Translate this page into:
Efficient removal potential of Microbacterium sp. strain 1S1 against arsenite isolated from polluted environment
⁎Corresponding author at: Department of Microbiology & Molecular Genetics, University of the Punjab, New Campus, Lahore 54590, Pakistan. rehman.mmg@pu.edu.pk (Abdul Rehman)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
A multiple metal resistant Microbacterium sp. strain 1S1, tolerated arsenite and arsenate upto 75 and 520 mM, was isolated from industrial wastewater. The arsenite surface adsorption and uptake into the bacterial cells, exposed to 15 mM arsenite, were confirmed through Scanning Electron Microscope (SEM), Energy Dispersive X-ray (EDX), and Fourier-transform infrared spectroscopy (FTIR) analyses. The cell physiology of strain 1S1 altered in arsenite exposure and the percent increase in GSH/GSSG ratio and NPSHs concentration was up to 40.0 and 78.50%. Furthermore, 240% increase in catalase provided evidence that arsenite induces hydrogen peroxide mediating oxidative stress. The bacterial cells growing in a rich medium with 15 mM arsenite were able to oxidize 98% arsenite after 96 h of incubation, and the inactivated biomass of the bacterium at 1 g per liter removed 99% of 15 mM arsenite after 10 h incubation. The harboring of multiple resistance strategies and appreciable arsenic oxidizing potential make strain 1S1 an impending foundation for green chemistry to exterminate environmental arsenite.
Keywords
Arsenic
Microbacterium sp. 1S1
As-resistance
Antioxidant enzymes
Bioremediation
1 Introduction
Arsenic (As) is a metalloid and ubiquitous in nature (Wang et al., 2017). Arsenic level is increasing in our surrounding day by day due to two main sources. One is natural source i.e. weathering of rocks and another is anthropogenic source which includes mining and agricultural practices by human beings (Andres and Bertin, 2016; Mujawar et al., 2019). Arsenic is found commonly in two oxidation states in the environment; one is arsenite (As3+) and other is arsenate (As5+). Arsenite is 100 times more toxic than arsenate due to high mobility or bioavailability (Branco et al., 2009).
Arsenite is considered toxic because of its carcinogenic nature. It affects living things including human beings because it induces the amplification of gene and transforms the eukaryotic cell structure (Bobrowicz et al., 1997). In living organisms As3+ reacts with the sulfhydryl group of the protein and inactivates the enzymes involved in different physiological processes, while As5+ substitutes the phosphorus in DNA due to homologous structure (Patra et al., 2004).
Microorganisms have different types of resistance mechanisms against arsenic and other heavy metal ions. Such resistance mechanisms can be explored and used in decontamination of drinking water or contaminated soil having arsenic or other heavy metal ions (Andres and Bertin, 2016). The resistance mechanisms in microorganism are oxidation and reduction which are responsible for arsenic transformation (Liao et al., 2011). In oxidation, As3+ can be converted into As5+ i.e. from more toxic form to less toxic form through aioA and aioB genes. On the other hand, arsenic reduction happens due to the presence of arsC gene which is a cytoplasmic gene (Yamamura and Amachi, 2014). There is also a third mechanism called arsenic methylation in which methyl groups enter into the inorganic arsenic through a protein encoded by arsM gene. Basically, arsenic oxidation is a detoxification process.
In the present study, various approaches were employed to investigate the dynamic changes in the strain 1S1 against arsenite stress. Antioxidant enzymes, glutathione and non-protein thiols contents were evaluated for strain 1S to combat oxidative stress produced under metal exposure. The metal–microbe interaction and arsenite uptake by bacterial cells were also confirmed through SEM, EDX, and FTIR analyses. Moreover, arsenite oxidizing potential of strain 1S1 to eradicate environmental arsenite was evaluated at lab scale and from the real wastewater.
2 Materials and methods
2.1 Sample collection and isolation of arsenic resistant bacteria
For isolation of arsenic resistant bacteria, wastewater samples were collected from drainage of chemical industries situated in the area of District Sheikhupura, Pakistan. This is a hub of industries. The screw capped autoclaved bottles were used for samples collection. The physicochemical features like temperature, pH, total dissolved solids (TDS), and arsenic concentration were measured in wastewater samples (Butt and Rehman, 2011).
For the isolation of arsenic resistant bacteria 100 μl of wastewater sample was spread on Luria-Bertani (LB) agar plates containing 5 mM arsenic stress. LB agar plates were prepared by dissolving 5 g NaCl, 10 g tryptone, 5 g yeast extract in 1 L of distilled water, pH was adjusted at 7 and then 15 g agar was added in 1 L flask. LB medium autoclaved at 121 °C for 15 min and growth of bacterial colonies was observed after overnight incubation at 37 °C.
2.2 Arsenite oxidation determination
Isolated strain was streaked on the LB-agar plate supplemented with 5 mM arsenite stress. After 3 days of incubation silver nitrate solution (0.1%) was used to flood the bacterial colonies. After that, results were noted according to Simeonova et al. (2004).
2.3 Cross metal resistance
The minimum inhibitory concentration (MIC) of arsenite against the bacterium was determined by streaking the strain on LB agar plates with different arsenic stress, 5, 10, 20, 30 mM up to 100 mM and cultured at 37 °C for 24 h. The isolated bacterial strain was also processed for its capability to resist other toxic metal ions like arsenate, lead, cadmium, chromium, nickel, mercury, zinc, selenium and cobalt. For this purpose various concentrations of different heavy metals were used by supplementing them into LB-agar plates and incubating the streaked plates overnight at 37 °C for 24 h (Naureen and Rehman, 2016). MIC is the minimum amount of chemical that inhibits the growth of microorganisms by adding the chemical into the media.
