Translate this page into:
Exploring the phytochemicals of Platycodon grandiflorus for TMPRSS2 inhibition in the search for SARS-CoV-2 entry inhibitors
⁎Corresponding author. arunbgurung@gmail.com (Arun Bahadur Gurung)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Platycodon grandiflorus (Jacq.) A. DC. (Campanulaceae) is commonly known as a balloon flower whose rhizomes have been widely utilized in traditional Chinese medicine (TCM) and in various Japanese prescriptions for the treatment of respiratory diseases, diabetes, and inflammatory disorders. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19) global pandemic requires priming of the virus's spike (S) protein by cleavage of the S proteins by a multi-domain type II transmembrane serine protease, transmembrane protease serine 2 (TMPRSS2) to gain entry into the host cell. The current research aims at the screening of active phytocompounds of P. grandiflorus as potential inhibitors of cellular TMPRSS2 using molecular docking and molecular dynamics simulations approach. In silico toxicity analyses show that out of a total of 34 phytocompounds selected for the study, 12 compounds obey Lipinski’s rule of five and have favourable pharmacokinetic properties. The top three lead molecules identified here were Apigenin, Luteolin and Ferulic acid which exhibited binding energies of −7.47 kcal/mol, −6.8 kcal/mol and −6.62 kcal/mol respectively with corresponding inhibition constants of 3.33 µM, 10.39 µM and 13.95 µM. The complexes between the lead molecules and the receptor were held by hydrogen bond interactions with key residues such as Gly383, Gly385, Glu389, Lys390, Asp435, Ser436, Ser441, Cys465 and Lys467, and hydrophobic interactions with surrounding residues. The stability of the protein–ligand complexes was evaluated during 100 ns molecular dynamics (MD) simulation by analysing key geometric properties such as RMSD, RMSF, radius of gyration, total solvent accessible surface area and the number of hydrogen bonds. The binding free energies analysis using MD simulations revealed that the compounds and TMPRSS2 have favourable thermodynamic interactions, which are primarily driven by van der Waals forces. As a result, the selected bioactive phytochemicals from P. grandiflorus that target the cellular TMPRSS2 could offer an alternative treatment option against SARS-CoV-2 infections.
Keywords
TMPRSS2
SARS-CoV-2
COVID-19
Platycodon grandiflorus
Molecular docking
Phytochemicals
Bioactive compounds
1 Introduction
Platycodon grandiflorus (Jacq.) A. DC. is the only single species of the Platycodon genus, (Campanulaceae), commonly known as balloon flower. China, Korea, Japan, Mongolia, and Russia are all home to this species. P. grandiflorus is quite common in China, and its cultivation sites may be found throughout the country (Zhang et al., 2015). The rhizomes of P. grandiflorus are popularly known as Jiegeng or Lingdanghua in China, Doraji in North Korea, Kikyo in Japan, and Huridunzhaga in Mongolia (China Pharmacopoeia Committee, 2005). P. grandiflorus is a perennial plant that grows to a height of 20–120 cm. The leaves are green, ovate, elliptic, or lanceolate, and measure 2–7 0.5–3.5 cm2. It has a blue or purple bloom that is 1.5–4.5 cm2 in size. Because of its importance as traditional medicine and food resource, P. grandiflorus roots and rhizomes are usually collected around August (Huang, 2008). P. grandiflorus thrives in sunny herbal communities in thickets and is only seldom seen in woods below 2000 m in height, owing to its high ecological flexibility (Zhang et al., 2015). The rhizomes of P. grandiflorus have been widely utilized in traditional Chinese medicine (TCM) with outstanding therapeutic benefits for the treatment of cough, excessive phlegm, and sore throat (China Pharmacopoeia Committee, 2005). Its rhizomes are traditionally used in various Japanese prescriptions to treat suppuration, chronic rhinitis, chronic tonsillitis, and other ailments (Zhang et al., 2015). The roots of P. grandiflorus were utilized to treat bronchitis, asthma, pulmonary tuberculosis (TB), diabetes, and inflammatory disorders in Korea after being grown for four years (Lee, 1973; Takagi and Lee, 1972). Furthermore, this plant may be utilized as a food source and is prepared into delectable meals in East Asian nations including China, Japan, and Korea (China Pharmacopoeia Committee, 2005).
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) depends on the host protein angiotensin-converting enzyme 2 (ACE2) receptor for entry into the host cells (Hoffmann et al., 2020). The viral entry necessitates not only binding to the ACE2 receptor but also priming of the virus's spike (S) protein by cleavage of the S proteins at the S1/S2 and S2 sites by a multi-domain type II transmembrane serine protease, transmembrane protease serine 2 (TMPRSS2) (Baughn et al., 2020). This cleavage process is necessary for the virus to fuse with the host cell membrane and enter the cell (Hoffmann et al., 2020; Matsuyama et al., 2020). TMPRSS2 and ACE2 are co-expressed in several tissues such as the lung, heart, stomach, smooth muscle, liver, kidney, neurons, and immune cells (Fathema et al., 2021; Fuentes-Prior, 2021; Gemmati et al., 2020). Besides, both are abundantly present in type II pneumocyte cells (Kaur et al., 2021). Given the critical function of TMPRSS2 in viral entry and the lack of approved treatments for addressing the current coronavirus disease 2019 (COVID-19) pandemic, drug repurposing strategies to block this protease have received much consideration. Camostat, nafamostat, and gabexate, which are clinically authorized pharmacologic treatments of pancreatitis in Japan, can also suppress the activity of TMPRSS2 (Yamaya et al., 2020). Given the importance of host TMPRSS2 in the pathogenesis of SARS-CoV-2 infections, our current research focuses on the binding potential of a few selected active compounds of Platycodon grandiflorus to human TMPRSS2 using molecular docking, as well as probing the structural changes in the target protein induced by binding of the active compounds using molecular dynamics simulations approach. The phytochemicals proposed in the study are most likely to suppress TMPRSS2 function, preventing the SARS-CoV-2 virus from infecting host cells.
2 Materials and methods
2.1 Protein preparation
The three-dimensional structure of TMPRSS2 was obtained from the research collaboratory for structural bioinformatics (RCSB) protein data bank (PDB) using PDB ID: 7MEQ which was determined using the X-ray diffraction technique with a resolution of 1.95 Å. The hetero atoms-ions, water molecules and co-crystallized ligands were removed from the protein structure for docking studies and H-atoms were inserted using AutoDock Tools-1.5.6 (Morris et al., 2009).
