Comprehensive computational analysis reveals human respiratory syncytial virus encoded microRNA and host specific target genes associated with antiviral immune responses and protein binding
⁎Corresponding authors. rizwan@gcuf.edu.pk (Muhammad Rizwan Javed), sidra.aslam29@yahoo.com (Sidra Aslam)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
Objective
About half-century ago, human respiratory syncytial virus (hRSV) was observed in infants and children under age of five years. As the mechanism of its pathogenesis inside the host is still lacking, in this in-silico study we hypothesized that RSV might create miRNAs, which could target the genes associated with host cellular regulatory pathways, thus provide persistent refuge to virus.
Methodology
Pre-miRNAs in RSV genome (accession no. NC_001803.1) were extracted through VMir software, and the identification of putative pre-miRNAs and mature miRNAs was accessed using iMiRNA-SSF and FOMmiR tool, respectively. Later, prediction of host specific target gene was accompanied by RNAhybrid tool. Moreover, bioinformatics analysis was done for the validation of target genes using microarray dataset (GSE80179). Lastly, Drug-gene interaction database was used to explore the small drug like candidate against RSV infection.
Results
Searching RSV genome for their pre-miRNAs yielded 15 pre-miRNAs like sequence with length vary for each pre-miRNA sequences. FOMmiR tool revealed a total of 7 mature miRNAs from 6 real pre-miRNA hairpins. Functional enrichment analysis of targeted genes, revealed their involvement in many biological pathways which facilitate their pathogenesis in host. The microarray dataset (GSE80179) was analyzed to validate altered expression level of target genes and found genes linked with pathways such as T-cell activation, apoptosis, NF-kappa B signaling, cell differentiation and autophagy. CYCS, CTLA4, and BTK were chosen as possible targets of 21 drugs. A total of 21 drugs were explored using DGIdb that might have potential to treat RSV patient.
Conclusion
This study updates the information and yield a new perspective in context of understanding the pathogenesis of RSV. Our study presents novel miRNAs and their targeted hub genes, which upon experimental validation could facilitate in developing new therapeutics against RSV infection. In vivo and in vitro investigation of miRNAs and pathway interaction is essential to delineate the specific roles of the novel miRNAs, which may help to reveal the mechanisms behind the pathogenesis. Based on the hub genes and miRNAs, experimental models may be designed in terms of the detection of pathogenesis, evaluation of risk, and determining the targeted therapies of RSV infections.
Keywords
In silico analysis
Respiratory syncytial virus
miRNA
Hub genes
Pathway analysis
Gene expression profiling
Candidate drug
- miRNA
-
MicroRNA
- pri-miRNA
-
Primary miRNA
- pre-miRNA
-
Precursor miRNA
- mRNA
-
Messenger RNA
- 3′ UTR
-
3′ Untranslated region
- MFE
-
Minimum free energy
- RSV
-
Respiratory Syncytial Virus
- PPI
-
Protein-Protein Interaction
- DEGs
-
Differential Expressed Genes
- MCC
-
Maximal Clique Centrality
- MNC
-
Maximum Neighborhood Component
- GEO
-
Gene Expression Omnibus
- DAVID
-
Database for annotation, visualization, and integrated discovery
- GO
-
Gene Ontology
- KEGG
-
Kyoto Encyclopedia of Genes and Genomes
- MF
-
Molecular function
- CC
-
Cellular components
- BP
-
Biological process
- STRING
-
Search Tool for the Retrieval of Interacting Genes/Proteins
- MCODE
-
Molecular Complex Detection
Abbreviations
1 Introduction
Viruses has become new threat to the mankind and thus drawn more attention, because of their potential to cause pandemic. Respiratory syncytial virus (RSV) also known as Human respiratory syncytial virus (hRSV) contributes to the infection of respiratory tract. The very first time, RSV was discovered in a colony of chimpanzee with symptoms of respiratory illness (Taleb et al., 2018). Short time later in Baltimore, the same symptoms were examined in children (Chanock and Finberg, 1957). The overall frequency of hospitalization of children is common, particularly those younger than 5 years of age (Graham, 2017). Pneumonia, hypoxia, blockage in any part of the airway, dyspnea, bronchiolitis, and wheezing are commonly seen in infected individuals. Moreover, in the early stages of life, RSV patient has been suffered from asthma (Graham, 2017; Sato et al., 2005). RSV is considered as a key factor of lower respiratory tract infections (LRTI) in children worldwide, with approximately 33 million cases and 160,000–190,000 deaths annually, accounting for 6.7 percent of all infants deaths under the age of one year (Nair et al., 2010). Hence the highest mortality rate reinforced the attentions of researchers to untangle the intricate molecular mechanism underlying and to uncover more enthralling and promising molecular candidates with effective prognostic value.
RSV diversity and variation in the reinfection is a significant factor to design various immune-diagnostic approaches, antiviral treatments, and preventive measures. A good grasp of the genetic variety of accessible viral sequences, as well as their precise genotype assignment, should aid in this analysis. RSV genome encodes eleven proteins that are encoded by >15,000 nucleotides. From these eleven protein, nine are structural and remaining protein are non-structural (Graham et al., 2015; Collins et al., 1986) along with the attachment protein, small hydrophobic protein, and fusion protein as surface protein (Rey-Jurado and Kalergis, 2017). However, the exact mechanism underlying this viral infection is poorly known. It has been pointed out that COVID-19 share symptoms with RSV, however no clear study has made on it yet. In this era of coronavirus, researchers investigated that children who claimed to have COVID-19 were basically have infection due to other viruses, mainly from RSV (Mansbach et al., 2020). The bell is ringing slightly, therefore the need of the hour is to develop a sound understanding behind the pathogenesis of RSV infection.
Micro-RNAs (miRNAs) are emerged as leading players in the viral infection due to post-translational regulatory activity in gene expression of many biological functions (Zhang et al., 2006). miRNAs come forward as the targets of interest due to their involvement in metabolic pathways. Certain piece of evidences revealed that miRNAs may have critical functions to play in viral infection (Fani et al., 2018; Sullivan and Ganem, 2005). In the past, miRNAs of human were administered by targeting the associated genes for combating the pathogenesis of infections (Hariharan et al., 2005). Multiple studies report the existence miRNA behind the pathogenesis of Ebola, Zika, and dengue viruses (Li and Zou, 2019). Moreover, it is becoming a fantastic research subject for various molecular biology researchers all around the world. More research on miRNAs regarding their contribution in viral infection revealed the potential miRNAs shown to target specific host genes (Ghosh et al., 2009; Grundhoff and Sullivan, 2011).
