Translate this page into:
Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in south India
*Corresponding author at: Department of Biochemistry, Kasturba Medical College, Manipal University, Manipal-576104, India. Tel.: +91 8202922326; fax: +91 08202571927 drpragnarao@gmail.com (Pragna Rao)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Coronary artery disease (CAD) also known as coronary heart disease is one of the leading causes of death and disability worldwide. Genetic and environmental factors play an important role in the pathogenesis and progression of CAD. The aim of this study was to evaluate the combined contribution of 3 gene polymorphisms to the risk of CAD and gene–gene interaction in the south Indian population. In this case-control study, 200 cases of CAD and 200 healthy controls were recruited. We studied 3 well known genetic polymorphisms of MTHFR (C677T; rs1801133), PON1 (Q192R; rs662) and ACE (I/D: rs4646994) in relation to CAD in South Indian population. Polymerase chain reaction (PCR) was carried out and followed by the restriction fragment length polymorphism and agarose gel electrophoresis. Genotypes of MTHFR C677T, CT and CT+TT, and PON1 Q192R QR were associated with the risk of CAD (C677T CT+TT vs CC: odds ratio [OR] = 3.3, 95% confidence intervals [CI] = 1.8–6.2; p = 0.00001), (CT vs CC: OR = 3.2, 95%CI = 1.8–5.6; p = 0.00003), and (Q192R QQ vs QR: OR = 2.1, 95%CI = 1.1–3.9; p = 0.03). The allele frequencies for T vs C: OR = 3.1, 95%CI = 1.8–5.3; p = 0.00001 and R vs Q: OR = 1.3, 95% CI = 1.0–1.7 p = 0.03. Multifactor dimensionality reduction (MDR) analysis was carried out with the combination of three genes and the results indicate that MDR analysis showed that, PON1 gene polymorphism formed a significant model in predicting the CAD risk in south Indian population.
Keywords
MTHFR
PON1
ACE
Coronary artery disease
South Indian population
1 Introduction
Atherosclerosis plays a key role in the pathogenesis of coronary artery disease (CAD), a leading cause of death, accounting for approximately 30% of deaths worldwide and has become a priority health problem in India. Currently, genetic and environmental factors play an important role in the pathogenesis and progression of CAD (Liu et al., 2014; Ahmed, 2012). Clinical manifestations of CAD comprise a host of acute and chronic conditions such as stable angina, acute coronary syndromes and heart failure. Various lines of evidence have suggested that genetic factors contribute significantly to the risk of CAD (Liu and Qiu, 2013) as conventional risk factors are insufficient to explain the etiology of CAD in all cases (Vijaya Lakshmi et al., 2011). The formation of athermanous plaques and plaque rupture that leads to thrombotic occlusion of the coronary arteries that are multifactorial with multiple processes contributes to this pathologic cascade. These include inflammation, lipid disorders, renin angiotensin system, diabetic vasculopathy, hyperhomocysteinemia and others. Genetic epidemiologic studies have suggested certain genetic variants, including polymorphisms in the genes like methylenetetrahydrofolate reductase (MTHFR), paraoxonase1 (PON1) and angiotensin converting enzyme (ACE) genes to have a strong association with CAD and related outcomes.
MTHFR is one of the metabolic pathways for CAD and is a key regulatory gene of the remethylation pathway. The C677T polymorphism (rs1801133) in MTHFR has been implicated in vascular disease (Masud and Qureshi, 2011). MTHFR catalyzes the conversion of 5′, 10′-methylenetetrahydrofolate reductase to 5′-methylenetetrahydrofolate reductase, the major circulating form of folate, which is required for a number of metabolic pathways, including the methylation of homocysteine to methionine (Wang et al., 2013). Individuals with C677T polymorphism (rs1801133) have reduced MTHFR activity and elevated homocysteine levels, which is associated with CAD (Danesh and Lewington, 1998).Controversy exists regarding the association of C677T polymorphism and CAD (Klerk et al., 2002; Lewis et al., 2005). Human PON1 is a calcium-dependent glycoprotein that is synthesized in the liver and secreted into the plasma associated with high density lipoproteins. PON1 prevents LDL lipid peroxidation and protects against atherosclerosis and coded by the HUMPONA, located on 7q 21.3, along with two adjacent genes, PON2 and PON3 (Macharia et al., 2012). The Q192R (rs662) polymorphism in the coding region of PON1 gene is an amino acid exchange from glutamine (Q) to arginine (R) in the A575G region and is associated with variation in its hydrolysis activity (Macharia et al., 2012). PON1 activity has been consistently reported to be significantly diminished in atherosclerosis and other cardiovascular diseases (Mohamed et al., 2013; Agrawal et al., 2009). ACE gene is present on the surface of vascular endothelial cells, generates the potent vasoconstrictor angiotensin II from angiotensin I and inactivates the vasodilator bradykinin. Angiotensin II modulates NO synthesis in cardiovascular tissue, and NO modulates the action of angiotensin II (Abdelhedi et al., 2013). A polymorphic variant in intron 16 (ACE-I/D) of the ACE gene, characterized by an insertion (I) or deletion (D) of a repeat sequence of 287 noncoding base pairs (Zintzaras et al., 2008), has been shown to be associated with hypertension and other cardiovascular risk factors (Abdelhedi et al., 2013). The aim of the present case-control study was to examine the association of C677T MTHFR, A575G PON1 and ACE I/D polymorphisms individually and in combination with coronary artery disease in South Indian population.