2.2 Ligand preparation:
The information on thirty-four phytochemicals of P. grandiflorus was obtained through a literature search (Zhang et al., 2015). The structures of the phytochemicals along with Nafamostat (control) were retrieved from the PubChem database (Kim et al., 2016) and their structures were optimised using Merck molecular force field (MMFF)94 force field (Halgren, 1996).
2.3 Calculation of drug-like properties
The phytochemicals were evaluated for drug-likeness using Lipinski's rule of five (ROF) (Lipinski, 2004). The physicochemical properties of the compounds were determined using the DataWarrior tool version 4.6.1. (Sander et al., 2015).
2.4 Validation of the docking process
Initially, the docking procedure was validated by re-docking the co-crystal ligand of TMPRSS2, 4-carbamimidamidobenzoic acid (GBS). The re-docked ligand was used to evaluate if the root mean square deviation (RMSD) between the docked and native position is within ≤2.0 Å.
2.5 Molecular docking
Molecular docking studies were performed with TMPRSS2 protein and the active compounds of P. grandiflorus using the AutoDock 4.2 program (Morris et al., 2009). This docking program operates using a Lamarckian genetic algorithm (LGA) (Morris et al., 1998). The active sites were input and a grid parameter file for the protein was generated by fixing the number of grid points on the x, y, and z axes to 70 × 70 × 70 with a grid box centred at x:-7.6961, y:-7.7232, z:18.3699 centred at the co-crystal ligand. AutoDock grids were calculated for regularly spaced points at intervals of 0.375 Å contained within a cube based on the active sites of TMPRSS2 protein. The population size was set to 250 and the individuals were initialized randomly. The maximum number of energy evaluations was set to 106, and the maximum number of generations was 1000. Other docking parameters were set to the default values. The Lamarckian genetic algorithm was chosen to determine the best conformers in fifty independent trials of each compound. The LigPlot+ v.1.4.5 tool (Laskowski and Swindells, 2011) was used to study the molecular interactions between the compounds and the target protein.
2.6 Molecular dynamics simulation
The lead compounds discovered from the docking experiments, as well as the reference molecule, were submitted to Molecular Dynamics (MD) simulations to get a better understanding of the protein–ligand interactions. The PRODRG server (Schüttelkopf and Van Aalten, 2004) was used to build ligand topologies, whereas the Gromos96 43 B1 force field was used to generate protein topologies in Groningen Machine for Chemical Simulations (GROMACS) 5.0.6 (Hess et al., 2008). The three-site transferable intermolecular potential (SPC216) water model was used to solve a triclinic box, which was then neutralized with counterions. All of the problematic connections were eliminated further by running the system through a 10,000-step steepest descent algorithm with a force limit of <1000 kJ/mol (Bavi et al., 2016). Following that, the equilibration was carried out using the Number of particles, Volume and Temperature (NVT) (Berendsen et al., 1984) and Number of particles, Pressure and Temperature (NPT) (Parrinello and Rahman, 1981) methods at 100 ps at 300 K and 100 ps at a pressure of 1 bar maintained by a Parrinello-Rahman barostat and allowing the movement of counterions and water molecules while constraining the protein backbone. The long-range electrostatic interaction was computed using Particle Mesh Ewald (PME) (Darden et al., 1993), with a cut-off distance of 12 Å for Coulombic and van der Waals interactions. MD simulations were run for 100 ns, with coordinate data being saved every 2 fs. The Xmgrace plotting tool was used to analyze the corresponding findings.
2.7 MM/PBSA binding energy analysis
The binding free energy (ΔGbind) of the compounds was determined using the LARMD program (Yang et al., 2020) which employs the equation below Eq. (1). where ΔEbind corresponds to the binding energy, TΔSsol is the solvation entropy and TΔSconf represents the conformational entropy. The enthalpy was calculated using the Molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) or molecular mechanics generalised Born surface area (MM-GBSA) method (Hou et al., 2011), while the entropy was calculated using an empirical method (Hao et al., 2009; Pan et al., 2008).
3 Results:
We chose 34 major bioactive compounds discovered in P. grandiflorus extracts for this investigation which belong to different classes such as Triterpenoid saponins (Platycodin A, Platycodin C, Platycodin D, Platycodin D2, Deapioplatycodin D and Platycodin D3), Flavonoids (Luteolin-7-O-glucoside, Apigenin-7-O-glucoside, Apigenin, (2R,3R)-taxifolin, Luteolin, Quercetin-7-O-glucoside. Quercetin-7-O-rutinoside, Platyconin), Phenolic acids (Flavoplatycoside, Caffeic acid, 3,4-dimethoxycinnamic acid, Ferulic acid, Isoferulic acid, m-coumaric acid, p-coumaric acid, p-hydroxybenzoic acid, α-resorcylic acid, 2,3-dihydroxybenzoic acid, 2-hydroxy-4-methoxybenzoic acid, Homovanillic acid, Chlorogenic acid), Polyacetylene (Lobetyolinin, Lobetyolin) and Sterols (Betulin, β-sitosterol, δ-7-stigmastenone-3, Spinasterol, α-spinasteryl-3-O-β-D-glucoside) (Table 1). These molecules were subjected to ROF filters (molecular mass <500 Da, hydrogen bond donors <5, hydrogen bond acceptors <10, and a log P octanol–water partition coefficient <5) yielding 12 orally bioactive compounds (Table 2). The chemical structures of the filtered compounds utilised for molecular docking studies are depicted in Fig. 1. A redocking experiment shows an RMSD value of 1.060 Å between the native and docked positions of the cocrystal ligand (Fig. 2). An RMSD value <2.0 Å suggests that molecular docking parameters used in the study can precisely predict the binding poses of the compounds. The molecular docking results of the filtered bioactive compounds of P. grandiflorus as well as the control (Nafamostat) against the target receptor TMPRSS2 are displayed in Table 3. Three compounds were shown to have the best docking with TMPRSS2: apigenin, luteolin, and ferulic acid. Apigenin binds to TMPRSS2 with a binding energy of −7.47 kcal/mol and inhibition constant of 3.33 µM forming four hydrogen bonds with Gly383, Gly385, Glu389 and Cys465 and hydrophobic interactions with Gly259, Trp384, Ala386, Thr387, Glu388, Asn398, Ala399, Ala400, Asp435, Cys437, Asp440 and Ala466 (Fig. 3A). Luteolin binds to TMPRSS2 with a binding energy of −6.8 kcal/mol and inhibition constant of 10.39 µM and establishes four hydrogen bonds with Lys390, Ser436 and Ser441 and hydrophobic interactions with Glu389, Asp435, Cys437, Gln438, Thr459, Ser460, Trp461, Gly464, Cys465 and Gly472 (Fig. 3B). Ferulic acid interacts with TMPRSS2 with a binding energy of −6.62 kcal/mol and inhibition constant of 13.95 µM and establishes four hydrogen bonds with Glu389, Asp435 and Lys467 as well as hydrophobic interactions with Ala386, Thr387, Glu388, Asn433, Ser436, Cys437, Asp440, Cys465 and Ala466 (Fig. 3C). The control drug, Nafamostat binds to TMPRSS2 with a binding energy of −9.19 kcal/mol and inhibition constant of 0.184 µM and establishes six hydrogen bonds with His296, Asp345 and Gly464 and hydrophobic interactions with Val280, Cys281, Cys297, Leu302, Asp435, Ser436, Cys437, Gln438, Ser441, Ser460, Trp461, Gly462 and Cys465 (Fig. 3D).