Numerous studies have been conducted on the RSV infection, but currently there is no sufficient evidence to prove the existence of miRNAs in RSV genome. To tackle this issue, we conducted computational analysis for identifying putative miRNAs in RSV genome. Our findings provide potential RSV-produced miRNAs and their targeted genes, which facilitate the pathogenesis and prolonged refuge of virus in host. Experimental validation of these miRNAs could facilitate in developing new therapeutics against RSV infection.
2 Materials and methods
2.1 Retrieval of RSV genome and prediction of pre-miRNA
National Center for Biotechnology Information (NCBI) is a freely available public data-hub (Pruitt and Maglott, 2001). By using RSV as a search term, RSV genome (accession no. NC_001803.1) was retrieved from NCBI in FASTA format. In the current work, the entire genome of RSV was subjected to the an ab intio based program, VMir software, to search out the viral genome for the presence of stem loop structures in primary miRNAs (pri-miRNA) and precursor miRNAs (pre-miRNA) and to distinguish putative candidates stretches having ability to develop a secondary stem loop structure (Grundhoff, 2011). The entire genome was subjected VMir software by setting all parameters (minimum hairpin size: 50 nt, minimum hairpin score: 100, and window of adjustable size: 500 nt)
2.2 Identification of real and pseudo miRNAs
Identification of real miRNAs from pseudo miRNAs in a preliminary step to figure out their mature sequences. In this regard, pre-miRNA sequences obtained from VMir were submitted to iMiRNA-SSF to differentiate real miRNAs from pseudo miRNAs as for example hairpin sequences having same stem-loops (Chen et al., 2016). iMiRNA-SSF is concerned with the prediction of pre-miRNA by integrating negative sets having different distribution. Later, RNAFold software was employed for the prediction of secondary structure of the real miRNA. The real pre-miRNA having hairpin size at least 50nt were analyzed using RNAFold algorithm. Moreover, the positional entropy along with minimum free‐energy (MFE) of real pre-miRNAs were created (Fig. 1).
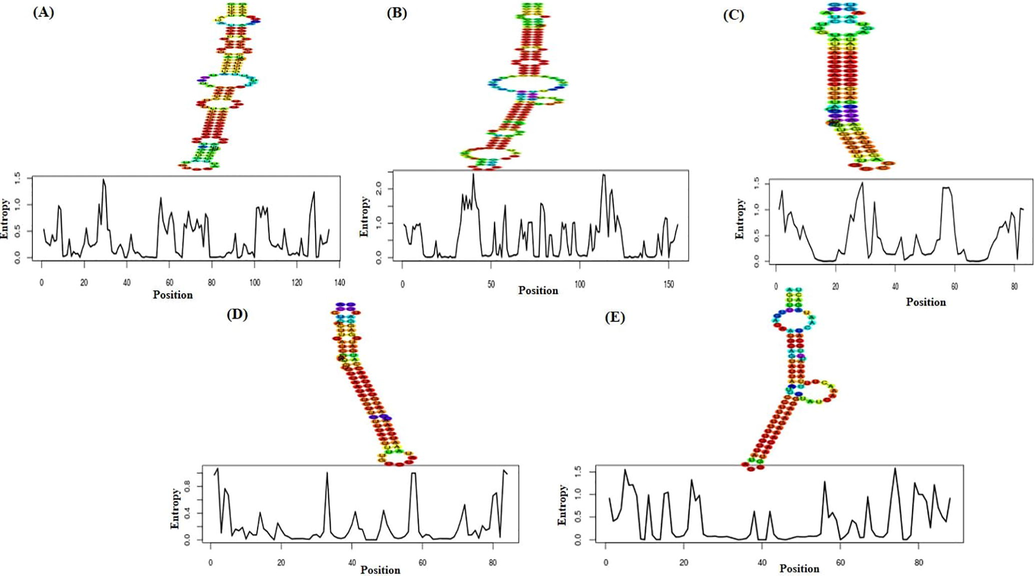
- Minimum free‐energy (MFE) value in RNAfold webserver and the MFE secondary structure and positional entropy of 5 real pre-miRNAs were created. (A) MR28; (B) MR52; (C) MR65; (D) MR121; (E) MR180.
2.3 Identification of mature miRNAs
The identification of mature miRNAs is very important to find their targeted host genes. With the purpose of extracting the mature miRNA position from the pre-miRNA hairpins, FOMmiR tool (Shen et al., 2012) was used. This tool use fixed-order Markov model for distinguishing the mature miRNAs and also provides information regarding the strand of mature miRNAs.
2.4 Prediction of host specific target gene
miRNA exert their effect by targeting three prime untranslated regions (3′-UTR) of messenger RNA (mRNA). Therefore, the prediction of target host gene in RSV genome could assist in the understanding of disease pathogenesis along with interaction among host and pathogens. Regarding that, RNAhybrid (with default parameters and cutoff of p-value < 0.05 to filter target genes) was employed in order to identify the host specific target genes of predicted miRNAs (Krüger and Rehmsmeier, 2006). The mature miRNA predicted using FOMmiR tool were used as input. The MFE percentage in RNAhybrid was kept 75% for the identification host specific target gene of mature miRNAs.
2.5 Construction of PPI network
Protein-protein interaction (PPI) network were constructed to determine the functional interactions between the resulted target genes. Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) was used for the functional interactions among target genes (Mering et al., 2003). The resulted genes were subjected to the Cytoscape_v3.8.2 (Shannon et al., 2003). Molecular Complex Detection (MCODE) plugin from cytoscape was utilized for distinguishing the module that best represent the clusters of target genes (Cline et al., 2007). In MCODE, the modules were considered significant having number of node > 4 and the score ≥ 4.