2 Materials and methods
2.1 Study design
This is a case-control study carried out on 400 subjects from South India region. The study cohort consists of 200 patients with documented CAD and 200 unrelated healthy control subjects. The cases were recruited between January 2007 and January 2012 from the patients, who visited Kamineni hospitals in Hyderabad/Telangana region. Healthy controls (n = 200) were recruited from the general population without any complications.
2.2 Angiographic assessment and participant recruitment
All patients were admitted with acute coronary syndrome and underwent coronary angiogram (CAG). Patients with ⩾70% stenosis in one or more coronary arteries were considered to have CAD. Our study recruited 200 patients with age ranging from 50 to 70 years who have triple vessel disease and underwent coronary artery bypass graft surgery (CABG). We excluded patients with septicemia, liver cirrhosis, renal failure, colitis, cardiomyopathy, congenital heart disease, rheumatic heart disease, neurological and cancer problems.
The control group consisted of 200 (139 males and 61 females) age and gender-matched healthy subjects from the same region, and with prior normal lipid profile. Clinical data were obtained from all patients using a standard case report form (CRF). Our study complied with the Declaration of Helsinki and was approved by the institutional review board, Kamineni hospitals, Hyderabad and informed consent was obtained from all the patients and controls.
2.3 Anthropometric measurements
Body mass index (BMI) was calculated as weight in kg and height in m2 (kg/m2). Height was measured using a wall-mounted stadiometer, and body weight was measured using a digital scale. BMI of each subject was calculated.
2.4 Blood samples
Peripheral blood (5 mL) was collected from all the patients and controls (n = 400) in EDTA tubes. The collected blood was divided into two portions; 2 mL for DNA extraction and molecular analysis and 3 mL for lipid profile in biochemical analysis.
2.5 Lipid profile
Serum sample was collected in the plain tube to measure the total cholesterol (TC), triglycerides (TG), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C) and very low density lipoprotein cholesterol (VLDL-C) by using commercially available kits.
2.6 Molecular analysis
Genomic DNA was extracted using the conventional salting out extraction method (Khan et al., 2013) and quantified by using NanoDrop 2000 (Thermo Fisher Scientific, MA, USA).
2.7 Primers
Genotyping for MTHFR (rs1801133) and PON1 (rs662) was performed by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP). ACE (rs4646994) I/D polymorphism was screened by direct PCR amplification. The details of the primers and PCR products are documented in Table 1. Successful amplification was confirmed by electrophoresis on an ethidium bromide impregnated 2% agarose gel. PCR reactions were performed in a thermal cycler with an initial denaturation at 95 °C for 5 min followed by 35 amplification cycles with each cycle consisting of denaturation at 95 °C for 30 s, 68 °C for rs1801133 (MTHFR gene) polymorphism (50 °C for rs662 polymorphism in PON1 gene and 59 °C for ACE gene) and 30 s, 72 °C for 45 s and the final extension with 72 °C for 5 min. PCR products were digested for 2 h with HinfI for MTHFR gene (Fermentas, USA) at 37 °C (2.5 μL of distilled water with 10 units of enzyme for 15 μL PCR product and 2 μL buffer in a final volume of 20 μL). For PON1 gene, PCR products were digested for 16 h with AlwI (Fermentas, USA) at 37 °C (2.5 μL of distilled water with 10 units of enzyme for 15 μL PCR product and 2 μL buffer in a final volume of 20 μL).
Gene
Mutation
rs number
Amino acid substitution
Forward primers
Reverse primers
PCR fragments
Annealing temperature
Enzyme
Digested fragments
MTHFR
Exon 4
rs1801133
Ala-Val
TGAAGGAGAAGGTGTCTGAGGGA
AGGACGGTGCGGTGAGAGTG
198bp
68 °C
HinfI
C: 198bp
T: 175/23bp
PON1
Exon 6
rs662
Glu-Arg
AAACCCAAATACATCTCCCAGAAT
GCTCCATCCCACATCTTGATTTTA
214bp
60 °C
AlwI
A: 214bp
G: 190/24bp
ACE
Intron 16
rs4646994
–
CTGGAGACCACTCCCATCCTTTCT
GATGTGGCCATCACATTCGTCACGAT
490bp
59 °C
–
I: 490bp
D: 190bp
2.8 Statistical analysis
Data were analyzed using the statistical software for windows (SPSS version 19.0, Chicago, IL, USA) and are represented as mean ± standard deviation (SD). Differences in baseline characteristics between CAD patients and healthy subjects were assessed by χ2 test for categorical variables and independent sample t-test was used to test for continuous variables. Allele and genotype frequency differences between patients and subjects were tested for each SNP using a chi-square test. Odds ratios (ORs) and 95% confidence intervals were calculated by multinomial logistic regression for the allele and genotype frequencies. A p value < 0.05 was considered statistically significant. Multifactor dimensionality reduction (MDR) analysis was carried out with the selected genes to find the correlation.
