrophanous brown pileus surface. In addition, it can grow well in groups of 2–4 fungi (but not in crowded groups) in cultivated regions or hardwood forests on wood debris or manure (dung) (Kuo, 2011). There are 38 nucleotides of P. conopilea strains that have been analyzed in NCBI (data not shown). According to https://www.gbif.org/species/10364983 (8/20/2021), P conopilea has recorded 2343 occurrences (not including Saudi Arabia) worldwide. P. conopilea grows well in spring and fall and has been collected from several regions in Hungary and USA (Ganga and Manimohan, 2019). The presence of these mushrooms in the local environment encourages their use for the production of bioactive compounds; for example, P. conopilea is a natural resource of drosophilid A (Riquelme et al., 2020).
In the present study, A. pediades and A. pusiola were also collected from the same location in Al-Baha. There are species of Agrocybe (but not A. pediades and A. pusiola) that have been collected in Saudi Arabia in previous studies from the Al-Taif, Abha, and Jizan regions (Abou-Zeid and Altalhi, 2006; Manzelat, 2019). Agrocybe is distinguished by basidiospores with a reduced to broad germ-pore, collybioid to tricholomatoid basidiomata (spore-producing structure with a fleshy club shape) with rusty to dark spore-print, and a hymeniform (spore-bearing surface) pileipellis (the superior layer of hyphae in the cap). A. pediades have basidia (sporangium or spore-producing structure) with four sterigmata (the apical extension of the basidium) and pileus (caps) with appendiculate annular remnants residues (Niveiro et al., 2020). A. pusiola contains numerous nuclei when stained with acetocarmine, and has a veil coated with gelatin (Arnolods, 2006). The nucleotides deposited NCBI related to A. pediades reached 1504 whereas there were not more than 2 nucleotides for A. pusiola according to research using https://www.ncbi.nlm.nih.gov/taxonomy (8/20/2021). According to research using https://www.gbif.org/species/search (8/20/2021), there are more than 20,500 occurrences of Agrocybe including 127 species (although they have a global widespread, they have not yet been recorded in Saudi Arabia) where there has been 4826 and 150 recorded occurrences of A. pediades and A. pusiola, respectively, among all occurrences of Agrocybe.
The only mushroom species we could get from Al-Taif was C. macrospora. This does not mean that Al-Taif is free of other mushrooms, but as time and location are crucial factors in detecting the mushrooms, the mushrooms you may find today may not be found tomorrow in the same location. They can grow singly or in groups on manured soil, burnt land, forests, and grasslands. Its front view is ellipsoid to somewhat lemon-shaped; its side view is ellipsoid, slightly spread ventrally or slightly amygdaliform, germ-pore central, about 3.0 μm wide; and its wall is yellow to brown, thick in water becoming reddish-brown in alkaline conditions (Prydiuk, 2014). The worldwide distribution of C. macrospora is 158 occurrences, most of them are in Europe, and there are not any occurrences in Saudi Arabia (https://www.gbif.org/species/2529858 (8/20/2021). According to https://www.ncbi.nlm.nih.gov/Taxonomy, the nucleotides deposited NCBI related to C. macrospora are only 2 nucleotides refer to 3 strains of C. macrospora.
P. demidoffii is an important and dangerous parasitic fungal species on several juniper species. There is no case report in Saudi Arabia about fruiting body fungus. However, there are many international reports of P. demindoffii parasites on many plant species such as Juniperus excelsa and Juniperus foetidissima. (Dogan and Karadelev, 2006). In China, Su and colleagues developed a method to produce the mycelial biomass of P. demidoffii as a resource of medicinal and therapeutic products (Su et al., 2010). Pyrofomes spp. are characterized by their slightly red to reddish-brown basidiocarps that have no cystidia (large cell on the basidiospore-carps) and setae with thick-walled, amputate, variable pseudoamyloid (dextrinoid), cyanophilous basidiospores.