2.6 Selection of hub genes
CytoHubba plugin in Cytoscape was used for distinguishing hub genes among host specific target genes. A total of twelve topological analysis methods are available in the cytoHubba. Several topological algorithm in cytoHubba predicts and explore the essential nodes and subnetworks in a given network. To further improve the specificity and sensitivity, 25 hub-forming proteins were identified on the basis of local approaches, including Maximal Clique Centrality (MCC), and Maximum Neighborhood Component (MNC), as well as three centrality methods involving closeness, betweenness, and degree respectively. Later, the topmost fifty genes ranked by MCC, MNC, degree, betweeness and closeness were selected. Finally, the FunRich tools was used to explore interaction among hub genes and their associated neighboring genes (Pathan et al., 2015).
2.7 Analysis of targeted genes at functional level
At functional level, database for annotation, visualization, and integrated discovery (DAVID) was used to perform GO enrichment analysis and KEGG pathway analysis (Dennis et al., 2003). The target genes were subjected to DAVID for the prediction of the function of genes at three level: Molecular function (MF), Biological process, and Cellular component (CC).
2.8 Gene expression microarray analysis
Gene Expression Omnibus (GEO) database in National Center for Biotechnology Information (NCBI) is a freely available public database, enclosing the gene profiles (Barrett et al., 2012). Expression profile of GSE80179 based on GPL10558 platform (Illumina HumanHT-12 V4.0 expression beadchip) were retrieved from NCBI-GEO database. Dataset includes 52 healthy controls and 27 RSV infected person.
2.9 Drug-gene interaction
Using Drug gene interaction database (DGIdb), related dugs of hub genes were chosen that acted as enthralling and promising target (Griffith et al., 2013). Only drugs that have recently been approved by the Food and Drug Administration were included in this study's final drug list. Finally, STITCH tool was used to visualize the interaction network among gene and their associated drug candidates (Kuhn et al., 2007). Moreover, the overall methodology that are used for prediction of miRNA along with their target genes is outlined in Fig. 2.
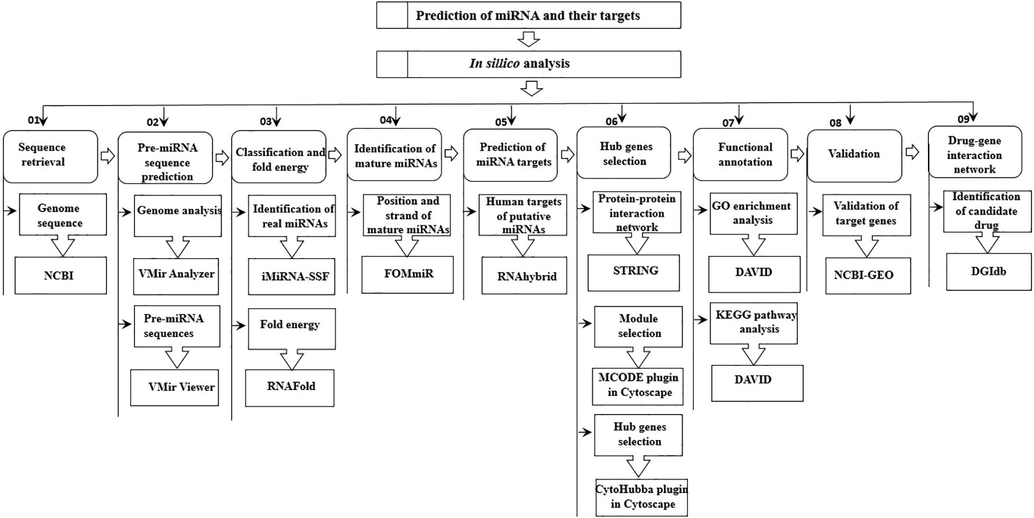
- Graphical synopsis of representing the overall strategy used in the prediction of miRNA and their targets.
3 Results
3.1 Retrieval of RSV genome and prediction of real pre-miRNA
Searching RSV genome for their pre-miRNAs yielded 15 pre-miRNAs like sequence with length vary for each pre-miRNA sequences. Vmir analyzer initially identified 161 sequences as novel pre-miRNAs (Fig. 3(A)). These 161 pre-miRNA candidates were passed via a filter (minimum hairpin size: 50 nt, minimum hairpin score: 100, and window of adjustable size: 500 nt). Only 15 hairpins met the cutoff and these 15 miRNAs were selected for further analysis (Fig. 3(B)).When these 15 pre-miRNAs were subjected to iMiRNA-SSF, 6 real and 9 pseudo miRNAs were predicted. These 6 real-miRNAs were used for further analysis while 9 pseudo-miRNAs were excluded. These 6 real miRNAs were further validated for their minimum free‐energy (MFE) value in RNAfold webserver. In total, 5 potential pre-miRNAs with MFE ≤ -20 kcal/mol, were identified as thermodynamically stable. MD63 is a real pre-miRNA however, it has been noted that MFE value was −9.20 kcal/mol hence MD63 was not used for further analysis. Furthermore, MFE secondary structure and positional entropy of 5 real pre-miRNAs were created (Table 1).
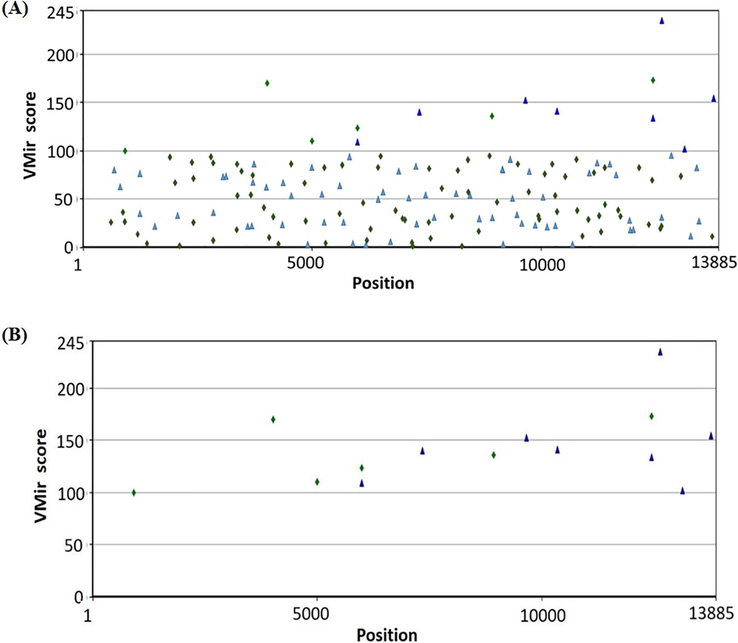
- (A)VMir analysis of the RSV genome which shows that all hairpins are widely dispersed across the viral genome.(B) Customized view of predicted pre-miRNA after filtering (minimum hairpin size: 50 nt, minimum hairpin score: 100, and window of adjustable size: 500 nt).