3 Results
3.1 Characteristics of the study group
Clinical and baseline characteristics of CAD patients and healthy controls are shown in Table 2. The mean age of the CAD patients and the controls was similar. The mean age was not different between the two groups and was 56.4 ± 2.6 for the patients and 55.4 ± 2.8 for the controls. Male formed 70% of the patients and 68% of the control group. There was no age difference between males and females in the two groups. Rate of risk factors for CAD was markedly higher in the patient group; diabetes 45% vs 0% (p < 0.0001), hypertension 44% vs 6% (p < 0.0001), smoking 32.5% vs 10.5% (p < 0.0001) and family history of CAD 24% vs 0% (p < 0.0001). Lipid profile analysis showed significantly higher TC, TG, LDL-C and VLDL-C and lower HDL-C in patients compared to control subjects (p < 0.0001) (Figs. 1 and 2, Table 2).
Profile
Cases (n = 200)
Healthy controls (n = 200)
p-Value
Age (mean ± SD)
56.4 ± 2.6
55.4 ± 2.8
p = 0.29
Sex (Male: Female)
140:60
136:64
p = 0.36
BMI (kg/m2)
27.2 ± 3.6
26.3 ± 3.4
p = 0.42
Hypertension
88 (44%)
12 (6%)
p < 0.0001
Diabetes
90 (45%)
0 (0)
p < 0.0001
Smoking
65 (32.5%)
21 (10.5%)
p < 0.0001
Alcoholism
42 (21%)
18 (9%)
p < 0.0001
Family history of MI
48 (24%)
0 (0)
p < 0.0001
TC (mg/dL)
235.0 ± 13.7
162.9 ± 16.2
p < 0.0001
TG (mg/dL)
173.5 ± 12.5
135.1 ± 10.5
p < 0.0001
HDL-C (mg/dL)
33.1 ± 4.2
38.3 ± 5.4
p < 0.0001
LDL-C (mg/dL)
167.5 ± 14.5
93.7 ± 17.9
p < 0.0001
VLDL (mg/dL)
34.2 ± 2.5
26.8 ± 2.0
p < 0.0001
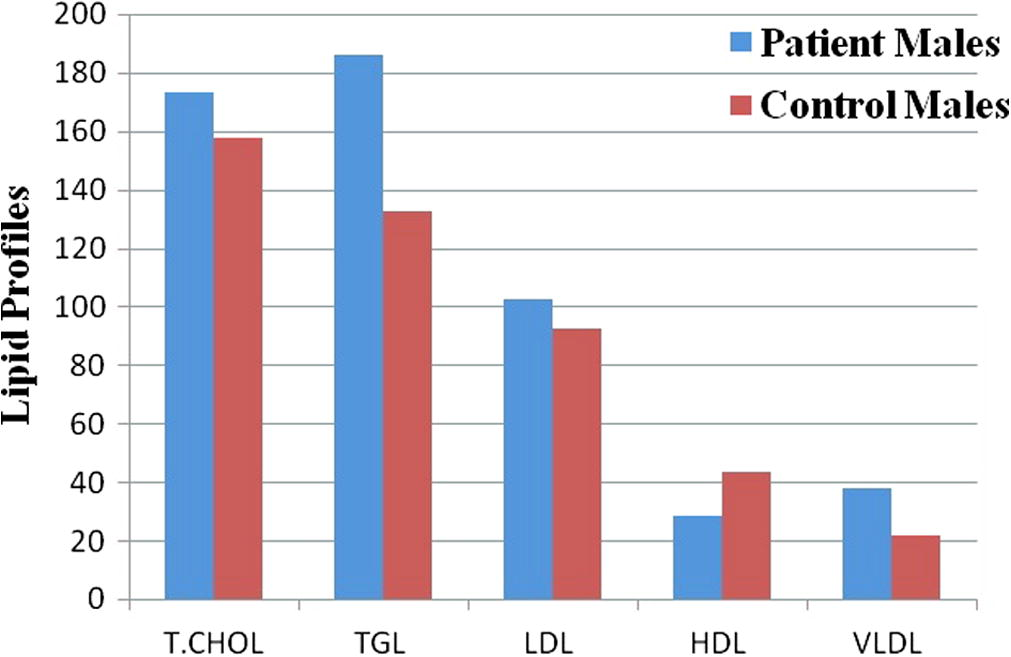
Lipid profile for CAD and control male subjects. T.CHOL – Total Cholesterol; TGL – Triglycerides; LDL – Low Density Lipoprotein Cholesterol; HDL – High Density Lipoprotein Cholesterol and VLDL – Very Low Density Lipoprotein Cholesterol.