Compounds
Name
Class
PubChem CID
1
Platycodin A
Triterpenoid saponins
46173910
2
Platycodin C
Triterpenoid saponins
46173919
3
Platycodin D
Triterpenoid saponins
162859
4
Platycodin D2
Triterpenoid saponins
53317652
5
Deapioplatycodin D
Triterpenoid saponins
70698266
6
Platycodin D3
Triterpenoid saponins
70698293
7
Luteolin-7-O-glucoside
Flavonoids
5280637
8
Apigenin-7-O-glucoside
Flavonoids
5280704
9
Apigenin
Flavonoids
5280443
10
(2R,3R)-taxifolin
Flavonoids
439533
11
Luteolin
Flavonoids
5280445
12
Quercetin-7-O-glucoside
Flavonoids
5381351
13
Quercetin-7-O-rutinoside
Flavonoids
44259247
14
Platyconin
Flavonoids
90659256
15
Flavoplatycoside
Phenolic acids
10416329
16
Caffeic acid
Phenolic acids
689043
17
3,4-dimethoxycinnamic acid
Phenolic acids
717531
18
Ferulic acid
Phenolic acids
445858
19
Isoferulic acid
Phenolic acids
736186
20
m-coumaric acid
Phenolic acids
637541
21
p-coumaric acid
Phenolic acids
637542
22
p-hydroxybenzoic acid
Phenolic acids
135
23
α-resorcylic acid
Phenolic acids
528564
24
2,3-dihydroxybenzoic acid
Phenolic acids
19
25
2-hydroxy-4-methoxybenzoic acid
Phenolic acids
10210429
26
Homovanillic acid
Phenolic acids
1738
27
Chlorogenic acid
Phenolic acids
1794427
28
Lobetyolinin
Polyacetylene
5459227
29
Lobetyolin
Polyacetylene
6369123
30
Betulin
Sterols
72326
31
β-sitosterol
Sterols
222284
32
δ-7-stigmastenone-3
Sterols
5748344
33
Spinasterol
Sterols
5281331
34
α-spinasteryl-3-O-β-D-glucoside
Sterols
12960498
Compounds
Name
MW
cLogP
HBA
HBD
TPSA
RB
1
Platycodin A
1267.37
−3.5801
29
16
459.35
17
2
Platycodin C
1267.37
−3.5801
29
16
459.35
17
3
Platycodin D
1225.33
−4.0647
28
17
453.28
15
4
Platycodin D2
1387.47
−5.9018
33
20
532.43
18
5
Deapioplatycodin D
1093.21
−2.711
24
15
394.36
12
6
Platycodin D3
1387.47
−5.9018
33
20
532.43
18
7
Luteolin-7-O-glucoside
448.379
7.00E-04
11
7
186.37
4
8
Apigenin-7-O-glucoside
432.38
0.3464
10
6
166.14
4
9*
Apigenin
270.239
2.3357
5
3
86.99
1
10*
(2R,3R)-taxifolin
304.253
0.9579
7
5
127.45
1
11*
Luteolin
286.238
1.99
6
4
107.22
1
12
Quercetin-7-O-glucoside
464.378
−0.4991
12
8
206.6
4
13
Quercetin-7-O-rutinoside
610.519
−1.4095
16
10
265.52
6
14
Platyconin
1421.23
−4.9099
37
21
596.03
23
15
Flavoplatycoside
612.535
−1.9418
16
10
265.52
6
16*
Caffeic acid
180.159
0.7825
4
3
77.76
2
17*