| Sr. no# | miRNA name | Orientation | Sequence | Genome location | Size, nt | MFE, kcal/mol |
|---|---|---|---|---|---|---|
| 1 | RSV-MR28 | Reverse | CUUCUAUAUCUAUUGAGUUGACAGAUAUGAUACUAUCUUUUUUCUUGGGAUCUUUAGGUGAUGUGAAUUUGCCCUUUAUUGAUUCUAGGAAUUUGGUAGCUCUGUUGUUUGCAUCUUCUCCAUGGAAUUCAGGAG | 2361–2495 | 135 | −34.60 |
| 2 | RSV-MR52 | Reverse | AUUUCCUCCAAUGAUUAUUUGCCCCAUGUGGAUAUUUUUAUUAACUUAUUUGAGUACUAGAUCUGAUGAACAAUGACUUGGGAUGAUCUGAGGCUCCAGAUGGGUUUUGUUUGAUUAGUUGAACCACAGAGGGUUGGUGAUUAUGAUGGUGAAGU | 4060–4214 | 155 | −39.30 |
| 3 | RSV-MR65 | Reverse | GCAUCUUGUAUGAUUGCAGUUGUUAGUGUGACUUUGUGGUUUGCCGAGGCUAUGAAUAUGAUGGCUGCAAUUAUAAGUGAAGU | 4812–4894 | 83 | −24.80 |
| 4 | RSV-MR121 | Reverse | ACCUCACUUGAUCGAUACUGUGUUAAUAUGUUGUUUAAUUUUGUGUAUAAAUUAAACCAAUGUAUUAACCAUGAUGGAGGAUGU | 9061–9044 | 84 | −21.80 |
| 5 | RSV-MR180 | Reverse | AGUUUUGACAAUGCAGUAUUAAUUCCUUUUUUUGUUAUAGGGUAACAAAGAAAGGGUAUCAAACUUUUAAUAUUUGCAUCAAUAGACU | 14601–14688 | 88 | −21.70 |
3.2 Identification of mature miRNAs
FOMmiR tool revealed a total of 7 mature miRNAs from real pre-miRNA hairpins. The sequences of mature miRNAs that have been predicted are shown in Fig. 4 in which red upper case letters represents the mature miRNAs sequence. Either one or both of the strand in a real pre-miRNA sequence can perform function as mature miRNA. From these 7 mature miRNAs, 2 mature miRNAs were present on 5′arm of stem loop hairpin structure while 5 were present on 3′arm of stem loop hairpin structure (Table 2).
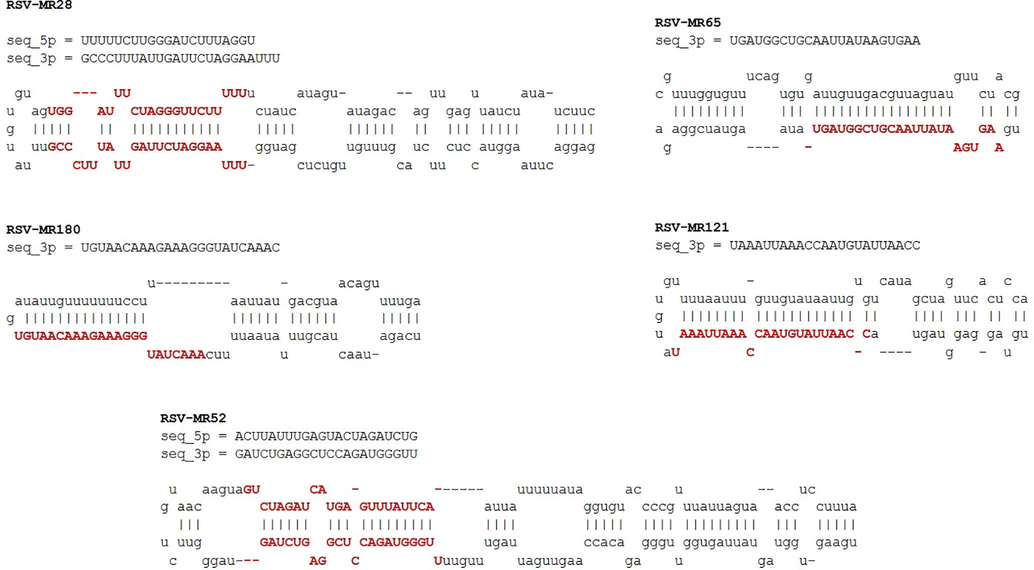
- mature miRNAs predicted using FOMmiR tool. Red upper-case letters indicate mature miRNA sequences.
| Sr. no# | miRNA name | 3′/5′ | Length of mature miRNAs | Predicted miRNA sequence |
|---|---|---|---|---|
| 1 | RSV-MR28 | 5′ | 21 | UUUUUCUUGGGAUCUUUAGGU |
| 2 | RSV- MR28 | 3′ | 24 | GCCCUUUAUUGAUUCUAGGAAUUU |
| 3 | RSV-MR52 | 5′ | 22 | ACUUAUUUGAGUACUAGAUCUG |
| 4 | RSV-MR52 | 3′ | 22 | GAUCUGAGGCUCCAGAUGGGUU |
| 5 | RSV-MR65 | 3′ | 23 | UGAUGGCUGCAAUUAUAAGUGAA |
| 6 | RSV-MR121 | 3′ | 23 | UAAAUUAAACCAAUGUAUUAACC |
| 7 | RSV-MR180 | 3′ | 24 | UGUAACAAAGAAAGGGUAUCAAAC |
3.3 Prediction of host specific target gene
RNAhybrid was used for the identification host specific target genes of RSV’s miRNAs. Through RNAhybrid, a total of 569 targeted genes were obtained (Supplementary File 1: Table S1). From these 569 targeted genes, total 494 genes were discovered to be unique in human. These 494 genes were further utilized in analysis because the present study was aimed to identify the targeted gene of RSV in humans.