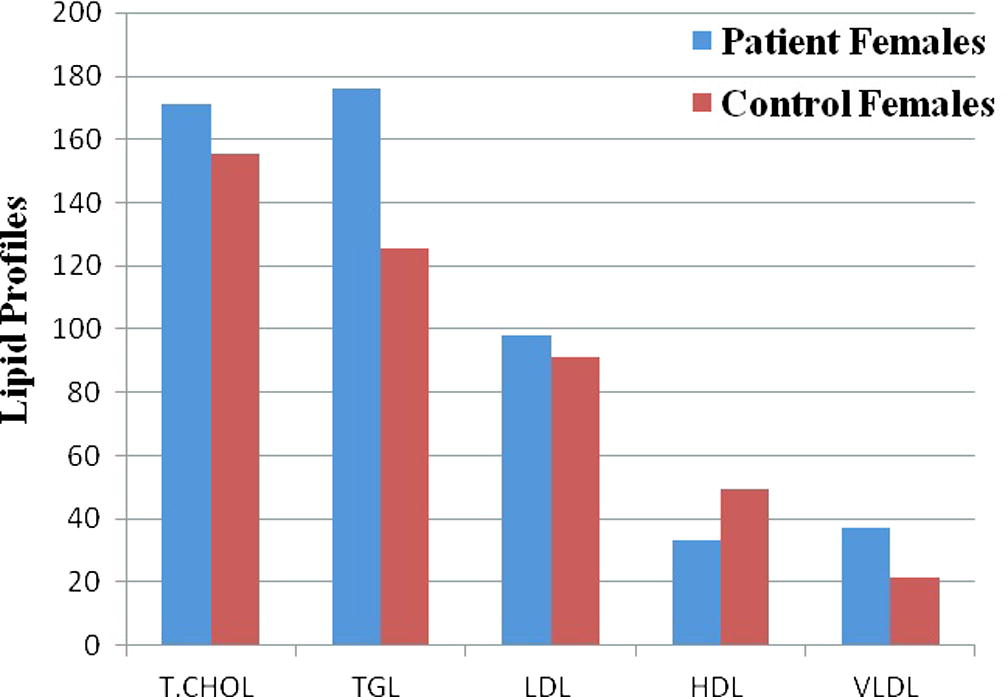
Lipid profile for CAD and control female subjects. T.CHOL – Total Cholesterol; TGL – Triglycerides; LDL – Low Density Lipoprotein Cholesterol; HDL – High Density Lipoprotein Cholesterol and VLDL – Very Low Density Lipoprotein Cholesterol.
3.2 Genotype frequencies
The genotype distribution of the three variants was in Hardy–Weinberg equilibrium in both groups. The genotype and allele distribution of MTHFR (rs1801133), PON1 (rs662) and ACE (rs4646994) variants are summarized in Table 3. There was a statistically significant difference in the genotype distribution in the rs1801133 polymorphism between the patients and controls (p = 0.00003). Similarly, the allele frequencies were significantly different (p = 0.00001). The risk of developing the disease was found to be 3.1-fold for T allele carriers, signifying a potential association. The genotype frequency for PON1 (rs662) polymorphism was also found to be significantly different between the patients and controls with more prevalent GG (p = 0.01), and AA+GG (p = 0.01) genotypes, and the G (p = 0.03) allele of rs662 polymorphisms in CAD patients. Bold values indicates the significantly associated when we perform the odds ratio.
SNP Genotype/allele
CAD patients (n = 200)
Healthy controls (n = 200)
Odds ratio (95%CI)
p-Value
677C→T
CC
148 (74%)
181 (90.5%)
CT
50 (25%)
19 (9.5%)
3.2 (1.8, 5.6)
0.00003
TT
2 (1%)
0 (0)
0.1 (0.04, 0.5)
0.002*
CT+TT
52 (26%)
19 (9.5%)
3.3 (1.8, 6.2)
0.00001
C
346 (0.87)
381 (0.95)
T
54 (0.13)
19 (0.05)
3.1 (1.8, 5.3)
0.00001
192Q→R
QQ
56 (28%)
67 (33.5%)
QR
103 (51.5%)
110 (55%)
1.1 (0.7, 1.7)
0.61
RR
41 (20.5%)
23 (11.5%)
2.1 (1.1, 3.9)
0.01
QR+RR
144 (72%)
133 (66.5%)
1.2 (0.8, 1.9)
0.23
Q
215 (0.54)
244 (0.61)
R
185 (0.46)
156 (0.39)
1.3 (1.0, 1.7)
0.03
I→D
II
65 (32.5%)
60 (30%)
ID
91 (45.5%)
105 (52.5%)
0.8 (0.5, 1.2)
0.33
DD
44 (22%)
35 (17.5%)
1.1 (0.6, 2.1)
0.60
ID+DD
135 (67.5%)
140 (70%)
0.8 (0.5, 1.3)
0.58
I
221 (0.55)
225 (0.56)
D
179 (0.45)
175 (0.44)
1.0 (0.7, 1.3)
0.77
Our results could not find a significant association between ACE I/D polymorphism and CAD in our study population. Table 4 represents the genotype and allele distribution of MTHFR (rs1801133), PON1 (rs662) and ACE (rs4646994) variants based on gender. In male subjects, the odds ratios for having CAD with T allele and CT genotype in MTHFR were 3.4 (CI = 1.2–9.7, p = 0.00008) and 2.7 (CI = 1.4–5.3, p = 0.002), and with G allele and AG genotype of PON1 1.4 (CI = 1.0–2.0, p = 0.02) and 2.6 (CI = 1.2–5.5, p = 0.009). While in female subjects only the T allele and CT genotype in MTHFR were associated with CAD [OR 3.4 (CI = 1.2–9.7); p = 0.001] and [OR 3.8 (CI = 1.3–11.4); p = 0.009]. Power analysis revealed that power of 76% can be achieved by genotyping only 24% of the total number of samples used in our study.
