Translate this page into:
Comprehensive analysis of antibiotic and heavy metal resistance, and virulence factors in Aeromonas veronii CTe-01: Implications for global antimicrobial resistance
⁎Corresponding author at: Calle Fermín Tangüis, 150 Urb. San Miguel Ica, Peru. jtantalean@unica.edu.pe (Juan C. Tantaleán)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Tellurite resistance in A. veronii: First report of tellurite resistance in Aeromonas veronii, isolated from a wastewater treatment plant. Antibiotic resistance profile: A. veronii CTe-01 exhibits resistance to multiple antibiotics, including penicillin, ampicillin, and erythromycin. Genomic analysis: Comprehensive genomic analysis reveals the presence of beta-lactamases, heavy metal resistance genes, and a functional type III secretion system. Dual role in the environment: A. veronii CTe-01 shows potential as both a pathogen and a heavy metal remediator in aquatic environments. Novel findings: Identification of novel genes and molecular mechanisms contributing to antibiotic and heavy metal resistance in A. veronii.
Abstract
Objectives
This study aimed to characterize Aeromonas veronii CTe-01 focusing on its resistance to heavy metal and antibiotics.
Methods
A. veronii CTe-01 was characterized using standard microbiological and molecular techniques. Antibiotic susceptibility and heavy metal resistance were tested per standard protocols. Genomic analysis, including plasmid characterization, was conducted in silico.
Results
A. veronii CTe-01 showed resistance to various heavy metals and antibiotics. Multiple resistance genes were identified, including those for beta-lactamases, heavy metal resistance, and type III secretion system components. The bacterium carries a 9 kb plasmid with repA/repB replication genes, parA/parB partitioning genes, and a type II toxin-antitoxin system for stability.
Conclusions
A. veronii CTe-01 is a genetic reservoir for antibiotic resistance, heavy metal resistance genes, and virulence factors. The study offers insights into its dual role as a pathogen and heavy metal remediator in aquatic environments.
Keywords
Aeromonas veronii
Heavy metal resistance
Antibiotic resistance
Genomics
- bp
-
base pairs
- kb
-
Kilobase pair
- kDa
-
Kilodaltons
- TeR
-
tellurium resistance
- HgR
-
mercury resistance
- AgR
-
silver resistance
- CrR
-
chromium resistance
- CoR
-
cobalt resistance
- PGAP
-
Prokaryotic Genome Annotation Pipeline
- NODE
-
contiguous sequence of bases in an assembly graph
- Locus Tag
-
specific genomic region within an organism’s genome
Abbreviations
1 Introduction
The Aeromonads group includes about 36 species (Fernández-Bravo and Figueras, 2020), with Aeromonas hydrophila, Aeromonas caviae, Aeromonas dhakensis, Aeromonas veronii, and Aeromonas salmonicida being economically and pathologically significant (Fernández-Bravo and Figueras, 2020; Skwor et al., 2014). These species are primary causes of infections and mortality in fish (Janda and Abbott, 2010; Walczak et al., 2017), and other aquatic animals (Hu et al., 2023). Besides, some are emerging pathogenic bacteria in humans (Janda and Abbott, 2010; Zhou et al., 2019). The pathogenicity factors related to these bacteria are: aer, hly, act, ast, alt (hemolysin), fla (flagellin), ser (serine protease), exu (DNase) (Fernández-Bravo and Figueras, 2020), and type III secretion systems (T3SS), which inject toxins and effectors into host cells (Silver et al., 2007).
Members of Aeromonas genus are resistant to multiple antibiotics and the related genes include β-lactams (blaTEM, blaSHV, blaCepH) (Sun et al., 2021), quinolone (qnrAB), tetracycline (tetACE) (Nawaz et al., 2006), sulfonamide (sul1), macrolide (mphA-mrx-mphR), aminoglycosides (aadA2, aac(30)-IIa), chloramphenicol (catB3), among others (Dahanayake et al., 2020). Similarly, heavy metal-resistant Aeromonas strains possess genes for resistance to copper (copA), mercury (merA), cobalt/zinc/cadmium efflux protein (czcA), and chromium (chrR) (De Silva et al., 2018).
In the environment, virulence factors and resistance genes can be transferred between bacteria through horizontal gene transfer (HGT). Aeromonas spp. carry mobile genetic elements (MGEs) like plasmids, with segregation mechanisms involving repA/repB genes (Dobiasova et al., 2016), transposons, bacteriophages, integrons, and insertion sequences (Piotrowska and Popowska, 2015). These elements drive bacterial evolution and adaptation to adverse conditions (Aminov, 2011).