3,4-dimethoxycinnamic acid
208.212
1.3339
4
1
55.76
4
18*
Ferulic acid
194.185
1.0582
4
2
66.76
3
19*
Isoferulic acid
194.185
1.0582
4
2
66.76
3
20*
m-coumaric acid
164.16
1.1282
3
2
57.53
2
21*
p-coumaric acid
164.16
1.1282
3
2
57.53
2
22*
p-hydroxybenzoic acid
138.122
0.799
3
2
57.53
1
23
α-resorcylic acid
496.91
9.314
4
0
44.76
10
24*
2,3-dihydroxybenzoic acid
154.121
0.4533
4
3
77.76
1
25
2-hydroxy-4-methoxybenzoic acid
583.719
3.9151
5
4
81.95
16
26*
Homovanillic acid
182.174
0.7271
4
2
66.76
3
27
Chlorogenic acid
354.31
−0.7685
9
6
164.75
5
28
Lobetyolinin
558.575
−2.5943
13
9
218.99
12
29
Lobetyolin
396.434
−0.7572
8
6
139.84
9
30
Betulin
442.725
6.7202
2
2
40.46
2
31
β-sitosterol
414.715
7.8552
1
1
20.23
6
32
δ-7-stigmastenone-3
412.699
7.9989
1
0
17.07
6
33
Spinasterol
412.699
7.603
1
1
20.23
5
34
α-spinasteryl-3-O-β-D-glucoside
574.84
5.7659
6
4
99.38
8
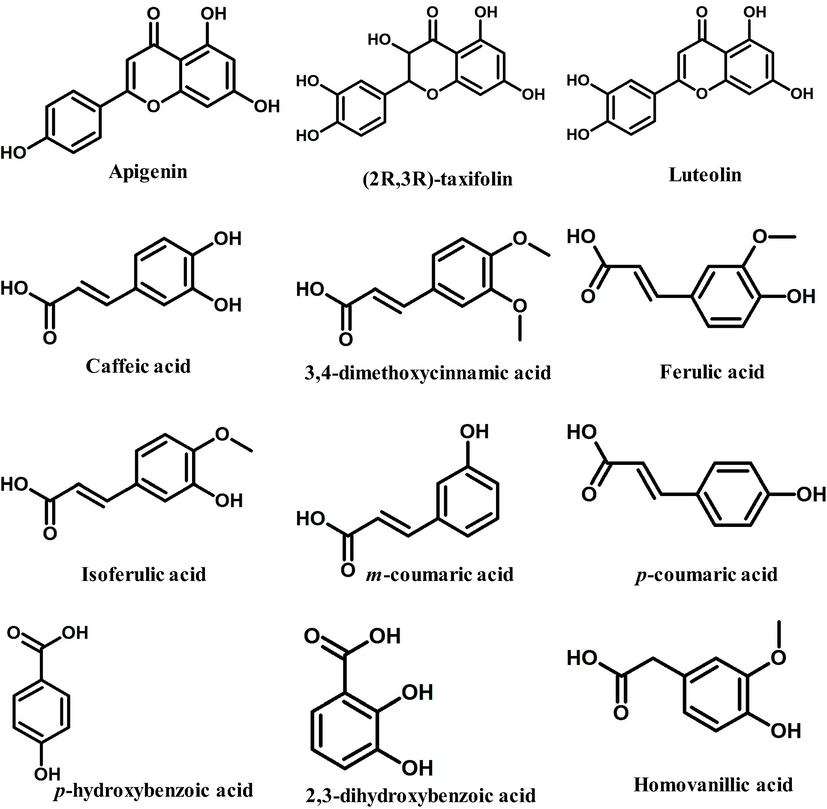
The drug-like bioactive compounds from P. grandiflorus used for molecular docking experiment.
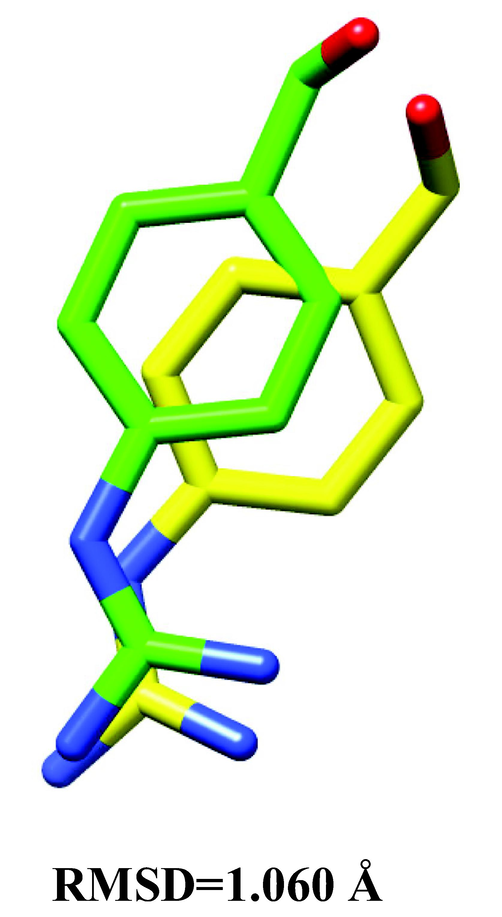
Superposition of the native conformation (yellow) of the co-crystal ligand (GBS) with its docked pose (green) revealed an RMSD of 1.060 Å.
Compounds
Name
Binding energy (kcal/mol)
Inhibition constant (µM)
9
Apigenin
−7.47
3.33
10
(2R,3R)-taxifolin
−6.34
22.67
11
Luteolin
−6.8
10.39
16
Caffeic acid
−6.38
21.13
17
3,4-dimethoxycinnamic acid
−6.41
19.91
18
Ferulic acid
−6.62
13.95
19
Isoferulic acid
−6.27
25.57
20
m-coumaric acid
−5.92
45.46
21
p-coumaric acid
−6.41
20.09
22
p-hydroxybenzoic acid
−3.94
1290
24
2,3-dihydroxybenzoic acid
−4.02
1130
26
Homovanillic acid
−4.61
419.57
Nafamostat (Control)
−9.19
0.184
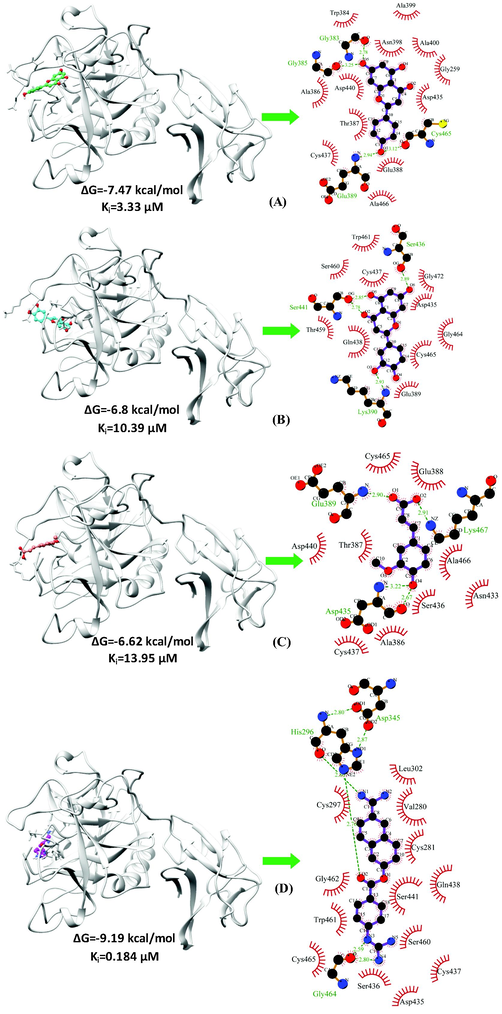
The binding conformation and molecular interactions of the compounds-(A) Apigenin (B) Luteolin (C) Ferulic acid and (D) Nafamostat with the target receptor TMPRSS2. On the left panel, the drug-receptor is depicted as a grey ribbon and compounds-Apigenin (spring green), Luteolin (cyan), Ferulic acid (salmon) and Nafamostat (magenta) are shown in ball and stick representations. On the right panel are displayed the molecular interactions between the compounds and the receptor with the hydrogen bonds shown in green and hydrophobic interactions shown in red semi-arcs.