2.4 Morphological and biochemical characterization
After the isolation of arsenic resistant bacteria, various biochemical tests were performed. These are gram staining, spore staining, catalase test, oxidase test, growth on blood, and growth on chocolate and MacConkey agar. Other tests like citrate utilization test, SIM agar test, indole test, nitrate reduction test, MR and VP test were also performed for bacterial identification. The bacterium was also characterized at molecular level. Briefly, genomic DNA of Microbacterium sp. 1S1 was isolated, which was then subjected to 16S rRNA gene amplification through primers (Elahi and Rehman, 2019). The amplified products were mailed for sequencing to Macrogen, Korea and the sequence obtained was then aligned by utilizing BLAST analysis. Moreover, a dendrogram was created by using the MEGA7 program on the basis of homology.
2.5 Growth conditions and effect of metal on bacterial strain
The culturing environment (i.e. optimum pH and temperature) of isolated strain was accessed by growing at different temperature (25, 30, 37, and 42) and pH (5, 7, 9, and 11) in LB broth (Naureen and Rehman, 2016). Effect of arsenite on the isolated bacterial strain was determined by culturing the bacterium in LB broth supplementing with 1.33 mM (100 μg/ml) arsenite into the medium. The growth of bacterium was monitored by taking the optical density at 600 nm after every 4 h for up to 36 h.
2.6 Bacterial oxidizing potential
Strain 1S1 was accessed for its potential to oxidize arsenic through culturing in 250 ml flasks containing 100 ml LB broth. For this, 3 flasks supplemented with arsenite 15 mM were maintained. Out of 3 flasks, one acted as control, having the same amount of metal but without bacterial cells. Three parameters i.e. temperature, pH, and arsenite concentration (mM) were considered to optimize the oxidation potential of bacterium. Temperature (20 °C, 25 °C, 30 °C, 37 °C, and 42 °C), pH (3, 5, 7, 9 and 11) and arsenic concentration (15, 30, 45, 60, and 75 mM ml−1) were maintained during the experiment. The bacterium arsenic oxidizing ability was determined with a suitable time period of 24 h upto 96 h. To harvest bacterial cells from each and every flask 5 ml sample was taken out and centrifuged at 3000 rpm for 5 min. Amount of arsenic was assessed through the P S Analytical Millennium Excalibur Method (Sher et al. 2020a).
2.6.1 Bacterial inactivated biomass arsenic removal
Briefly, bacterial culture was grown, centrifuged to obtain pellet, and pellet was incubated at 70 °C (2–3 times) to inactivate the cells. Initially, 1 g/L bacterial biomass was mixed in 1 L of arsenic solution of 1500 ml flasks containing 15 mM As+3 stress. The flasks were incubated at 37 °C on the shaker for 10 h. Arsenic concentration was estimated before and after incubation through atomic absorption spectrophotometer (Sher et al., 2020a). The bacterial inactivated biomass bioremediation efficiency (E) was calculated by given formulas. q represents bacterial biomass in grams, Ci and Cf indicate initial and final amount of arsenic, m designates biosorbent mass used in the assay, and V represents mixture volume.
2.7 Arsenite removal from industrial wastewater by Microbacterium sp. strain 1S1
The experiment to ascertain the potential of bacterium to eradicate arsenic from the real industrial wastewater at room temperature (25 °C ± 2) along with arsenite concentration (15 mM) was performed according to Elahi and Rehman (2019).
2.8 Measurement of antioxidant enzymes and glutathione contents
Behavior of antioxidant enzymes of bacterial strain 1S1 was evaluated in arsenite presence. Briefly, the strain was grown in 100 ml minimal salt medium and then was placed in a shaking incubator at 37 °C containing 10 mM As3+ stress. After 48 h of incubation, cells were harvested through centrifugation at 14,000 rpm for a time period of 10 min. The obtained pellets were weighed, dissolved in phosphate buffer, and finally were sonicated. The sonicated pellets were centrifuged at 14,000 rpm for 10 min and aliquots obtained were used for antioxidant enzymes assays. Peroxidase (POX) enzyme reaction was performed according to Reddy et al. (1995) while the catalase, ascorbate peroxidase (APOX) and superoxide dismutase (SOD) reactions were performed according to protocols of Luck (1974), Ratkevicius et al. (2003), and Marklund and Marklund (1974), respectively.
The level of GSH, GSSG, and NPSHs was estimated with and without arsenite for strain 1S1 according to protocol mentioned in Shamim and Rehman (2015).
2.9 SEM, EDX and FTIR analysis
To perform scanning electron microscopy, bacterial samples were treated and prepared as described in Khan et al. (2016). Briefly, microbial culture in the absence and presence of 15 mM arsenite was prepared and a drop of suspension was mounted onto the aluminum stub and treated as described by Khan et al. (2016). The gold film was used to wrap samples with a sputter coater (Denton, Desk V HP) and examined via Scanning Electron Microscope (Nova NanoSEM 450) attached with Oxford Energy Dispersive X-ray (EDX) analysis system. The Fourier-transform infrared spectroscopy (FTIR) was used to get Infrared spectra for strain 1S1 in arsenite challenge and samples were prepared according to protocols mentioned in Deokar et al. (2013). To understand the underlying mechanism of metal-microbe interaction, it is important to validate the metal presence inside the cells of bacteria. EDX analysis was performed to achieve this objective.