Genotype/allele
CAD
N (%)Healthy controls
N (%)Odds ratio (95% CI)
p-Value
Males
Females
Males
Females
Males
Females
Males
Females
677C→T
CC
102 (73.9)
46 (74.2)
124 (89.2)
56 (91.8)
Reference
Reference
Reference
Reference
CT
34 (24.7)
16 (25.8)
15 (10.8)
5 (8.2)
2.7 (1.4, 5.3)
3.8 (1.3, 11.4)
0.002
0.009
TT
2 (1.4)
0 (0)
0(0)
0 (0)
6.0 (0.2, 127.9)
1.2 (0.02, 62.5)
0.18
0.92
CT+TT
36 (26.1)
16 (25.8)
15 (10.8)
5 (8.2)
2.9 (1.5, 5.6)
3.8 (1.3, 11.4)
0.001
0.009
C
238 (0.86)
108 (0.87)
263 (0.95)
117 (0.96)
Reference
Reference
Reference
Reference
T
38 (0.14)
16 (0.13)
15 (0.05)
5 (0.04)
2.7 (1.5, 5.2)
3.4 (1.2, 9.7)
0.0008
0.01
192Q→R
AA
35 (25.4)
21 (33.9)
45 (32.4)
22 (36)
Reference
Reference
Reference
Reference
AG
70 (50.7)
33 (53.2)
78 (56.1)
32 (52.5)
1.1 (0.6, 1.9)
1.0 (0.4, 2.3)
0.6
0.8
GG
33 (23.9)
8 (12.9)
16 (11.5)
7 (11.5)
2.6 (1.2, 5.5)
1.1 (0.3, 3.8)
0.009
0.7
AG+GG
103 (74.6)
41 (66.1)
94 (67.6)
39 (64)
1.4 (0.8, 2.3)
1.1 (0.5, 2.3)
0.19
0.7
A
140 (0.51)
75 (0.60)
168 (0.60)
76 (0.62)
Reference
Reference
Reference
Reference
G
136 (0.51)
49 (0.40)
110 (0.40)
46 (0.38)
1.4 (1.0, 2.0)
1.0 (0.6, 1.8)
0.02
0.7
I→D
II
45 (32.6)
20 (32.3)
40 (28.8)
20 (32.8)
Reference
Reference
Reference
Reference
ID
61 (44.2)
30 (48.4)
75 (53.9)
30 (49.2)
0.7 (0.4, 1.2)
1 (0.4, 2.2)
0.2
1
DD
32 (23.2)
12 (19.3)
24 (17.3)
11 (18)
1.1 (0.6, 2.3)
1.0 (0.3, 3.0)
0.6
0.8
ID+DD
93 (67.4)
42 (67.7)
99 (71.2)
41 (67.2)
0.8 (0.5, 1.3)
1.0 (0.4, 2.1)
0.4
0.9
I
151 (0.55)
70 (0.56)
155 (0.56)
70 (0.57)
Reference
Reference
Reference
Reference
D
125 (0.45)
54 (0.44)
123 (0.44)
52 (0.43)
1.0 (0.7, 1.4)
1.0 (0.6, 1.7)
0.8
0.8
3.3 Correlation of MTHFR genotypes with serum homocysteine levels (SHL)
Forty two patients exhibited CC genotype while 45 had CT genotype and 2 were TT genotype patients. The Mean ± SD of SHL for CC genotype was 12.95 ± 1.83 μmol/L, for CT genotype 19.34 ± 3.13 μmol/L, for TT genotype 27 ± 1.41 μmol/L. The comparisons of SHL levels with the genotypes in patients are presented in Fig. 3. The levels of SHL were abnormal in both TT and CT genotype patients and were within normal limits in CC genotype patients. Furthermore, SHL were higher in the TT than CT genotype patients. These results indicate that C677T polymorphism might possibly be associated with increased homocysteine levels and may therefore influence individual susceptibility to CAD.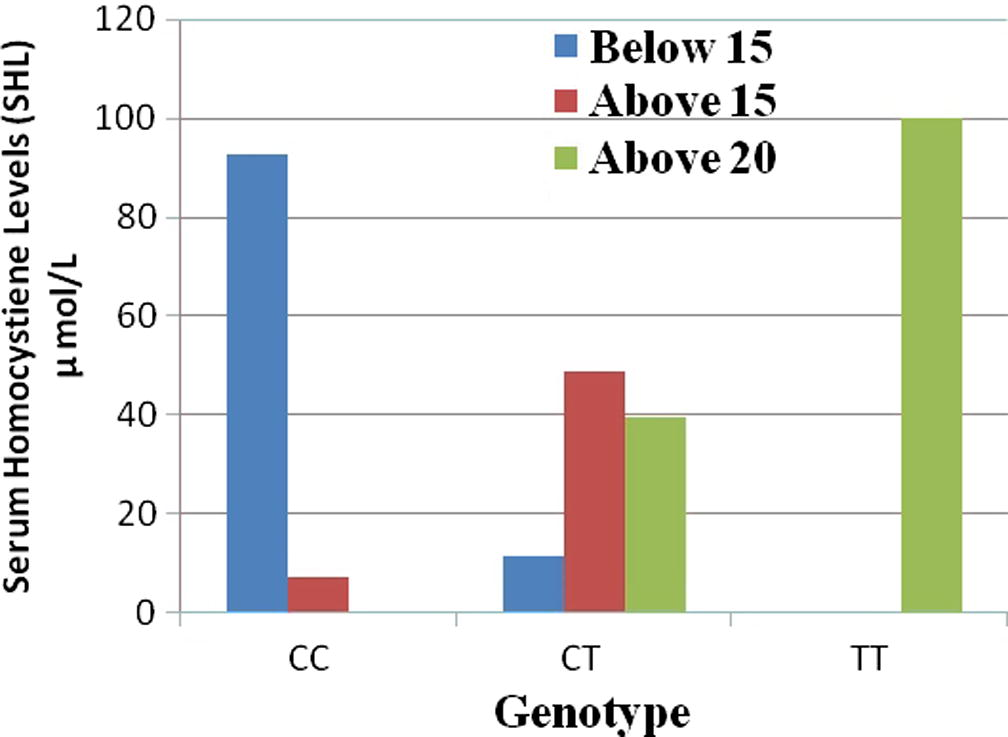
Serum homocysteine levels of MTHFR genotypes in CAD patients.