MD simulations of the unbound TMPRSS2 and its complexes with the top three ranking compounds-Apigenin, Luteolin, Ferulic acid, and the control (Nafamostat) were performed for 100 ns, and various geometrical properties were determined from their trajectories (Table S1). The root-mean-square deviation (RMSD) is a valuable metric for assessing the atomic position structural deviation and protein structural stability. The average RMSD of backbone atoms of TMPRSS2, TMPRSS2_Apigenin, TMPRSS2_Luteolin and TMPRSS2_Ferulic acid and TMPRSS2_Nafamostat complexes were 0.33161855 ± 0.020691134 nm, 0.282429554 ± 0.043508183 nm, 0.32489471 ± 0.045273601 nm, 0.304615439 ± 0.032035141 nm and 0.300230713 ± 0.028235882 nm respectively (Table S1). The binding of the compounds induces a reduction in the structural flexibility of the target enzyme (Fig. 4). The average RMSD values of Apigenin, Luteolin, Ferulic acid and the Nafamostat were 0.247362992 ± 0.062295857 nm, 0.576125057 ± 0.214772452 nm, 0.730537652 ± 0.21814513 nm and 0.507619867 ± 0.078623572 nm indicating that the compounds have favourable binding orientations in the target enzyme's binding pocket. To analyse the local fluctuations in the target enzyme before and after compound binding, an average of the residual fluctuations in TMPRSS2 was computed and plotted as the root-mean-square fluctuation (RMSF) (Fig. 5). The RMSF figure revealed significant residual variations in several locations in the target enzyme, with high amplitudes of fluctuations in Ser204 (0.5562 nm), Gly205 (0.5075 nm), Gln253 (0.5027 nm), Ser254 (0.4391 nm), Arg255 (0.7522 nm), Ser339 (0.4562 nm), Lys340 (0.5289 nm), Glu388 (0.6266 nm), Glu389 (0.6227 nm) and Lys390 (0.6265 nm). The radius of gyration (Rg) of unbound TMPRSS2 and TMPRSS2 docked complexes were determined to measure their structural compactness (Fig. 6). The Rg values TMPRSS2, TMPRSS2_Apigenin, TMPRSS2_Luteolin and TMPRSS2_Ferulic acid and TMPRSS2_Nafamostat complexes were 2.072465355 ± 0.014329819 nm, 2.080123706 ± 0.012810064 nm, 2.06185013 ± 0.013976506 nm, 2.067286903 ± 0.020060061 nm and 2.074381249 ± 0.01411647 nm respectively. The TMPRSS2_Luteolin and TMPRSS2_Ferulic acid complexes show slightly lower Rg values whereas TMPRSS2_Apigenin and TMPRSS2_Nafamostat show higher Rg values when compared to unbound TMPRSS2. The Rg plot analysis, in this case, reveals that TMPRSS2 protein undergoes conformational changes in response to the binding of compounds, resulting in changes in structural compactness. The solvent-accessible surface area (SASA) of a protein is the part of the protein that interacts directly with the solvent molecules around it. The SASA plot for unbound TMPRSS2 and TMPRSS2 docked complexes was generated during the 100 ns MD simulation. (Fig. 7). The unbound TMPRSS2, TMPRSS2_Apigenin, TMPRSS2_Luteolin, TMPRSS2_Ferulic acid and TMPRSS2_Nafamostat complexes show average SASA values of 150.9597053 ± 3.249067344 nm2, 152.919972 ± 2.82381675 nm2, 152.9353017 ± 3.806978439 nm2, 153.5488102 ± 5.332797744 nm2 and 153.0682637 ± 3.424944207 nm2 respectively. A decrease in total SASA after interaction with the compounds was observed due to the conformational changes in the protein. Intramolecular hydrogen bonding plays a significant role in determining a protein's stability and overall structure. The stability of unbound TMPRSS2 and TMPRSS2 docked complexes was confirmed by monitoring hydrogen bonds established throughout the simulation (Fig. S1.A). The average number of intramolecular hydrogen bonds in TMPRSS2, TMPRSS2_Apigenin, TMPRSS2_Luteolin, TMPRSS2_Ferulic acid and TMPRSS2_Nafamostat complexes were 225.1298701 ± 9.051691382, 215.3916084 ± 8.497675536, 218.1968032 ± 8.978431365, 225.8011988 ± 9.14677203, 223.5194805 ± 8.42637942 respectively. The compounds- Apigenin, Luteolin, Ferulic acid and the Nafamostat showed an average number of intermolecular hydrogen bonds of 1.196803197 ± 0.937139141, 1.545454545 ± 1.018912076, 0.754245754 ± 0.907493502 and 1.632367632 ± 1.051052467 respectively which aids in the stabilization of the protein–ligand complexes (Fig. S1.B).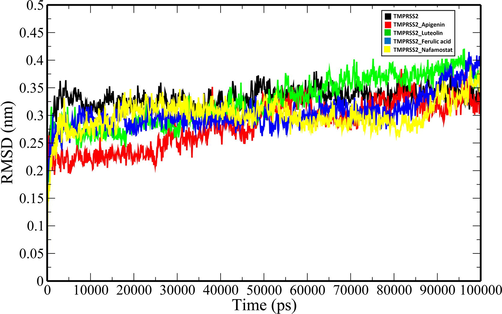
RMSD plot of unbound TMPRSS2 (black) and docked TMPRSS2 backbone atoms (TMPRSS2_Apigenin: red; TMPRSS2_Luteolin: green; TMPRSS2_Ferulic acid: blue and TMPRSS2_Nafamostat).