3.4 Construction of protein–protein interaction network and module analysis
PPI network of targeted genes obtained from STRING were subjected to the MCODE plugin of cytoscape which provided significant 12 modules. From these modules, the top seven functional clusters of modules (Supplementary File 2: Fig. S1) were selected based on the cutoff criteria of node > 4 and the score is ≥ 4 (Supplementary File 1: Table S2).
3.5 Selection of hub genes
Using five methods available in the cytoHubba, the topmost fifty genes were selected, ranked by MCC, MNC, degree, betweenness, and closeness methods in order to elucidate the potential candidate hub genes. We looked for intersections between these five techniques and used a Venn diagram to find significant hub genes (Fig. 5). The 18 most significant genes were SNRNP200, ATG5, IKZF4, BSN, FAIM2, EED, CYCS, SLC6A1, IKBKG, NRXN2, IKZF1, DNMT3A, DRD2, CELF3, MAP2K7, CTLA4, RBM8A, and BTK, all of which interacted with one another. Further, these 18 genes were mapped into PPI network using STRING database (Fig. 6). The network comprised of 21 edges and 18 nodes respectively. Later, we used the FunRich algorithm to further evaluate the identified driver genes along with their genetic interaction. FunRich analyzes genes along with their neighboring interactional gene using a variety of reputable databases (Fig. 7).
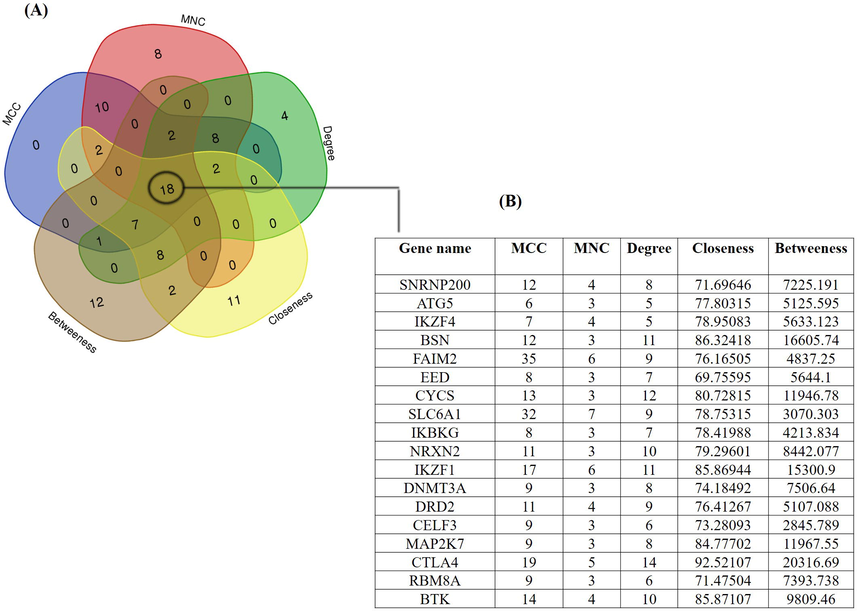
- (A) Venn diagram comparing gene ranked by MCC, MNC, Betweeness, closeness, and degree in CytuHubba. (B) Information of 12 commonly identified genes.
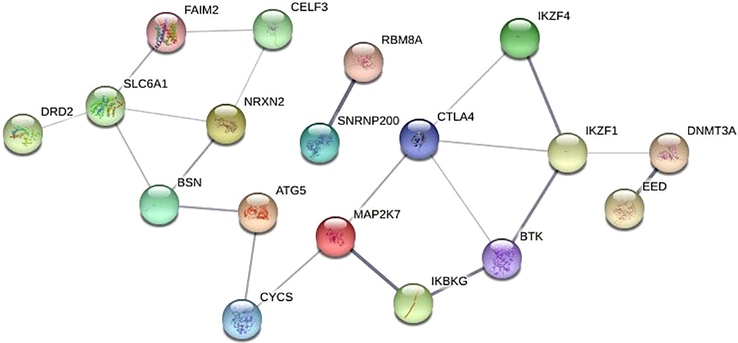
- Overview of PPI Network constructed using STRING database. The network includes 21 edges (interaction) between 18 nodes respectively.
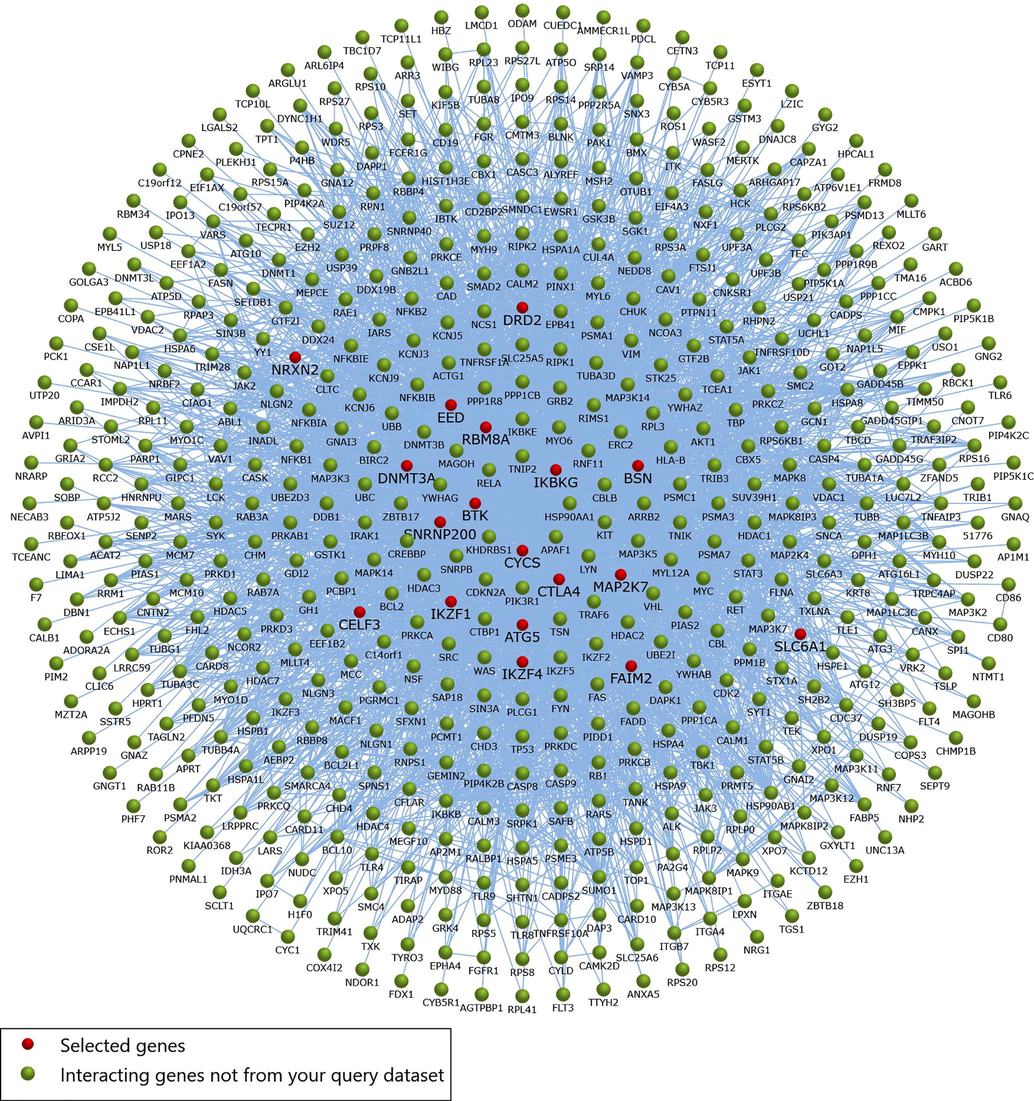
- Interaction network of genes to their related neighboring genes. Red dots represent that selected hub genes while the green dots represents interacting genes not from the query dataset. Blue lines the interaction among genes to neighboring genes.
3.6 Functional enrichment analysis
GO enrichment and KEGG pathways analysis of target genes were performed to analyze the gene function in terms of biological processes, cellular components, and molecular function as well as their associated pathways. GO enrichment analysis of target genes showed that in BP category genes are associated with the regulation of gene expression by genetic imprinting, synapse assembly, RNA splicing, mRNA processing and regulation of alternative mRNA splicing, via spliceosome. In terms of CC, the genes are enriched in catalytic step 2 spliceosome. For MF, category the genes are mainly concentrated in metal ion binding. KEGG enrichment pathway analysis revealed that genes are enriched in primary immunodeficiency, apoptotic process, B cell receptor signaling pathway, regulation of gene expression, T cell receptor signaling pathway, and osteoclast differentiation.