3.4 Interaction of nuclear genes by MDR analysis
The effect of interaction of three nuclear genes are analyzed in the pathogenesis of CAD by MDR. The list of attributes included three genes (ACE, MTHFR and PON1). All attributes in these datasets are categorical. Graphical representation of MDR is shown in Fig. 4. By MDR model analysis MTHFR can be used as a disease marker because of its high sensitivity value. Further radial graphic representation by MDR analysis was carried out between three genes and the interaction appears to be between PON1 and ACE (Fig. 5).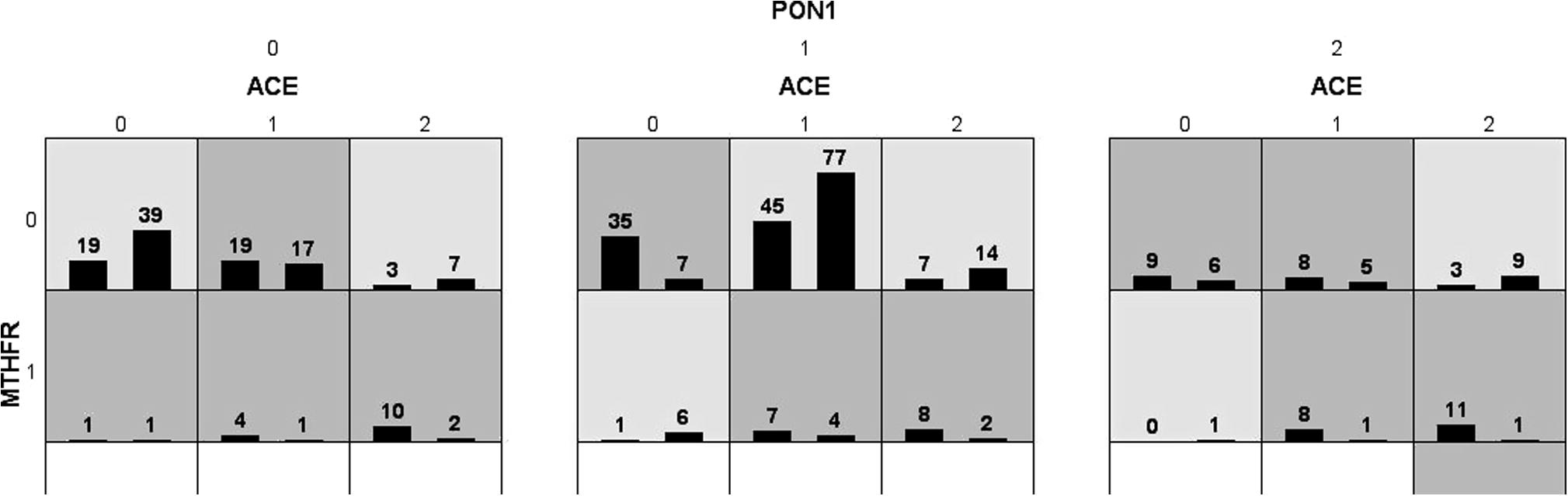
Interaction of MTHFR, PON1 and ACE genes between cases and controls by MDR analysis by graphical representation.
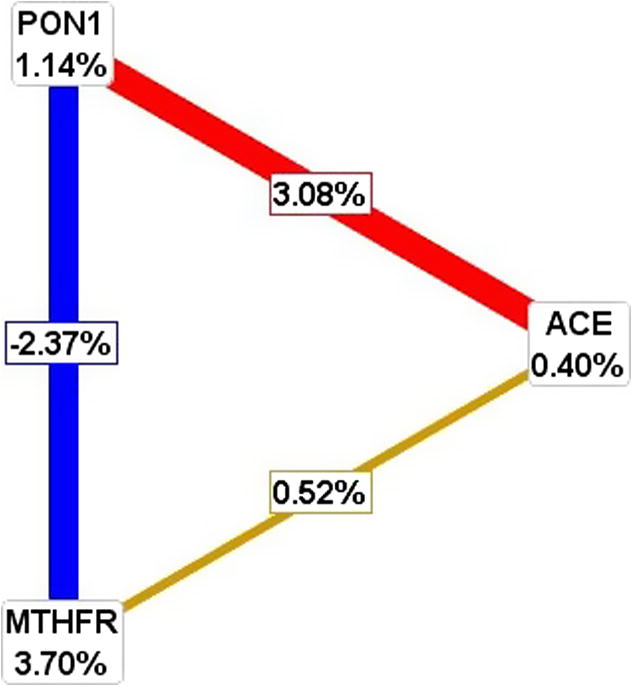
Radial graphic representation of interaction of ACE, MTHFR, PON1 genes by MDR analysis.