A. veronii has been isolated from aquatic sources, soil, food, warm-blooded, and cold-blooded animals (Fernández-Bravo and Figueras, 2020; Janda and Abbott, 2010). Strains from different sources exhibit varied profiles of virulence, heavy metal resistance, and antimicrobial resistance (Hu et al., 2023). In wastewater, the coexistence with diverse bacterial species enhances the potential for gene exchange between clinical and environmental origins (Aminov, 2011).
Genomic sequencing and in silico analysis are powerful tools for predicting genes related to metabolism, pathogenicity, resistance, and more, aiding our understanding of microbes roles in nature (Elarabi et al., 2023; El-Beltagi et al., 2023; Halema et al., 2024). Genomic analysis of A. veronii MS-18–37 predicted genes for heavy metal, fluoroquinolone, and multidrug resistance (Abdelhamed et al., 2019). Similarly, the draft genome of A. veronii CTe-01 revealed genes for heavy metal and antibiotic resistance (Tataje-Lavanda et al., 2019).
This study aimed to characterize the antibiotic and heavy metal resistance, along with the pathogenic properties of A. veronii CTe-01. We also investigated potential MGEs on its genome, an aspect not yet widely explored in Aeromonas spp. Our findings suggest that A. veronii serves as a reservoir of virulence, heavy metal, and antibiotic resistance genes that could spread to other bacteria in aquatic sources as wastewater.
2 Material and methods
2.1 Bacterial strains and growth conditions
A. veronii CTe-01 and E. coli K-12 BW25113 were grown in LB medium at 37 °C for 24 h. Strain stocks were stored at −20 °C in LB medium with 30 % glycerol.
2.2 Molecular identification
The identity of A. veronii CTe-01 was confirmed through sequence analysis of the 16S rRNA, rpoD, and gyrB genes from the draft genome (VATZ00000000.2), compared with the GenBank database using BLASTn for nucleotide collection and highly similar sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch). The sequences are available under accession codes MK876839.1, VATZ01000092.1:14350–16215, and VATZ01000001.1:16817–19228, respectively.
2.3 Biochemical and physiological characterization
In addition to biochemical characteristics assessed with the VITEK® 2 Compact system (Tataje-Lavanda et al., 2019), further tests included citrate utilization; fermentation of glucose, lactose, and sucrose; catalase and oxidase tests; lysine decarboxylation and deamination; acetoin production; urease, gelatinase, and ornithine decarboxylase activities; indole production; motility; and esculin hydrolysis.
2.4 Determination of minimal inhibitory concentration (MIC) for heavy metals and antibiotics
The MICs (µg/mL) of various metal(loid) salts for A. veronii CTe-01 were determined as follows: HgCl2, AgNO3, K2Cr2O7, and K2CrO4 (0–100); K2TeO3 (0–200); CdCl2 and ZnSO4·7H2O (0–300); CoCl2·6H2O (0–400); NiSO4·6H2O and CuSO4 (0–600). Triplicate assays were conducted at 37 °C in 2 mL LB medium, without agitation. Antibiotic susceptibility and MICs were assessed using the MicroScan WalkAway 96 Plus system (Siemens).
2.5 Molecular characterization of plasmid pCTe-01
The plasmid pCTe-01 was purified with the QIAprep Spin Miniprep kit (QIAGEN) and its size estimated by agarose gel electrophoresis (1 %) in TAE buffer, using a Supercoiled DNA ladder (New England Biolabs) as a standard. The plasmid was then digested with BamHI, BsTNI, MboI, and PstI, according to the manufacturer's instructions (New England Biolabs). Fragment sizes were determined by 1.5 % agarose gel electrophoresis using a 1 kb Plus DNA ladder (Thermo Fisher Scientific) as a standard.
2.6 Sequencing plasmid pCTe-01 and improving the draft genome of A. veronii CTe-01
Draft genome of A. veronii CTe-01 [VATZ00000000.1], and plasmid pCTe-01 were additionally sequencing at Macrogen, Inc. (South Korea) using the Illumina platform with 101-bp paired-end reads (TruSeq Nano DNA Kit) [BioSample: SAMN11620977]. Quality control included FastQC v0.11.9 and Trimmomatic v0.39 (Bolger et al., 2014) for the reads of three libraries. De novo genome assembly using SPAdes v3.13.1 improved metrics compared to the previous version (N50: 103,789; L50: 12). Contig identity was verified with KmerFinder v3.2, showing similarity to A. veronii strain X12 (NZ_CP024933.1). Contiguator2 (Galardini et al., 2008) organized the contigs into 60 mapped and 809 unmapped contigs. Plasmid pCTe-01 was manually assembled and circularized, incorporating unmapped contigs with coverage over 10 000 bp. The updated draft genome has been deposited in GenBank for annotation with NCBI PGAP v5.1 after automatically removing small contigs and contaminants (VATZ00000000.2).