2.10 Statistical analysis
For each experiment at least three separate flasks were maintained. Each time three readings were taken, their mean, and standard error of the mean were calculated.
3 Results
3.1 Wastewater characteristics and arsenite resistant bacterial isolation
Physicochemical characteristics like temperature, pH, TDS, color and arsenic amount were measured for the environmental sample. The sample characteristics were i.e. temperature 25 °C, pH 6.5, TDS 15500 mg/l and color was muddy. The arsenic concentration, determined from the sample, was 85 µg/l. A lot of bacterial colonies appeared on LB agar supplemented with 5 mM arsenite stress. By increasing the arsenite concentration in agar the number of colonies was decreasing and at the end just one bacterial strain appeared on 75 mM arsenite stress, was selected for further research work. The isolated strain was named as 1SI with MIC of 75 mM corresponding with arsenite.
3.2 Strain identification
The strain isolated in this study was gram positive, rod-shaped and yellow pigmented. Morphological and biochemical characteristics of the isolate are given in Table S1. The growth of bacterial strain on the LB agar plate can be seen in Fig. S1. The 16S rRNA gene sequence of bacterium showed 99% homology with 16S rRNA gene sequence of Microbacterium sp. and was deposited to GenBank under accession number of CP043430. Microbacterium sp. strain 1S1 has also been submitted at First Fungal Culture Bank of Pakistan under accession number FCBP-B-731.
3.3 Arsenite oxidizing test
To check the oxidizing ability of isolated strain, brown precipitation appeared on agar plates having bacterial colonies by applying the AgNO3 (Fig. S2).
3.4 Cross metal resistance
The MIC of isolated strain against other commonly found metal ions is in following concentration Pb (5.5 mM/ml), Cd (4 mM/ml), Cr (5 mM/ml), Ni (4 mM/ml), Hg (6 mM/ml), Se (3 mM/ml), and Co (8 mM/ml).
3.5 Growth conditions and arsenite impact on bacterium
Isolated strain 1S1 showed maximum cell density at 37 °C and pH of 7. The maximum cell density of the strain was measured after 28 h of incubation. The cell density of strain 1S1 culture was higher in control (without arsenite) as compared to the strain 1S1 culturing in arsenite stress (Fig. S3).
3.6 Metal decontamination potential
The bacterial strain 1S1 was grown for 96 h at its optimum culturing conditions (pH 7; 37 °C) and arsenite oxidizing ability was measured at suitable time period of 24 h up to 96 h. Bacterial efficiency of arsenite oxidation was measured after 24, 48, 72, and 96 h was 40, 65, 85, and 98%, respectively (Fig. 1a).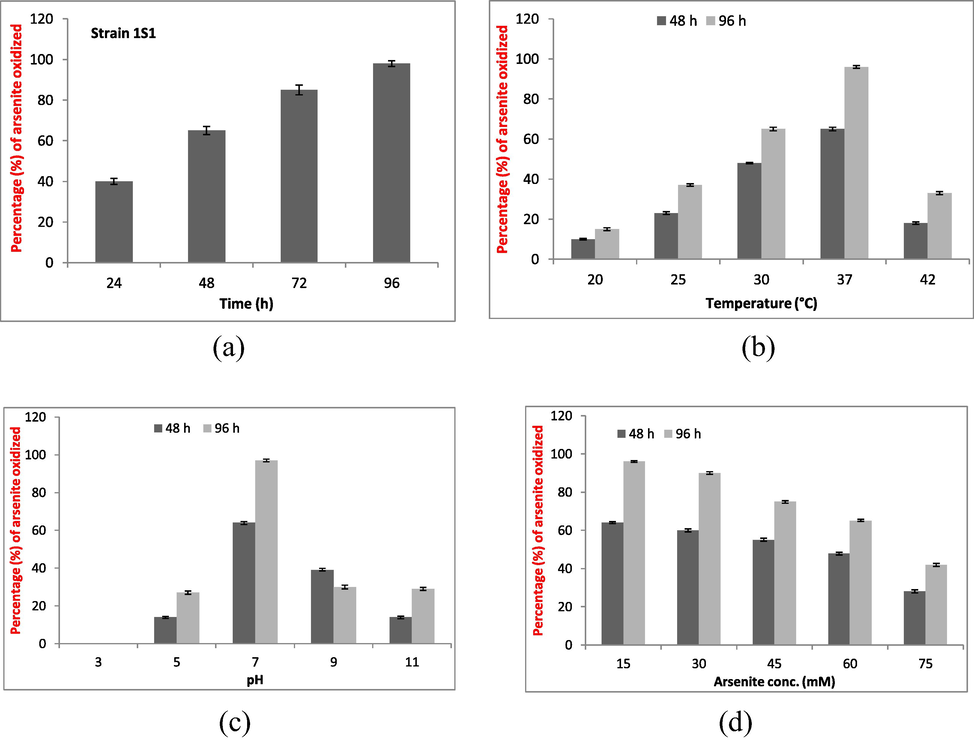
Strain 1S1 was grown at its optimum growth conditions in 250 ml flasks containing 100 ml LB broth with arsenite @15 mM. Bacterium oxidizing potential was checked at time period i.e. 24, 48, 72 and 96 h (a), temperature i.e. 20 °C, 25 °C, 30 °C, 37 °C, and 42 °C (b), pH i.e. 3, 5, 7, 9, and 11 (c), and arsenic concentration i.e.15, 30, 45, 60, and 75 mM (d) after 48 and 96 h of time period. Each treatment was repeated thrice (n = 3).