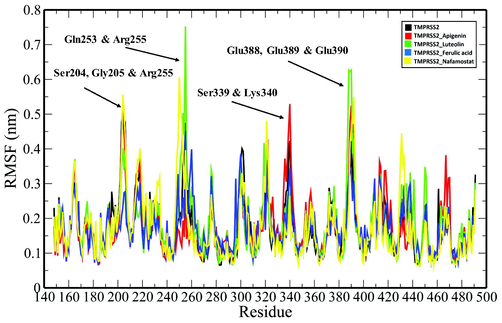
RMSF plot of unbound TMPRSS2 (black) and TMPRSS2 docked complexes (TMPRSS2_Apigenin: red; TMPRSS2_Luteolin: green; TMPRSS2_Ferulic acid: blue and TMPRSS2_Nafamostat) where residues with large amplitude of fluctuations are marked and labelled.
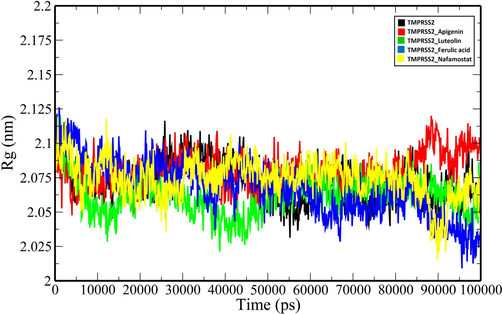
Rg plot of unbound TMPRSS2 and TMPRSS2 docked complexes (TMPRSS2_Apigenin: red; TMPRSS2_Luteolin: green; TMPRSS2_Ferulic acid: blue and TMPRSS2_Nafamostat).
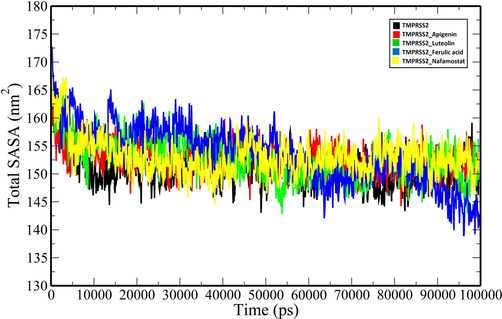
Total SASA plot of unbound TMPRSS2 and TMPRSS2 docked complexes (TMPRSS2_Apigenin: red; TMPRSS2_Luteolin: green; TMPRSS2_Ferulic acid: blue and TMPRSS2_Nafamostat).
The binding free energies from MD simulation studies were found to be negative for Apigenin (ΔPB = -12.37 kcal/mol, ΔGB = -12.37 kcal/mol), Luteolin (ΔPB = -6.40 kcal/mol, ΔGB = -10.05 kcal/mol), Ferulic acid (ΔPB = -2.40 kcal/mol, ΔGB = -0.24 kcal/mol) and Nafamostat (ΔPB = -9.30 kcal/mol, ΔGB = -24.91 kcal/mol) (Table S2). The van der Waals energy make a significant contribution to the binding free energy except for Ferulic acid where the electrostatic energy component has predominant contributions. The key residues contributing towards the binding interaction between Apigenin and TMPRSS2 include Gly259 (−1.25 kcal/mol), Cys437 (−1.22 kcal/mol), Asn398 (−1.06 kcal/mol), Ala466 (−1.01 kcal/mol), Val434 (−0.95 kcal/mol), Ala399 (−0.82 kcal/mol), Asp435 (−0.80 kcal/mol), Thr387 (−0.74 kcal/mol), Ile381 (−0.68 kcal/mol) and Ala400 (−0.54 kcal/mol) (Fig. S2.A). In case of TMPRSS2_Luteolin complex, residues such as Cys465 (−1.69 kcal/mol), Gln438 (−1.47 kcal/mol), Cys437 (−1.04 kcal/mol), Gly462 (−0.96 kcal/mol), Gly464 (−0.96 kcal/mol), Glu389 (−0.91 kcal/mol), Trp461 (−0.78 kcal/mol), Lys390 (−0.75 kcal/mol), Ser436 (−0.63 kcal/mol), Thr459 (−0.58 kcal/mol) contribute majorly to the total binding energy (Fig. S2.B). The top ten residues contributing towards the binding interaction between Ferulic acid and TMPRSS2 include Lys401 (−1.40 kcal/mol), Leu378 (−1.26 kcal/mol), Leu403 (−0.65 kcal/mol), Phe429 (−0.47 kcal/mol), Val402 (−0.17 kcal/mol), Arg252 (−0.07 kcal/mol), Trp380 (−0.06 kcal/mol), Lys449 (−0.06 kcal/mol), Arg255 (−0.05 kcal/mol) and Arg409 (−0.05 kcal/mol) (Fig. S2.C). In case of TMPRSS2_Nafamostat complex, residues such as Gln438 (−2.00 kcal/mol), Cys437 (−1.37 kcal/mol), Asp435 (−1.33 kcal/mol), Trp461 (−1.32 kcal/mol), Pro471 (−1.15 kcal/mol), Gly472 (−0.89 kcal/mol), Gly462 (−0.85 kcal/mol), Cys465 (−0.79 kcal/mol), Leu430 (−0.72 kcal/mol) and Gly432 (−0.68 kcal/mol) have higher contribution toward the total binding energy (Fig. S2.D).