P. demidoffii is characterized by the brightly rusty red color of the context tissue (Assefa et al., 2015). According to https://www.gbif.org/species/2546121 (8/20/2021) there were 139 occurrences of P. demidoffii globally, but not one record in Saudi Arabia. This fungus is frequently collected from many regions in Asia and North America. A total of 33 nucleotides that have been deposited in NCBI related to P. demidoffii (https://www.ncbi.nlm.nih.gov/taxonomy (8/20/2021).
Research effort, location, and time are important factors for recording new mushroom species in any country. For example, Hobart reported that it is surprising that T. fimbriatum was not recorded in the United Kingdom before 2012, although it is one of the most frequently recorded species of Tulostoma in Europe (Hobart, 2012). T. fimbriatum reportedly grows in open lands, forests, and cultivated lands on sandy soil (Karadelev and Rusevska, 2009).
In our work, all T. fimbriatum samples were collected from the surfaces of sandy soils in Al-Baha. T. fimbriatum is one of the most commonly distributed Tulostoma species worldwide and it has distinguishable features including fimbriate (having a narrow border) stoma, hyphal exoperidium (a protective layer that encloses spores), and its spores have verrucose and decoration of sub-reticulate (Esqueda et al., 2004).
There are 1934 occurrences of T. fimbriatum Fr. worldwide according to https://www.gbif.org/species/5243305 (8/20/2021), but no case has been recorded yet in the Middle East and Africa. According to https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1305709 (8/20/2021), there are five subspecies of T. fimbriatum: var. campestre, egranulosum, heterosporum, punctatum, and tuberculatum, and the total number of nucleotides deposited was 67.
In the present study, we did not find any fruiting body fungi from the Abha region during the above collection period, but it does not mean that the mushrooms cannot grow in the natural environment in the Abha region. We believe that more researches and surveillance are required during different seasons, along with surveys in several valleys where human activity is low.
5 Conclusion
The findings obtained from the present work reported the existence of eight species of Agaricomycotina recorded for the first time in Saudi Arabia. C. scabella, P. conopilea, A. pediades, P. demidoffii, and T. fimbriatum were collected from Al-Baha while C. macrospora, and C. palaeotropicum were collected from Al-Taif and Jeddah, respectively.
Acknowledgments
This Project was funded by the National Plan for Science, Technology, and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award Number (0002-001-01-17-2).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Survey of some mushrooms in Al-Taif governorate of Saudi Arabia. World J. Agric. Sci.. 2006;2(1):1-5.
- [Google Scholar]
- Adamczyk, J., 2016. Diversity of macromycete communities. In: The Ecological Role of Abandoned Agricultural Lands in Buffer Zones Around Landscape Parks in the Łódź Voivodeship, eds. S. Krysiak, J. Adamczyk, Wydawnictwo Uniwersytetu Łódzkiego, Łódź 2016; Wydawnictwo Uniwersytetu Łódzkiego.
- Molecular identification of some wild Nigerian mushrooms using internal transcribed spacer: polymerase chain reaction. Amb. Express. 2018;8(1):1-9.
- [Google Scholar]
- Antonín, V., Noordeloos, M.E., 2010. A Monograph of Marasmioid and Collybioid Fungi in Europ: IHW Verl. P. 480.
- A confusing duo: Calocybe cerina and Callistosporium pinicola (Agaricales) ACTA Mycol.. 2006;41(1):29-40.
- [Google Scholar]
- Characterization of P yrofomes demidoffii from E thiopian A fromontane forests. Forest Pathol.. 2015;45(4):263-273.
- [Google Scholar]
- Ecology and distribution of two parasitic fungal species (Pyrofomes demidoffii and Antrodia juniperina) on scale-leaf juniper trees in Turkey. Cryptogam., Mycol.. 2006;27(1):35-43.
- [Google Scholar]
- Anti-obesity effects of medicinal and edible mushrooms. Molecules. 2018;23(11):2880.