3.7 Factors versus arsenite oxidizing potential
The metal oxidizing potential of bacterium at various factors including initial arsenite concentration, pH, and temperature was worked out for a time period of 48 and 96 h of incubation.
3.7.1 Effect of temperature
The strain 1S1 showed maximum As+3 oxidation potential at 37 °C i.e. 65 and 96% As+3 oxidizing potential after 48 and 96 h (Fig. 1b). At 20 °C, As+3 oxidizing potential was 10% and 15% after 48 and 96 h while it was 23% and 37% at 25 °C after 48 and 96 h of incubation. At 30 °C, bacterium 1S1 showed 48 and 65% As+3 oxidation potential after 48 and 96 h. Microbacterium sp. has shown 18 and 33% As+3 oxidizing potential at 42 °C after 2 and 4 days (Fig. 1b).
3.7.2 pH effect
No As+3 oxidation was measured at pH 3 after 2 and 4 days while optimum As+3 oxidizing potential was measured at pH 7 after 48 h (64%) and 96 h (97%). At pH 5, 14 and 27% As+3 oxidizing potential was estimated after 48 and 96 h of incubation (Fig. 1c). At pH 9, the oxidizing potential was 39 and 30% after 48 and 96 h of incubation. The bacterium showed 14 and 29% As+3 oxidizing potential after 48 and 96 h at pH 11 (Fig. 1c).
3.7.3 Arsenite concentration effect
The highest arsenite oxidizing potential was measured at 15 mM As+3/L (96%) while the lowest was measured at 75 mM As+3/L (42%). At 30 mM As+3/L, strain 1S1 oxidizing potential was 60% after 48 h (Fig. 1d) and 90% after 96 h. At 45 mM As+3/L, the metal oxidizing potential was 55% after 48 h and 75% after 96 h. At 60 mM As+3/L, the bacterium arsenite oxidizing potential was 48 and 65% after 48 h and 96 h, respectively (Fig. 1d).
3.8 Arsenic biosorption through bacterium heat inactivated biomass
After heat treatment, the bioremediation efficiency (E) was ascertained for 10 h with a time period of 2 h. Microbacterium sp. showed arsenite removal efficiency (E) of 60, 75, 89, and 95% after 2, 4, 6, and 8 h of regular time period. Bacterium inactivated cells eradicated 99% arsenite from the medium after 10 h of incubation contact (Fig. 2a).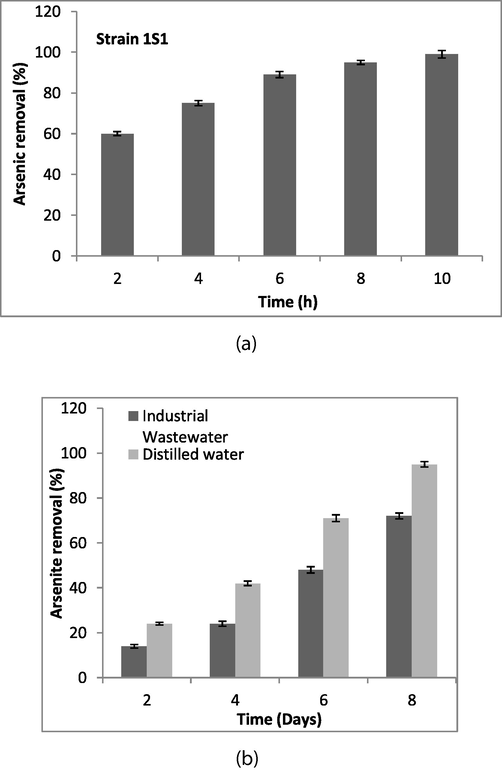
(a) The bacterial culture was grown, centrifuged to obtain pellet, and pellet was incubated at 70 °C (2–3 times) to inactivate the cells. Initially, 1 g/L bacterial biomass was mixed in 1 L of arsenic solution of 1500 ml flasks containing 15 mM As+3 stress. The flasks were incubated at 37 °C on the shaker for 2 to 10 h. Arsenic concentration was estimated before and after incubation through atomic absorption spectrophotometer (b) Arsenite removal potential of strain ISI from industrial wastewater was determined. For this, a set of two plastic containers was used; the first container contained wastewater (10 L) with 1.5 L culture, while the second container contained 10 L distilled water with 1.5 L culture. Each container was supplemented with 15 mM As+3 and incubated at room temperature (25 ± 2 °C). Ten milliliter samples were taken after 2, 4, 6, and 8 days of incubation, centrifuged for 10 min at 4000 rpm, and the samples supernatants were analyzed for arsenite estimation through atomic absorption spectrophotometer.
3.9 Arsenic amelioration from real wastewater
It was ascertained that bacterium has the potential to decrease 15 and 24% arsenite from real and distilled water after 48 h of incubation. After 4 days, bacterium was able to eradicate arsenite 24 and 42% from the real wastewater and distilled water. At day 6, bacterium was able to remove 48 and 72% arsenite from wastewater and distilled water and after incubation of 8 days, the arsenite eradication potential of Microbacterium sp. was 72 and 95% from real wastewater and distilled water (Fig. 2b).