3.7 Gene expression microarray analysis
Finally, validation of hub genes with DEGs of infected RSV patient was performed using microarray dataset. In the present study, microarray dataset GSE80179 was obtained from GPL10558 platform (Illumina HumanHT-12 V4.0 expression beadchip), which consists of 79 samples (52 healthy controls and 26 RSV patients). The hub genes that satisfy the criteria of adjusted P-value < 0.05 were distinguished as significant genes. From 18 hub genes, a total of six genes namely ATG5, EED, CYCS, CTLA4, SNRNP200, and BTK were distinguished as significant genes. Based on pathway analysis, a total of 3 hub namely CYCS, CTLA4, and BTK genes were used in further analysis.
3.8 Drug-gene interaction
A total of 21 drugs were explored using DGIdb that might have potential to treat RSV patient. CYCS, CTLA4, and BTK were chosen as possible targets of 21 drugs (Supplementary file 1: Table S3). As explained in the Table S3, that all these 21 drugs are FDA approved, hence might prove very fruitful to combat the disease condition. The predicted drugs for hub genes through the gene-drug interaction database were used for constructing drug-protein network using STITCH database. The STITCH database shown significant interaction network with compounds /drugs (Fig. 8). As shown in Fig. 8(A) may have downstream effect on CD86 molecule, CD80 molecule, FYN oncogene related to SRC, FGR, YES, lymphocyte-specific protein tyrosine kinase, adaptor-related protein complex 2, v-akt murine thymoma viral oncogene homolog 1, forkhead box P3, signal transducer and activator of transcription 5B, inducible T-cell co-stimulator ligand, and signal transducer and activator of transcription 5A. BTK may have downstream effect on ibrutinib (440.5 g/mol), phospholipase C, gamma 2 (phosphatidylinositol-specific), Dasatinib, general transcription factor IIi , myeloid differentiation primary response 88, SH3-domain binding protein 5 (BTK-associated), toll-like receptor 4, inositol phosphates, toll-like receptor 9, and v-rel reticuloendotheliosis viral oncogene homolog (Fig. 8(B)). CYCS may have downstream effect on cytochrome c-1, apoptotic peptidase activating factor 1, BCL2-like 1, caspase 9, cytochrome c oxidase subunit IV isoform 2, ubiquinol cytochrome c reductase, ubiquinol cytochrome c reductase core protein I, adenine nucleotide containing three phosphate groups, ubiquinol cytochrome c reductase binding protein, and B-cell CLL/lymphoma 2 (Fig. 8(C)).