4 Discussion
We have carried out a case-control study involving 200 cases and 200 controls with CAD in south Indian population. Thus, two of three studied polymorphisms of the MTHFR and PON1 genes were significantly associated with the risk of CAD. Both the alleles and genotypes were found to be significantly associated. This study shows lack of association with ACE I/D polymorphism but in combination of MDR analysis it is associated with the combination of PON1 and ACE gene, with CAD, is a complex disease with the contribution of multiple genetic and environmental risk factors. A number of studies on the impact of genetic polymorphism in ACE on the risk of coronary heart disease (CHD) have been conducted in various ethnic populations (O’Malley et al., 1999; Oei et al., 2005). In this study, no significant difference was observed in the distribution of genotype of ACE in CAD cases and controls. This confirms previous finding of Lindpaintner et al. (1995), Sobti et al. (2010) and Pandey et al. (2011). The other studies carried out on ACE and CHD risk among Indians have shown association of D allele with the risk of CHD (Ashavaid et al., 2002; Mishra et al., 2012; Dhar et al., 2012). The present results have shown the susceptible effect of polymorphism in ACE on CAD among south Indians to be involved with factors like smoking, alcohol, diabetes and hypertension.
Diabetes and dyslipidemia are two major risk factors for CAD, which are associated with oxidative stress (Ganesan et al., 2011). The analysis of PON1 192 QR polymorphism has revealed that the high 10 activity allele (R) is associated with a more atherogenic lipid profile than the low-activity allele (Q). In this study, PON1 192R allele frequency in CAD patients was found to be higher when compared with the controls (p = 0.03). RR genotype was found to be associated with two fold increase in the risk of CAD development. In our earlier study PON1 gene was proposed to be a candidate biomarker for T2DM and CAD patients (Ganesan et al., 2011). The present study investigated the role of this polymorphism in the development of CAD. The frequencies of the genotypes and alleles were significantly higher in patients compared with controls, indicating the association of PON1 gene Q192R polymorphism with the disease in both the formats (i.e., gene association study and the MDR analysis). The results of the present study were in accordance with observations of earlier investigators, which support that RR genotype is associated with CAD (Ahmed, 2012; Ganesan et al., 2011; Hassan et al., 2013).
MDR analysis was performed to interact the MTHFR, PON1 and ACE genes by the pathogenesis of CAD. Our study concludes that, PON1 can be used as a disease marker because of its high sensitivity value. ACE alone does not play any role in the disease risk but PON1 and ACE interaction increase the risk by 3.08%. There is a negative interaction between PON1 and MTHFR genes, but MTHFR alone has a risk of 3.70%. MDR analysis reveals that in overall entropy (randomness of disorder) MTHFR is showing 3.70%, PON1 is showing 1.14% and ACE is showing 0.4% which is negligible indicating MTHFR and PON1 are showing a major effect. Our study indicated that gene–gene interaction showed a significant model in predicting the CAD risk.
The significance of homozygous polymorphisms considered to be risk factors for CAD when analyzed: MTHFR and PON1. The genotypes with statistically significant higher frequencies in the CAD patients were MTHFR and PON1. Unlike many other studies, our analysis was not limited to examining whether a single polymorphism presents increased risk, but evaluated various polymorphisms and some of the most important gene–gene associations, some of them never studied before (Mendonca et al., 2009). Sanghera et al. (1997), Sanghera et al. (1998) reported that the PON1-192*R allele is a risk factor for CHD among Asian Indians in Singapore. Pati and Pati (1998) reported a higher frequency of the QR and RR genotypes among Indian CAD patients.
We believe that the methodology used in this study was appropriate and that our results may contribute to understanding the far from simple genetic aspects of complex and multifactorial diseases such as CAD. Our results suggest that the association of polymorphisms in the same gene does not increase the risk of the isolated polymorphism, but that the association of polymorphisms in genes belonging to different enzyme systems is linked to increased risk compared to the isolated polymorphisms. The study may explain the difficulty of interpreting the risk of different polymorphisms in CAD, and may open up new possibilities in this field.
In conclusion, our results suggest that the D allele of the ACE (rs4646994) may not be associated, whereas T allele of MTHFR (rs1801133) and R allele of PON1 (rs662) gene polymorphisms were associated with an increased risk of CAD in south Indian population. Our data strongly suggest that PON1 gene is associated with genotype analysis as well as in the combination of MDR analysis.
Acknowledgments
The authors are thankful to the Lady Tata Fellowship for sponsoring the funding for this research. Gratitude is expressed to all the participants who have participated in this study. We are thankful to the Kamineni hospitals for supporting our research work.
References
- Lack of association of NOS3 and ACE gene polymorphisms with coronary artery disease in Southern Tunisia. Biochem. Genet.. 2013;51:92-100.
- [Google Scholar]
- Paraoxonase 1 gene polymorphisms contribute to coronary artery disease risk among north Indians. Indian J. Med. Sci.. 2009;63:335-344.
- [Google Scholar]
- Letter by Ahmed regarding article, “Second internal thoracic artery versus radial artery in coronary artery bypass grafting: a long-term, propensity score-matched follow-up study”. Circulation. 2012;125:e629.