3.10 Antioxidant enzymes
The activities of antioxidant enzymes i.e. catalase, ascorbate peroxidase, peroxidase and superoxide dismutase were determined in strain 1S1 when exposed to 15 mM arsenite. A mixed response of such enzymes was ascertained in metal stressed bacterial cells. The POX and SOD activities were decreased while CAT (240%) and APX (11%) activities were enhanced in metal stressed culture (Table 1).
Antioxidant enzymes
U min−1 mg protein−1
Microbacterium sp. strain 1S1 (without arsenite)
Microbacterium sp. strain 1S1 (with arsenite)
Superoxide dismutase (SOD)
16.12 ± 0.22
6.24 ± 0.22
Catalase (CAT)
20.78 ± 0.14
70.70 ± 1.34
Peroxidase (POX)
0.822 ± 0.06
0.476 ± 0.05
Ascorbate peroxidase (APX)
0.456 ± 0.02
0.506 ± 0.01
3.10.1 Measurement of GSH and NPSHs
A diversified response of bacterium under 15 mM arsenite stress was obtained in term of GSH and GSSG glutathione and NPSHs content. The enhanced percent in GSH and GSSG ratio and NPSHs was 40.0 and 78.50%, respectively. The change in amount of reduced, oxidized, total glutathione, reduced and oxidized glutathione ratio and non-protein thiols in strain 1S1 upon exposure to arsenite in comparison to its absence has been shown in Table S2.
3.11 SEM, EDX, and FTIR analysis
No significant change in bacterial size was measured when the organism was treated with 15 mM arsenite as revealed by SEM analysis (Fig. 3a). The EDX results confirmed the surface absorption of As+3 into the microbial cells when challenged with 15 mM As+3 but no arsenite surface absorption was measured in bacterial culture having no arsenite stress (Fig. 3b).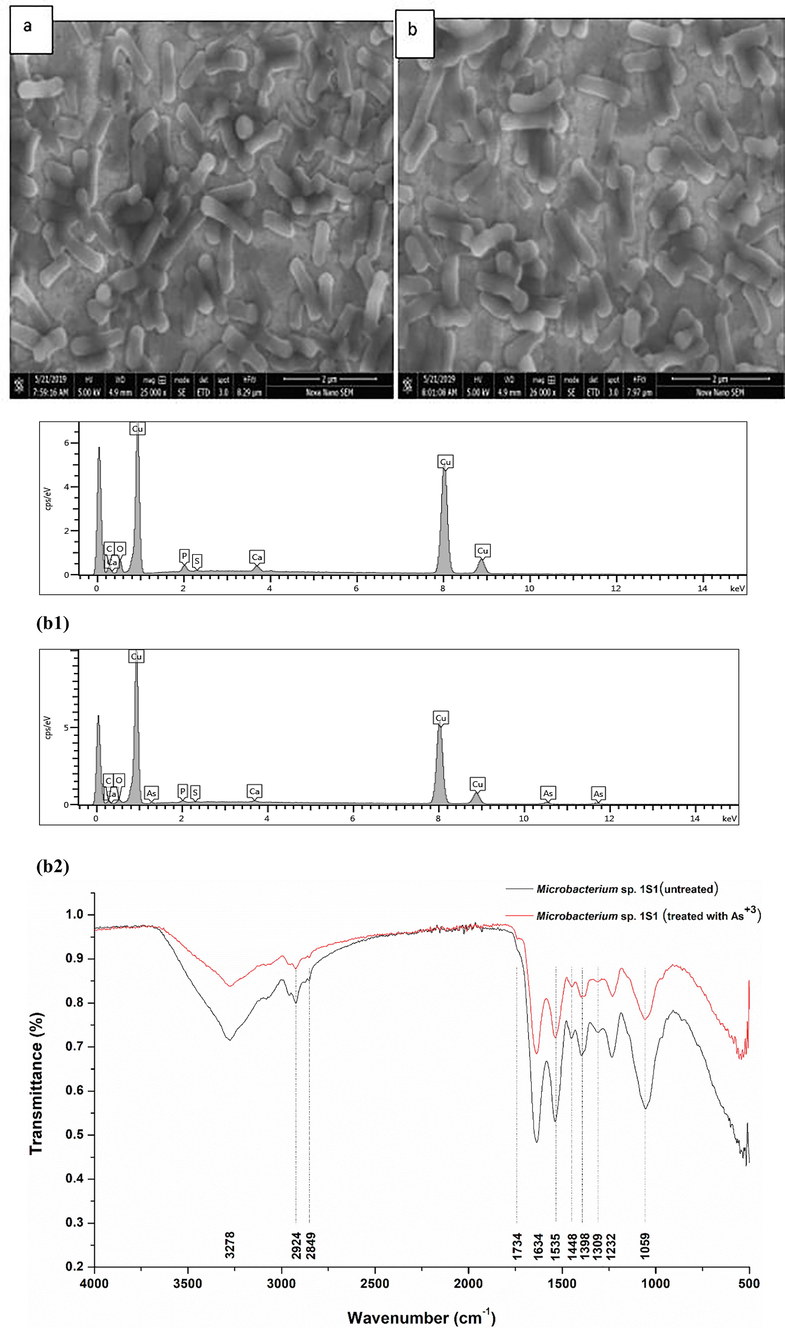
(a) SEM analysis; (a) Strain 1S1 cells in exponential growth phase without As+3 showing cocci shape morphology. (b) Strain 1S1 cells in exponential growth phase challenged with 15 mM As+3 showing no change in bacterial cells shape morphology, (b) (b1) Energy dispersive X-ray spectrum of Microbacterium sp. in exponential phase without 15 mM As+3, (b2) Energy dispersive X-ray spectrum of Microbacterium sp. in exponential phase when challenged with 15 mM As+3. (c) FTIR analysis of Microbacterium sp. strain 1S1, red line represents cells challenged with 15 mM As+3 and black line indicates control (no As+3).