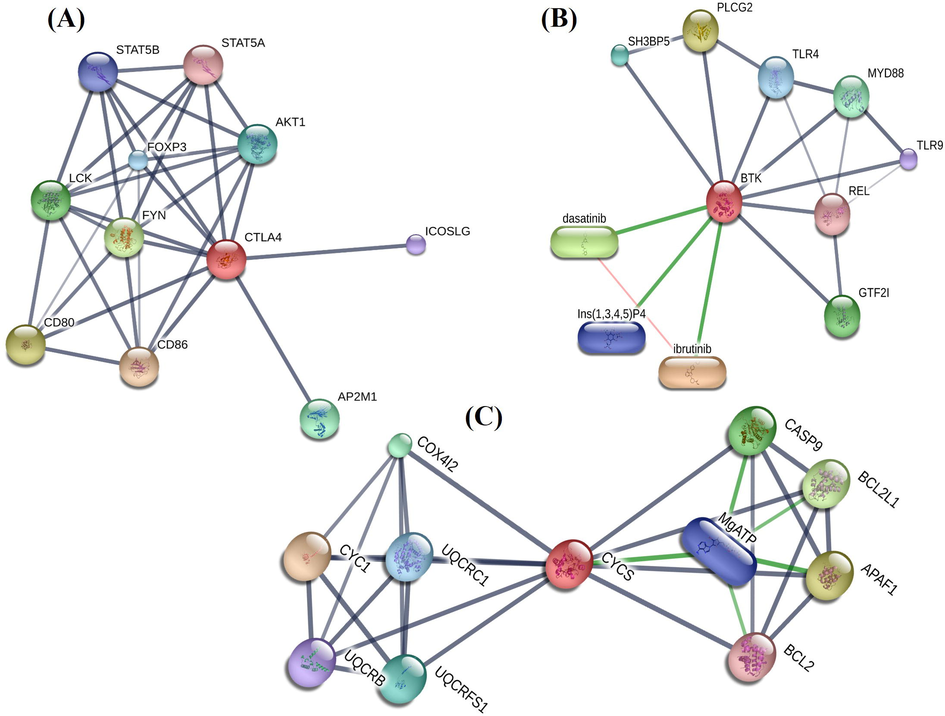
- (A) Targetable CTLA4 subnetwork. (B) Targetable BTK subnetwork. (C) Targetable CYCS subnetwork.
4 Discussion
Respiratory syncytial virus (RSV) is considered as a leading cause of acute lower respiratory tract infections. This virus mainly found to cause viral infection in children and infants in late-fall, winter, and spring (Brandt et al., 1973; Sato et al., 2005; Holberg et al., 1991). The symptoms of RSV vary from common cold to pneumonia and bronchiolitis infection (Brandt et al., 1973). This variability depends on the host factor, such as socio-economic status, early age, prematureness, and other underlying diseases including congenital heart anomaly, pulmonary anomalies, and immune-deficiency disorders (Walsh et al., 1997). Multiple mechanisms involved in RSV pathogenesis is poorly known. To emphasize whether RSV successfully targets and controls host genes, we proceeded with multiple scientific papers from various labs to learn more about the role of RSV's miRNAs in their pathogenesis. Our functional annotation of host specific target genes by RSV’s miRNAs might be helpful in understanding this targeted slicing on the pathogenesis of RSV.
Viruses retain multiple constantly evolving policies to subvert the host cellular environment. The interaction among cell-cycle machinery of host with the protein of virus is not necessarily favorable for the life cycle of host (Fan et al., 2018). However, in case of RSV there is no study available regarding the RSV’s interruptions in these processes. In the current analysis, we discovered many target proteins that are found to be involved in regulation of gene expression by genetic imprinting, apoptotic process, synapse assembly, regulation of alternative mRNA splicing, Fc epsilon RI signaling pathway, via spliceosome, catalytic step 2 spliceosome, T cell receptor signaling pathway, mRNA processing, osteoclast differentiation, primary immunodeficiency, RNA splicing, and B cell receptor signaling pathway. These whole findings strengthened the hypothesis that RSV selectively target the host-cellular machinery thus ensure their prolonged survival inside the host cell.
miRNAs carry out their functions by coupling with a target mRNA (Catalanotto et al., 2016; Rops et al., 2018). Two proteins named RBM8A, CELF3 are discovered to be directly involved in the mRNA processing. Similarly, two other genes namely EED, DNMT3A are involved in the regulation of gene expression. Majority of the viral infection leads to the cell death in host. By considering the current analysis, FAIM2, ATG5, CYCS, and IKBKG was discovered to be associated with the apoptotic process, hence these host genes found to be targeted by the RSV miRNAs associated with the apoptosis. Moreover, CTLA4, IKBKG, and MAP2K7 are involved in the T cell receptor signaling pathway while BTK and IKBKG are involved B cell receptor signaling pathway. These findings conclude that the miRNAs of RSV targeted these genes interfere the signaling pathways in host, that unfortunately lead to RSV infection in human. It is also noteworthy that target genes named NRXN2 and BSN are associated with pathways concerned with the synapse assembly in host. Recently, it has been investigated that RSV directly infected the dendrites cell hence inhibit the activation of T cell which ultimately leads to the impairment in the synapse assembly in host (González et al., 2008). Following that, it gives a clear evidence that RSV’s miRNAs are directly target the NRXN2 and BSN in human which cause impairment in synapse assembly. A few viruses such as HIV, dengue and measles virus are associated with the osteoclast differentiation. Viruses interfere with the bone hemostasis by the activation of osteoclast (Reddy et al., 2001; Modarresi et al., 2009; Huang et al., 2016). Our findings propose that BTK, IKBKG, and MAP2K7 are involved in the osteoclast differentiation. Hence the rate of RSV infection might be higher in the persons who already suffered from bone diseases.
Finally, validation of targeted hub genes was performed through microarray dataset (GSE80179). Validation of hub genes with differential expressed genes (DEGs) of infected RSV patient has led to the identification of six genes named ATG5, EED, CYCS, CTLA4, SNRNP200, and BTK. These genes are significant as the adj.p-value < 0.05. The pathway analysis of these genes revealed that CYCS, CTLA4, and BTK genes are associated with the apoptotic process, T cell receptor signaling pathway, and B cell receptor signaling pathway. Autophagy favors RSV replication through blocking cell apoptosis. Mechanistically, RSV induces mitophagy, which maintains mitochondrial homeostasis and therefore decreases cytochrome c release and apoptosis induction (Li et al., 2018). These findings validate the present conclusion that RSV significantly target the host signaling pathways and apoptotic machinery at higher level through their miRNAs. Furthermore, 21 drugs of CYCS, CTLA4, and BTK were found, which may have therapeutic potential to treat RSV infection. In the future, more experimental research and clinical trials are required to explore promising small drug like molecules that are safe and effective for treating RSV. Nonetheless, this research used variety of databases to offer novel and fresh insights regarding RSV pathophysiology and treatment.
This study presents a list of potential viral miRNAs from the RSV genome and predicts putative gene targets from the human host. (Supplementary file 1: Fig. S3). This study will serve as significant pioneer for the researchers who wants to identify the associated pathways involved in the pathogenesis of RSV. Additional experimental research on these putative miRNAs and their associated target genes lead to increase our knowledge to fight against RSV in future by means of novel therapeutic approaches.