- [Google Scholar]
- Gene polymorphism and coronary risk factors in Indian population. Clin. Chem. Lab. Med.. 2002;40:975-985.
- [Google Scholar]
- Plasma homocysteine and coronary heart disease: systematic review of published epidemiological studies. J. Cardiovasc. Risk. 1998;5:229-232.
- [Google Scholar]
- Polymorphism of ACE gene as the genetic predisposition of coronary artery disease in Eastern India. Indian Heart J..
- The relationship of ACE and CETP gene polymorphisms with cardiovascular disease in a cohort of Asian Indian patients with and those without type 2 diabetes. J. Diabetes Complications. 2011;25:303-308.
- [Google Scholar]
- The Q192R polymorphism of the paraoxonase 1 gene is a risk factor for coronary artery disease in Saudi subjects. Mol. Cell. Biochem.. 2013;380:121-128.
- [Google Scholar]
- Khan, I.A., Jahan, P., Hasan, Q., et al., 2013. Angiotensin-converting enzyme gene insertion/deletion polymorphism studies in Asian Indian pregnant women biochemically identifies gestational diabetes mellitus. J. Renin Angiotensin Aldosterone Syst. (Epub ahead of print).
- MTHFR 677C→T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA. 2002;288:2023-2031.
- [Google Scholar]
- Meta-analysis of MTHFR 677C→T polymorphism and coronary heart disease: does totality of evidence support causal role for homocysteine and preventive potential of folate? BMJ. 2005;331(7524):1053.
- [Google Scholar]
- A prospective evaluation of an angiotensin-converting- enzyme gene polymorphism and the risk of ischemic heart disease. N. Engl. J. Med.. 1995;332:706-711.
- [Google Scholar]
- Association between the receptor for advanced glycation end products gene polymorphisms and coronary artery disease. Mol. Biol. Rep.. 2013;40:6097-6105.
- [Google Scholar]
- Association between the -786T→C 1polymorphism in the promoter region of endothelial nitric oxide synthase (eNOS) and risk of coronary artery disease: a systematic review and meta-analysis. Gene. 2014;545(1):175-183.
- [Google Scholar]
- The growing importance of PON1 in cardiovascular health: a review. J. Cardiovasc. Med. (Hagerstown). 2012;12:443-453.
- [Google Scholar]
- Tetra primer ARMS-PCR relates folate/homocysteine pathway genes and ACE gene polymorphism with coronary artery disease. Mol. Cell. Biochem.. 2011;355:289-297.
- [Google Scholar]
- Gene–gene interaction affects coronary artery disease risk. Rev. Port. Cardiol.. 2009;28:397-415.
- [Google Scholar]
- Impact of renin–angiotensin–aldosterone system gene polymorphisms on left ventricular dysfunction in coronary artery disease patients. Dis. Markers. 2012;32:33-41.
- [Google Scholar]
- Mohamed, A.E., Hasen, A.M., Mohammed, G.F., et al., 2013. Real-Time PCR of cytomegalovirus and Epstein-Barr virus in adult Egyptian patients with systemic lupus erythematosus. Int. J. Rheum. Dis (Epub ahead of print).
- Lipoprotein-associated phospholipase A2 activity is associated with risk of coronary heart disease and ischemic stroke: The Rotterdam Study. Circulation. 2005;111:570-575.
- [Google Scholar]
- Angiotensin-converting enzyme and cardiovascular disease risk. Curr. Opin. Lipidol.. 1999;10:407-415.
- [Google Scholar]
- Association of angiotensin-converting enzyme, methylene tetrahydrofolate reductase and paraoxonase gene polymorphism and coronary artery disease in an Indian population. Cardiol. J.. 2011;18:385-394.
- [Google Scholar]
- Paraoxonase gene polymorphism and coronary artery disease in Indian subjects. Int. J. Cardiol.. 1998;66:165-168.
- [Google Scholar]
- Genetic polymorphism of paraoxonase and the risk of coronary heart disease. Arterioscler. Thromb. Vasc. Biol.. 1997;17:1067-1073.
- [Google Scholar]
- The codon 55 polymorphism in the paraoxonase 1 gene is not associated with the risk of coronary heart disease in Asian Indians and Chinese. Atherosclerosis. 1998;136:217-323.
- [Google Scholar]
- Association of ACE and FACTOR VII gene variability with the risk of coronary heart disease in north Indian population. Mol. Cell. Biochem.. 2010;341:87-98.
- [Google Scholar]
- Interactions of 5′-UTR thymidylate synthase polymorphism with 677C→T methylene tetrahydrofolate reductase and 66A→G methyltetrahydrofolate homocysteine methyl-transferase reductase polymorphisms determine susceptibility to coronary artery disease. J. Atheroscler. Thromb.. 2011;18:56-64.
- [Google Scholar]
- MTHFR C677T polymorphism and risk of congenital heart defects: evidence from 29 case-control and TDT studies. PLoS One. 2013;8:e58041.
- [Google Scholar]
- Angiotensin-converting enzyme insertion/deletion gene polymorphic variant as a marker of coronary artery disease: a meta-analysis. Arch. Intern. Med.. 2008;168:1077-1089.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jksus.2014.09.002.
Appendix A
Supplementary data
Supplementary data
Supplementary data
Supplementary data contains Tables 1–3.