The shifting of various peaks in FTIR analysis in Microbacterium sp. challenged with arsenite (15 mM) could be allocated to various functional groups which might be involved in arsenite adsorption or uptake into the microbial cells. FTIR analysis revealed that amide and hydroxyl moieties play an active role in arsenite adsorption by showing a displacement in the area of 3278–2851 cm−1. The shifting of peaks from 1741 to 1220 cm−1 in the bacterial cell wall is due to amide linkages of peptides and proteins. The possible cause behind the peaks shift from 1228 to 1038 cm−1 is stretching of C–O from an alcohol, carboxylic acid, and ester and stretching of C–N from an aliphatic amine indicating the interaction of arsenite ions with bacterial cell chemical moieties (Fig. 3c).
4 Discussion
Metal resistance mechanism is commonly found in microorganisms including bacteria i.e. both in gram positive as well as in gram negative bacterial strains but most frequently found in gram positive bacteria (Albarracín et al., 2008; Elahi and Rehman, 2019). Many researchers reported that arsenic resistance (Taoxidizing ability is most common in gram positive bacteria and only three gram positive genera i.e. Bacillus, Arthrobacter, and Microbacterium are well known for this capability (Mokashi and Paknikar, 2002; Prasad et al., 2009; Cavalca et al., 2010). It was reported that Microbacterium lacticum can tolerate arsenite 50 mM while the MIC of the isolated Microbacterium sp. strain 1S1 was 75 mM. The isolated strain was also able to resist other metal ions and the order of MIC is As+5 > As+3 > Se > Ni > Pb > Cr > Cd > Co=Hg. Similarly, the multiple metal resistance of Bacillus cereus and Acinetobacter junii in the following order As+3 > As+5 > Cu+2 > Pb+2 > Cd+2 > Cr+6 was reported by Naureen and Rehman (2016).
In biogeochemical cycling of arsenic, microorganisms play an important role in oxidation and reduction of arsenic and change its mobility and bioavailability in the environment (Glazer and Nikaido, 1995; Oremland and Stolz, 2003). The autotrophic microorganisms accept a pair of electrons from arsenite for the sake of energy in combination with carbon dioxide (Santini et al., 2000). Through the activity of arsenite oxidase heterotrophic bacteria convert arsenite into arsenate (Muller et al., 2003). In this present study, Microbacterium sp. 1S1 was isolated from chemical industry wastewater located in district Sheikhupura, Pakistan. The isolated bacterial strain is chemoautotrophic and accepts electrons from arsenite for the sake of energy gain. Many researches all over the world have already characterized various arsenic resistant bacteria having different capacity for arsenic detoxification (Valenzuela et al., 2009; Chang et al., 2010; Hamamura et al., 2013; Noreen and Rehman, 2016; Sher and Rehman, 2019; Sher et al., 2020a).
In the present study, strain 1S1 was able to oxidize arsenite 40, 65, 85, and 98% after 24, 48, 72, and 96 h (Fig. 1a) while Microbacterium sp. strain 1S1 inactivated biomass removed 99% of arsenite after 10 h. The bacterium has also shown its ability to remove 72% arsenite from the real wastewater indicating its propitious potential to eradicate metal ions from the contaminated sites (Fig. 2b). Sher et al. (2020b) reported that inactivated biomass of Micrococcus luteus strain AS2 removed 95% arsenite after 8 h. This laboratory has embarked upon characterizing and reporting the multiple metal resistant microorganisms with efficient potential to exterminate toxic metal ions from the environment. The Staphylococcus sp. strain AS6 was able to remove 93% arsenite through its inactivated biomass from the medium after 10 h of contact time (Sher et al., 2020a). Tapase and Kodam (2018) reported that Microvirga indica S-MI1b sp. nov., isolated from metal industry soil, was able to completely oxidize 15 mM of arsenite in 39 h. Fazi et al. (2016) reported that many of the communities but predominantly Alphaproteobacteria were able to oxidize arsenite up to 95%, in most of the experimental conditions (Table 2).
Sr. #
Bacterial strain
Arsenite resistance
(mM)Arsenite oxidationpotential
(%)Reference
1
Microbacterium sp.strainA33
50.0
–
Achour-Rokbani et al. (2010)
2
Microbacterium sp. GE1017
61.5
–
Kaushik et al. (2012)
3
M. paraoxydans strain 3109
69.2
–
Kaushik et al. (2012)
4
Microbacterium sp.CAS905i
10.0
–
Paul et al. (2014)
5
Micrococcus sp. EIKU8
25.0
86
Bhakat et al. (2019)
6.