4 Discussion
The SARS-CoV-2, the causative agent of COVID-19 depends on the host multi-domain type II transmembrane serine protease, transmembrane protease serine 2 (TMPRSS2) angiotensin-converting enzyme 2 (ACE2) receptor for the cleavage process of S proteins to enter the host cells. Platycodon grandiflorus (Jacq.) A. DC. which belongs to the Campanulaceae family has immense medicinal importance. The rhizomes of P. grandiflorus have not only been widely utilized in traditional Chinese medicine (TCM) but also has been traditionally used in various Japanese prescriptions. More recently, Kim et al. (2021) found that platycodin D (PD), a triterpenoid saponin rich in P. grandiflorus efficiently prevents SARS-CoV-2 infection via lysosome and transmembrane protease serine 2 (TMPRSS2)-driven entry. Therefore, the bioactive phytochemicals of P. grandiflorus can be explored as a source of novel chemical entities to block the replication of SARS-CoV-2. Given the importance of TMPRSS2 in viral entry and the dearth of effective therapeutics for the present COVID-19 pandemic, small compounds or the Food and Drug Administration (FDA)-approved drugs can be tested for their ability to suppress the enzyme's activity. Camostat, nafamostat, and gabexate which are clinically authorized pharmacologic treatments of pancreatitis in Japan have been also used to suppress the activity of TMPRSS2 (Yamaya et al., 2020) and these known inhibitors have IC50 values of 6.2 nM, 0.27 nM and 130 nM respectively (Shrimp et al., 2020). A recent study suggested that nafamostat was protective against SARS-CoV-2 pulmonary infections in two mouse models for COVID-19 (Li et al., 2021). Given the relevance of host TMPRSS2 in the pathogenesis of SARS-CoV-2 infections, our present research focuses on identifying active Platycodon grandiflorus compounds as potential TMPRSS2 inhibitors, which would then prevent SARS-CoV-2 from infecting host cells. We have used a diverse set of 34 phytochemicals of P. grandiflorus and applied drug filters to assess their oral bioavailability. The ability of the filtered drug-like compounds to bind and interact with the target receptor TMPRSS2 was investigated. The best three molecules interacting with the drug-receptor TMPRSS2 identified were Apigenin, Luteolin and Ferulic acid which bind to the active pocket with hydrogen bonds and hydrophobic interactions. 100 ns MD simulations were performed to evaluate the dynamic behaviour of the protein–ligand complexes and their stabilities were assessed in terms of RMSD, Rg, SASA and number of hydrogen bonds. Apigenin (4′,5,7-trihydroxyflavone), a flavone family member, is a dietary flavonoid that is both nontoxic and nonmutagenic and it has hydroxyl groups in both its B and C rings (Wang et al., 2020). Apigenin has antiviral properties in vitro and in vivo against a variety of viruses, including enterovirus 71 (EV71) (Lv et al., 2014; Zhang et al., 2014), hepatitis C virus (HCV) (Ahmed-Belkacem et al., 2014), Human Immunodeficiency Virus (HIV) (Kehinde et al., 2019), and adenoviruses (Kanerva et al., 2007). Apigenin inhibited HCV replication by lowering the levels of mature miR122 expression (Shibata et al., 2014) and suppressed FMDV (Foot and Mouth Disease Virus) infection at the post-entry stage by reducing internal ribosome entry site (IRES)-driven translational activity (Qian et al., 2015). Epstein-Barr virus (EBV) reactivation into the lytic cycle and virion generation by EBV-positive nasopharyngeal carcinoma (NPC) cells are both inhibited by apigenin (Wu et al., 2017). Luteolin (3,4,5,7-tetrahydroxyflavone) is a clear yellow crystal that belongs to the bioflavonoid family (Wang et al., 2020). By blocking the host proprotein convertase furin, luteolin can obstruct the later stages of the dengue virus (DENV) viral life cycle in infected cells (Peng et al., 2017), Epstein-Barr virus (Wu et al., 2016), Japanese encephalitis virus (Fan et al., 2016), HIV-1 (Mehla et al., 2011), Hepatitis B virus (Bai et al., 2016) and influenza A virus (Yan et al., 2019) are all inhibited by luteolin. The previous study suggests that the flavonoids-Apigenin and Luteolin inhibit SARS-CoV 3CLpro activity as well with IC50 values of 280.8 µM and 20.2 µM respectively (Ryu et al., 2010). Besides, luteolin has been found to block the entry of SARS-CoV (EC50 = 10.6 µM) into host cells by binding to the surface spike protein (Yi et al., 2004). While our findings are encouraging, additional in vitro or in vivo studies are needed to determine the anti-SARS-CoV-2 efficacy of the compounds. Further, in TMPRSS2¯cells, severe acute respiratory syndrome coronavirus 2 (SARS-CoV) S can use the endosomal cysteine proteases cathepsin B and L (CatB/L) for S protein priming (Simmons et al., 2005) and therefore it would be fascinating to see if these compounds have similar inhibitory activity against them. Because TMPRSS2 functions as a viral protein processing protease in the pathogenesis of other coronaviruses-SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), as well as influenza viruses (Mahoney et al., 2021), these lead compounds may be quite promising to be developed as broad-spectrum antivirals. The compounds could be subjected to structure–activity studies in order to enhance their potency and specificity.
5 Conclusion
The paucity of effective therapeutic medications for SARS-CoV-2 infections, as well as the rising mortality rate, necessitate the development of innovative drug candidate molecules with few adverse effects. Here, we explored the bioactive phytochemicals from a traditionally important medicinal plant Platycodon grandiflorus as a source of novel molecules for the inhibition of cellular TMPRSS2 activity, a key pharmacological target for preventing the SARS-CoV-2 entry into the host cells. Our study concludes that Apigenin, Luteolin and Ferulic acid are the most effective compounds for inhibition of the TMPRSS2 activity demonstrating that these agents may be used in COVID-19 treatment. These compounds should be subjected to further experimental studies to confirm their efficacies.
Acknowledgements
The authors would like to extend their sincere appreciation to the Researchers Supporting Project number (RSP2022R418), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention. Nucleic Acids Res.. 2014;42(14):9399-9409.
- [Google Scholar]
- Luteolin inhibits hepatitis B virus replication through extracellular signal-regulated kinase-mediated down-regulation of hepatocyte nuclear factor 4$α$ expression. Mol. Pharm.. 2016;13(2):568-577.
- [Google Scholar]
- Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clinic Proceed.. 2020;95(9):1989-1999.
- [Google Scholar]
- Molecular interactions of UvrB protein and DNA from Helicobacter pylori: Insight into a molecular modeling approach. Comput. Biol. Med.. 2016;75:181-189.
- [Google Scholar]
- Molecular dynamics with coupling to an external bath. J. Chem. Phys.. 1984;81(8):3684-3690.
- [Google Scholar]
- China Pharmacopoeia Committee, 2005 China Pharmacopoeia Committee, (Eds.), 2005. Pharmacopoeia of the People’s Republic of China, the first division of 2005 edition. China Chemical Industry Press, Beijing, pp. 291–292.
- Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys.. 1993;98(12):10089-10092.
- [Google Scholar]
- Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res.. 2016;220:112-116.
- [Google Scholar]
- COVID 19 in children: gastrointestinal, hepatobiliary and pancreatic manifestation. Mymensingh Med. J. MMJ. 2021;30:570-579.