5 Conclusion
In the present work, a new mechanism was proposed which explain that progression in pathogenesis of RSV might be due to the miRNA that target and upregulate the gene involved in multiple pathways in host. CYCS, CTLA4, and BTK has not been previously reported to be related to RSV, hence these genes might act as potential biomarkers for diagnosis of RSV infection at early stage. Moreover, CYCS, CTLA4, and BTK along with their candidate drugs might improve the treatment option against RSV infection in future. This finding can be further strengthened using appropriate experimental models to learn more about the host-pathogen interactions and combat emerging pathogens.
Acknowledgments
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP-2021/350), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res.. 2012;41:D991-D995.
- [CrossRef] [Google Scholar]
- Epidemiology of respiratory syncytial virus infection in Washington, DC: III. Composite analysis of eleven consecutive yearly epidemics. Am. J. Epidemiol.. 1973;98(5):355-364.
- [CrossRef] [Google Scholar]
- MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci.. 2016;17(10):1712.
- [CrossRef] [Google Scholar]
- Recovery from Infants with Respiratory Illness of a Virus related to Chimpanzee Coryza Agent (CCA). II. Epidemiologie Aspects of Infection in Infants and Young Children. Am. J. Epidemiol.. 1957;66(3):291-300.
- [CrossRef] [Google Scholar]
- IMiRNA-SSF: improving the identification of MicroRNA precursors by combining negative sets with different distributions. Sci. Rep.. 2016;6:19062.
- [CrossRef] [Google Scholar]
- Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc.. 2007;2(10):2366.
- [CrossRef] [Google Scholar]
- Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc. Natl. Acad. Sci.. 1986;83(13):4594-4598.
- [CrossRef] [Google Scholar]
- DAVID: database for annotation, visualization, and integrated discovery. Genome Biol.. 2003;4(9):1-11.
- [CrossRef] [Google Scholar]
- The Role of microRNAs in the Viral Infections. Curr. Pharm. Des.. 2018;24(39):4659-4667.
- [CrossRef] [Google Scholar]
- Breaking bad: how viruses subvert the cell cycle. Front. Cell. Infect. Microbiol.. 2018;8:396.
- [CrossRef] [Google Scholar]
- Cellular versus viral microRNAs in host–virus interaction. Nucleic Acids Res.. 2009;37(4):1035-1048.
- [CrossRef] [Google Scholar]
- Respiratory syncytial virus impairs T cell activation by preventing synapse assembly with dendritic cells. Proc. Natl. Acad. Sci.. 2008;105(39):14999-15004.
- [CrossRef] [Google Scholar]
- Vaccine development for respiratory syncytial virus. Curr. Opin. Virol.. 2017;23:107-112.
- [CrossRef] [Google Scholar]
- Computational prediction of viral miRNAs. Antiviral RNAi. Springer. 2011:143-152.
- [CrossRef] [Google Scholar]
- Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun.. 2005;337(4):1214-1218.
- [CrossRef] [Google Scholar]
- Risk factors for respiratory syncytial virus-associated lower respiratory illnesses in the first year of life. Am. J. Epidemiol.. 1991;133(11):1135-1151.
- [CrossRef] [Google Scholar]
- CLEC5A is critical for dengue virus-induced osteoclast activation and bone homeostasis. J. Mol. Med.. 2016;94(9):1025-1037.
- [CrossRef] [Google Scholar]
- RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res.. 2006;34(suppl_2):W451-W454.
- [CrossRef] [Google Scholar]
- STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res.. 2007;36(suppl_1):D684-D688.
- [Google Scholar]
- Respiratory syncytial virus replication is promoted by autophagy-mediated inhibition of apoptosis. J. Virol.. 2018;92(8):e02193-02117.
- [CrossRef] [Google Scholar]
- Severe coronavirus bronchiolitis in the pre–COVID-19 Era. Pediatrics. 2020;146(3)
- [CrossRef] [Google Scholar]
- STRING: a database of predicted functional associations between proteins. Nucleic Acids Res.. 2003;31(1):258-261.
- [CrossRef] [Google Scholar]
- Wnt/β-catenin signaling is involved in regulation of osteoclast differentiation by human immunodeficiency virus protease inhibitor ritonavir: relationship to human immunodeficiency virus-linked bone mineral loss. Am. J. Pathol.. 2009;174(1):123-135.
- [CrossRef] [Google Scholar]
- Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545-1555.
- [CrossRef] [Google Scholar]
- FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597-2601.
- [Google Scholar]
- RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res.. 2001;29(1):137-140.
- [CrossRef] [Google Scholar]
- Osteoclasts formed by measles virus-infected osteoclast precursors from hCD46 transgenic mice express characteristics of pagetic osteoclasts. Endocrinology. 2001;142(7):2898-2905.
- [CrossRef] [Google Scholar]
- Immunological features of respiratory syncytial virus-caused pneumonia—implications for vaccine design. Int. J. Mol. Sci.. 2017;18(3):556.
- [CrossRef] [Google Scholar]
- Interleukin-6 is essential for glomerular immunoglobulin A deposition and the development of renal pathology in Cd37-deficient mice. Kidney Int.. 2018;93:1356-1366.
- [CrossRef] [Google Scholar]
- Molecular epidemiology of respiratory syncytial virus infections among children with acute respiratory symptoms in a community over three seasons. J. Clin. Microbiol.. 2005;43(1):36-40.
- [CrossRef] [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.. 2003;13(11):2498-2504.
- [CrossRef] [Google Scholar]
- MicroRNA prediction using a fixed-order Markov model based on the secondary structure pattern. PLoS One. 2012;7(10):e48236
- [CrossRef] [Google Scholar]
- Human respiratory syncytial virus: pathogenesis, immune responses, and current vaccine approaches. Eur. J. Clin. Microbiol. Infect. Dis.. 2018;37(10):1817-1827.
- [CrossRef] [Google Scholar]
- Severity of respiratory syncytial virus infection is related to virus strain. J. Infect. Dis.. 1997;175(4):814-820.
- [CrossRef] [Google Scholar]
- Computational identification of microRNAs and their targets. Comput. Biol. Chem.. 2006;30(6):395-407.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101562.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