Microbacterium sp.strain1S1
75.0
98
This study
The metal resistant bacterium showed peaks under EDX upon exposure to arsenite (15 mM). On the other hand no peaks were noted in the bacterial culture which was not challenged with arsenite. Another research work reported that in strain RJB-2 arsenic peak was present when stressed with arsenic through EDX analysis (Banerjee et al., 2011). A similar EDX peaks pattern was observed in Klebsiella pneumoniae strain SSSW7 (Mujawar et al., 2019). The displacement of specific functional groups peaks in the presence of arsenite when compared with the peaks of non-stressed culture indicates the interaction of cations (metal ions) with anions (microbial cell wall). This underlying mechanism has been revealed by FTIR analysis. The bacterial cell wall harbors functional groups which could be carboxyl, hydroxyl, and amino group. The peak at 3278 cm−1 represents a strong bond between –NH and –OH groups of biomass. The peaks range between 1742 and 1242 can be allocated to –C=O in the given spectrum. In the given spectrum, the peaks between 1042 and 1059 can be assigned to C–O stretching of carboxylic acid and alcohols. Wu et al. (2010) and Singh et al. (2016) reported similar functional groups interaction with arsenite in E. coli, Arthrobacter sp., and B. aryabhattai, respectively.
Arsenic produces ROS in living cells (Hughes et al., 2011). The antioxidant enzymes play major role in cells protection from ROS damage (Jha et al., 2015). In this investigation, catalase synthesis was significant in comparison to the ascorbate peroxidase against ROS. Both enzymes i.e. ascorbate peroxidase and catalase have the same role to dismutase hydrogen peroxide (H2O2). The low level synthesis of superoxide dismutase in arsenic stressed bacterial culture in comparison to the control was due to inhibitory effect of arsenite on superoxide dismutase. The H2O2 toxicity can inhibit the superoxide dismutase activity as reported by Scandalios (1997). Jobby et al. (2016) found that Enterobacter sp. MUM2 upon exposure of 9 mM arsenite enhanced catalase activity 4.6 folds in comparison to the control and no considerable change in POX and APX activities was determined.
Microorganisms produce GSH against toxic metal ions. GSH has a great affinity for metals due to presence of cysteine residues (Qiu et al., 2014). GSH is a strong reducing agent and can reduce the other things and oxidizes itself. For example H2O2 reduction can happen with GSH and as a result H2O2 converts into H2O and GSH oxidizes into GSSH. In oxidative stress studies, one important factor is to work out the ratio of GSH/GSSG. It was noted that level of GSH, ratio of GSH/GSSG and non-protein thiols content were all increased in 15 mM arsenite presence as compared to its absence (Table S2). In Staphylococcus strain AS2 upon exposure to 15 mM arsenite the ratio of GSH/GSSG and non-protein thiols content were increased up to 43.9% and 72.72% as compared to the culture without 15 mM arsenite (Sher et al., 2020b).
5 Conclusions
In conclusion, a multiple metal resistant Microbacterium strain 1S1, isolated from industrial wastewater, resisted arsenite up to 75 mM. The microbe–metal interaction, adsorption on cell wall surface and subsequent uptake was confirmed SEM, EDX, and FTIR analysis under exposure of 15 mM arsenite. To understand the underlying mechanism of metal resistance under arsenite exposure antioxidant enzymes, GSH/GSSG ratio, and NPSHs levels which were enhanced up to 240, 40.0, and 78.50%. The strain 1S1 was able to oxidize 98% arsenite after 96 h at lab scale while inactivated strain 1S1 biomass removed 99%/10 h arsenite from the medium. Resistance strategies and an efficient arsenic bioremediation potential make Microbacterium sp. strain 1S1 a most suitable candidate to eradicate arsenite from the contaminated area.
Funding
No funding was received for this work from any organization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl. Environ. Microbiol.. 2010;76(3):948-955.
- [Google Scholar]
- Copper removal ability by Streptomyces strains with dissimilar growth patterns and endowed with cupric reductase activity. FEMS Microbiol. Lett.. 2008;288(2):141-148.
- [Google Scholar]
- Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J. Environ. Sci. Health A. 2011;46(14):1736-1747.
- [Google Scholar]
- Characterization of arsenic oxidation and uranium bioremediation potential of arsenic resistant bacteria isolated from uranium ore. Environ. Sci. Pollut. Res.. 2019;26(13):12907-12919.
- [Google Scholar]
- Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast. 1997;13(9):819-828.
- [Google Scholar]
- Identification of an aox system that requires cytochrome c in the highly arsenic-resistant bacterium Ochrobactrum tritici SCII24. Appl. Environ. Microbiol.. 2009;75(15):5141-5147.
- [Google Scholar]
- Isolation of arsenite-oxidizing bacteria from industrial effluents and their potential use in wastewater treatment. World J. Microbiol. Biotechnol.. 2011;27(10):2435-2441.
- [Google Scholar]
- Arsenic-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an arsenic polluted soil, and screening of potential plant growth-promoting characteristics. Syst. Appl. Microbiol.. 2010;33(3):154-164.
- [Google Scholar]
- Arsenic detoxification potential of aox genes in arsenite-oxidizing bacteria isolated from natural and constructed wetlands in the Republic of Korea. Environ Geochem. Health. 2010;32(2):95-105.
- [Google Scholar]
- Single-walled carbon nanotube coated antibacterial paper: preparation and mechanistic study. J. Mater. Chem. B.. 2013;1:2639-2646.
- [Google Scholar]
- Multiple metal resistance and Cr6+ reduction by bacterium, Staphylococcus sciuri A-HS1, isolated from untreated tannery effluent. J. King Saud Univ. Sci.. 2019;31(4):1005-1013.
- [Google Scholar]
- The arsenite oxidation potential of native microbial communities from arsenic-rich freshwaters. Microb. Ecol.. 2016;72(1):25-35.
- [Google Scholar]
- Microbial Biotechnology: Fundamentals of Applied Microbiology. New York, USA: WH Freeman and Company; 1995. p. :561-614.