- [Google Scholar]
- Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021:296.
- [Google Scholar]
- COVID-19 and individual genetic susceptibility/receptivity: role of ACE1/ACE2 genes, immunity, inflammation and coagulation. Might the double X-chromosome in females be protective against SARS-CoV-2 compared to the single X-chromosome in males? Int. J. Mol. Sci.. 2020;21:3474.
- [Google Scholar]
- Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem.. 1996;17:490-519.
- [CrossRef] [Google Scholar]
- Understanding the mechanism of drug resistance due to a codon deletion in protoporphyrinogen oxidase through computational modeling. J. Phys. Chem. B. 2009;113(14):4865-4875.
- [Google Scholar]
- GRGMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput.. 2008;4:435-447.
- [CrossRef] [Google Scholar]
- SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271-280.e8.
- [Google Scholar]
- Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model.. 2011;51(1):69-82.
- [Google Scholar]
- Flora of China. Beijing: Science Press; 2008.
- Chlorpromazine and apigenin reduce adenovirus replication and decrease replication associated toxicity. J. Gene Med. A cross-disciplinary. J. Res. Sci. gene Transf. Clin. Appl.. 2007;9(1):3-9.
- [Google Scholar]
- Targeting host cell proteases to prevent SARS-CoV-2 invasion. Curr. Drug Targets. 2021;22(2):192-201.
- [Google Scholar]
- The pharmacokinetic properties of HIV-1 protease inhibitors: A computational perspective on herbal phytochemicals. Heliyon. 2019;5(10):e02565
- [Google Scholar]
- PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202-D1203.
- [Google Scholar]
- Platycodin D, a natural component of Platycodon grandiflorum, prevents both lysosome-and TMPRSS2-driven SARS-CoV-2 infection by hindering membrane fusion. Exp. Mol. Med.. 2021;53(5):956-972.
- [Google Scholar]
- LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model.. 2011;51:2778-2786.
- [CrossRef] [Google Scholar]
- Pharmacological studies on Platycodon grandiflorum A. DC. IV. A comparison of experimental pharmacological effects of crude platycodin with clinical indications of platycodi radix (author’s transl) Yakugaku zasshi J. Pharm. Soc. Japan. 1973;93(9):1188-1194.
- [Google Scholar]
- The tmprss2 inhibitor nafamostat reduces sars-cov-2 pulmonary infection in mouse models of covid-19. MBio. 2021;12(4):e00970-21
- [Google Scholar]
- Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today. Technol.. 2004;1:337-341.
- [CrossRef] [Google Scholar]
- Apigenin inhibits enterovirus 71 replication through suppressing viral IRES activity and modulating cellular JNK pathway. Antiviral Res.. 2014;109:30-41.
- [Google Scholar]
- Mahoney, M., Damalanka, V.C., Tartell, M., Chung, D.H., Lourenco, A.L., Pwee, D., Bridwell, A.E.M., Hoffmann, M., Voss, J., Karmakar, P., others, 2021. A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. bioRxiv.
- Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci.. 2020;117(13):7001-7003.
- [Google Scholar]
- A flavonoid, luteolin, cripples HIV-1 by abrogation of tat function. PLoS One. 2011;6(11):e27915
- [Google Scholar]
- Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem.. 1998;19(14):1639-1662.
- [Google Scholar]
- AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem.. 2009;30:2785-2791.
- [CrossRef] [Google Scholar]
- Modeling the catalysis of anti-cocaine catalytic antibody: competing reaction pathways and free energy barriers. J. Am. Chem. Soc.. 2008;130(15):5140-5149.
- [Google Scholar]
- Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys.. 1981;52(12):7182-7190.
- [Google Scholar]
- Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antiviral Res.. 2017;143:176-185.
- [Google Scholar]
- Apigenin restricts FMDV infection and inhibits viral IRES driven translational activity. Viruses. 2015;7:1613-1626.
- [Google Scholar]
- Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med Chem.. 2010;18:7940-7947.
- [Google Scholar]
- DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model.. 2015;55:460-473.
- [CrossRef] [Google Scholar]
- PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr.. 2004;60(8):1355-1363.
- [Google Scholar]
- The flavonoid apigenin inhibits hepatitis C virus replication by decreasing mature microRNA122 levels. Virology. 2014;462:42-48.
- [Google Scholar]
- An enzymatic TMPRSS2 assay for assessment of clinical candidates and discovery of inhibitors as potential treatment of COVID-19. ACS Pharmacol. Transl. Sci.. 2020;3:997-1007.
- [Google Scholar]
- Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci.. 2005;102(33):11876-11881.
- [Google Scholar]
- Pharmacological studies on Platycodon grandiflorum A. DC. 3. Activities of crude platycodin on respiratory and circulatory systems and its other pharmacological activities. Yakugaku zasshi J. Pharm. Soc. Japan. 1972;92:969-973.
- [Google Scholar]
- Research progress of the antiviral bioactivities of natural flavonoids. Nat. Products Bioprospect.. 2020;10(5):271-283.
- [Google Scholar]
- Inhibition of Epstein-Barr virus reactivation by the flavonoid apigenin. J. Biomed. Sci.. 2017;24:1-13.
- [Google Scholar]
- Luteolin inhibits Epstein-Barr virus lytic reactivation by repressing the promoter activities of immediate-early genes. Antiviral Res.. 2016;132:99-110.
- [Google Scholar]
- Protease inhibitors: candidate drugs to inhibit severe acute respiratory syndrome coronavirus 2 replication. Tohoku J. Exp. Med.. 2020;251(1):27-30.
- [Google Scholar]
- Luteolin decreases the yield of influenza A virus in vitro by interfering with the coat protein I complex expression. J. Nat. Med.. 2019;73(3):487-496.
- [Google Scholar]
- LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform.. 2020;21:2206-2218.
- [Google Scholar]
- Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol.. 2004;78(20):11334-11339.
- [Google Scholar]
- Platycodon grandiflorus–An Ethnopharmacological, phytochemical and pharmacological review. J. Ethnopharmacol.. 2015;164:147-161.
- [Google Scholar]
- Apigenin inhibits enterovirus-71 infection by disrupting viral RNA association with trans-acting factors. PLoS One. 2014;9(10) e110429
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102155.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