- [Google Scholar]
- Parasola psathyrelloides (Psathyrellaceae), a new species from Kerala State, India. Phytotaxa. 2019;405(5):255-262.
- [Google Scholar]
- Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol.. 1985;22(2):160-174.
- [Google Scholar]
- Karadelev, M., Rusevska, K., 2009. Ecology and distribution of species from genus Tulostoma (Gasteromycetes) in the Republic of Macedonia. Plant, fungal and, 437.
- molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [Google Scholar]
- Kuo, M., 2011. The Genus Psathyrella. In (pp. http://www.mushroomexpert.com/psathyrella.html 20 November 2021).
- Mushroom biodiversity in India: prospects and potential. In: Paper Presented at the Proceedings of the 8th International Conference on Mushroom Biology and Mushroom Products (ICMBMP8). 2014.
- [Google Scholar]
- How to Identify Mushrooms: to Genus III: Microscopic Features. Mad River Press Inc.; 1977. p. :94-114.
- Morphological diversity of Agaricomycetes in Kuningan Botanical Garden, West Java, Indonesia. IOP Conf. Ser.: Earth Environ. Sci.. 2019;374(1):012016.
- [Google Scholar]
- Mushrooms in Saudi Arabia (local and imported) Int. J. Curr. Res. Biosci. Plant Biol.. 2019;6(8):8-12.
- [CrossRef] [Google Scholar]
- Martins, A., 2017. The numbers behind mushroom biodiversity. book: Wild Plants, Mushrooms and Nuts: Functional Food Properties and Applications (Eds. Isabel CF R. Ferreira, Patricia Morales, Lillian Barros) John Wiley & Sons Ltd. DOI, 10(9781118944653), 15-50.
- Anti-inflammatory properties of edible mushrooms: a review. Food Chem.. 2018;243:373-381.
- [Google Scholar]
- Niveiro, N., Uhart, M., Albertó, E., 2020. Revision of the genera Agrocybe and Cyclocybe (Strophariaceae, Agaricales, Basidiomycota) in Argentina. Rodriguésia, 71.
- Some rare and interesting Conocybe found in Vyzhnytsia National Nature Park (Ukrainian Carpathians) Mycobiota. 2014;4:1-24.
- [Google Scholar]
- The de novo production of halogenated hydroquinone metabolites by the Andean-Patagonian white-rot fungus Phylloporia boldo. Curr. Res. Environ. Appl. Mycol. (J. Fungal Biol.). 2020;10(1):198-205.
- [Google Scholar]
- Biodiversity of mushrooms in conservative forest of Batu Katak resort, Langkat regency, North Sumatra. In: Paper Presented at the IOP Conference Series: Earth and Environmental Science. 2019.
- [Google Scholar]
- Molecular identification and antimicrobial activities of some wild Egyptian mushrooms: Bjerkandera adusta as a promising source of bioactive antimicrobial phenolic compounds. J. Genet. Eng. Biotechnol.. 2021;19(1):1-11.
- [Google Scholar]
- Optimization of culture conditions for the Mycelial growth of Pyrofomes demidoffii. Food Sci.. 2010;31(23):146-150.
- [Google Scholar]
- Survey and identification of mushrooms in Erbil Governorate. Res. J. Environ. Earth Sci.. 2013;5(5):262-266.
- [Google Scholar]
- Usyk, M., Zolnik, CP., Patel, H., Levi, M.H., Burk, RD., 2017. Novel ITS1 fungal primers for characterization of the mycobiome. mSphere 2:e00488-17. https://doi.org/10.1128/mSphere .00488-17.
- Biodiversity of mushrooms in Patharia forest of Sagar (MP)-III. Int. J. Biodivers. Conserv.. 2014;6(8):600-607.
- [Google Scholar]
- Exploring the species diversity of edible mushrooms in Yunnan, Southwestern China, by DNA barcoding. J. Fungi. 2021;7(4):310.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102345.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