- Identification of antimony-and arsenic-oxidizing bacteria associated with antimony mine tailing. Microbes Environ.. 2013;28(2):257-263.
- [Google Scholar]
- Arsenic exposure and toxicology: a historical perspective. Toxicol. Sci.. 2011;123(2):305-332.
- [Google Scholar]
- Differential expression of antioxidant enzymes during degradation of azo dye reactive black 8 in hairy roots of Physalis minima L. Int. J. Phytoremediat.. 2015;17(4):305-312.
- [Google Scholar]
- Differential expression of antioxidant enzymes under arsenic stress in Enterobacter sp. Environ. Prog. Sustain. Energy. 2016;35(6):1642-1645.
- [Google Scholar]
- Arsenic hyper-tolerance in four Microbacterium species isolated from soil contaminated with textile effluent. Toxicol. Int.. 2012;19(2):188-194.
- [Google Scholar]
- Cadmium resistance and uptake by bacterium, Salmonella enterica 43C, isolated from industrial effluent. AMB Express. 2016;6(1):1-16.
- [Google Scholar]
- Arsenite-oxidizing and arsenate-reducing bacteria associated with arsenic-rich groundwater in Taiwan. J. Contam. Hydrol.. 2011;123(1–2):20-29.
- [Google Scholar]
- Methods in Enzymatic Analysis. New York: Academic Press; 1974. p. :885-894.
- Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur. J. Biochem.. 1974;47(3):469-474.
- [Google Scholar]
- Arsenic (III) oxidizing Microbacterium lacticum and its use in the treatment of arsenic contaminated groundwater. Lett. Appl. Microbiol.. 2002;34(4):258-262.
- [Google Scholar]
- Arsenite biotransformation and bioaccumulation by Klebsiella pneumoniae strain SSSW7 possessing arsenite oxidase (aioA) gene. BioMetals. 2019;32(1):65-76.
- [Google Scholar]
- Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. J. Bacteriol.. 2003;185(1):135-141.
- [Google Scholar]
- Arsenite oxidizing multiple metal resistant bacteria isolated from industrial effluent: their potential use in wastewater treatment. World J. Microbiol. Biotechnol.. 2016;32(8):133.
- [Google Scholar]
- Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance. Environ. Exp. Bot.. 2004;52(3):199-223.
- [Google Scholar]
- Characterization of arsenite-oxidizing bacteria isolated from arsenic-contaminated groundwater of West Bengal. J. Environ. Sci. Health, Part A. 2014;49(13):1481-1492.
- [Google Scholar]
- Purification and characterization of arsenite oxidase from Arthrobacter sp. BioMetals. 2009;22(5):711-721.
- [Google Scholar]
- Enhanced phytoremediation of toxic metals by inoculating endophytic Enterobacter sp. CBSB1 expressing bifunctional glutathione synthase. J. Hazard Mater.. 2014;267(1):17-20.
- [Google Scholar]
- Copper accumulation, synthesis of ascorbate and activation of ascorbate peroxidase in Enteromorpha compressa (L.) Grev. (Chlorophyta) from heavy metal-enriched environments in northern Chile. Plant Cell Environ.. 2003;26:1599-1608.
- [Google Scholar]
- Solubility and mobility of copper, zinc and lead in acidic environments. Plant Soil. 1995;171(1):53-58.
- [Google Scholar]
- A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl. Environ. Microbiol.. 2000;66(1):92-97.
- [Google Scholar]
- Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Oxidative Stress in Mitochondria. New York: Cold Springer Harbor Lobortory Press; 1997. p. :169-200.
- Antioxidative enzyme profiling and biosorption ability of Cupriavidus metallidurans CH34 and Pseudomonas putida mt2 under cadmium stress. J. Basic Microbiol.. 2015;55(3):374-381.
- [Google Scholar]
- Multiple resistance mechanisms in Staphylococcus sp. strain AS6 under arsenite stress and its potential use in amelioration of wastewater. J. King Saud Univ. Sci.. 2020;32(7):3052-3058.
- [Google Scholar]
- Phenotypic and genomic analysis of multiple heavy metal–resistant Micrococcus luteus strain AS2 isolated from industrial waste water and its potential use in arsenic bioremediation. Appl. Microbial Biotechnol.. 2020;104(5):2243-2254.
- [Google Scholar]
- Use of heavy metals resistant bacteria-a strategy for arsenic bioremediation. Appl. Microbial Biotechnol.. 2019;103(15):6007-6021.
- [Google Scholar]
- Microplate screening assay for detection of arsenite oxidizing and arsenate-reducing bacteria. FEMS Microbiol. Lett.. 2004;237:249-253.
- [Google Scholar]
- Arsenic mediated modifications in Bacillus aryabhattai and their biotechnological applications for arsenic bioremediation. Chemosphere. 2016;164:524-534.
- [Google Scholar]
- Assessment of arsenic oxidation potential of Microvirga indica S-MI1b sp. nov. in heavy metal polluted environment. Chemosphere. 2018;195:1-10.
- [Google Scholar]
- An oxidoreductase AioE is responsible for bacterial arsenite oxidation and resistance. Sci. Rep.. 2017;7:41536.
- [Google Scholar]
- The characteristics of Escherichia coli adsorption of arsenic (III) from aqueous solution. World J. Microbiol. Biotechnol.. 2010;26(2):249-256.
- [Google Scholar]
- Microbiology of inorganic arsenic: from metabolism to bioremediation. J. Biosci. Bioeng.. 2014;118(1):1-9.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102066.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