2.7 In silico predictions
Resistome analysis was performed using ARIBA v.2.146 with default parameters and databases including ARG-ANNOT (Gupta et al., 2014), CARD (McArthur et al., 2013), MEGARes (Doster et al., 2020), ResFinder (Zankari et al., 2012), and VFDB (Chen et al., 2016), using unfiltered reads, and KmerFinder software. The RASTtk (Aziz et al., 2008), KmerResistance v2.2, and Phaster (Arndt et al., 2016) were used to identify antimicrobial and heavy metal resistance genes, and phage and prophage sequences in contigs, using default parameters. Prophage DNA sequences were classified as intact (>90), questionable (70–90), or incomplete (<70). The pCTe-01 sequence was analyzed in silico with PGAP v5.1 and RASTtk, and subjected to virtual digestion using SnapGene 1.1.3. We also examined A. veronii CTe-01 genome for the presence of aer, hlyA, alt, and ast virulence genes.
3 Results
3.1 Characterization of A. veronii CTe-01
The identity of A. veronii was confirmed by 16S rRNA, rpoD, and gyrB sequence similarity. This strain is motile, non-lactose fermenting, catalase-negative, oxidase-negative, and indole-positive. It utilizes citrate and succinate, and efficiently ferments D-glucose, D-maltose, D-mannitol, D-mannose, and D-trehalose, with less efficiency for sucrose. Additionally, A. veronii CTe-01 exhibits L-proline-arylamidase, tyrosine-arylamidase, β-N-acetyl-glucosaminidase, and lysine decarboxylase activities.
This bacterium harbors the hemolytic aer gene, which is 1467 bp long and located between nucleotides 14 313 and 15 779, on NODE 18 (VATZ00000000.2). The Aer protein is predicted to be a 54 kDa polypeptide (https://web.expasy.org/compute_pi/). No similar sequences to other hemolytic genes as hlyA, alt, and ast genes were found on the gDNA sequence.
3.2 Characterization of plasmid pCTe-01
We identified a 9059 bp plasmid in A. veronii CTe-01 with a GC content of 54.8 %, confirmed by electrophoresis and in silico analysis (Fig. 1, Fig. 2). Gel electrophoresis fragment sizes (Fig. 1), matched predictions from in silico digestion using SnapGene v1.1.3. Digestion with BamHI and PstI enzymes yielded fragments of approximately 5586, 2752, 721 bp; and 2813, 2216, 1941, 1696, 393 bp, respectively. BsTNI produced a 2000 bp fragment and several smaller than 700 bp, while MboI yielded no fragments.
Agarose gel electrophoresis (1.5 % in TAE buffer) of plasmid pCTe-01 after restriction endonuclease digestion. Lane M, molecular weight standard (1 kb plus); lane 1, undigested pCTe-01; lanes 2–5, digestions carried out using BamHI, BsTNI, MboI, and PstI enzymes, respectively.
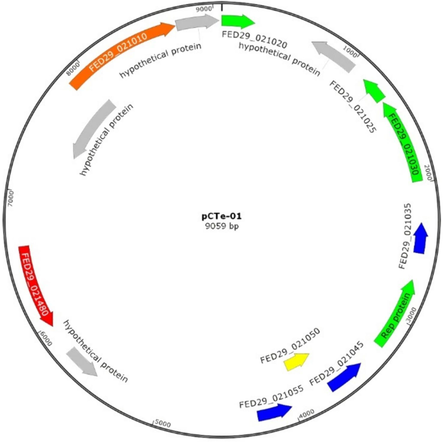
Circular diagram of plasmid pCTe-01. Colored arrows indicate coding sequences (CDS): green, ParAB and RepAB; blue, Rel; yellow, antitoxins; red, Sel1 family proteins; orange, integrase domain proteins; and gray, hypotheticals.
In silico predictions indicated that plasmid pCTe-01 replicates with RepA/RepB proteins and segregates using ParA-like and ParB proteins. Stabilization proteins identified include a type II toxin-antitoxin (TA) system (RelE/ParE, RelB/StbD, and RelB/DinJ) (Table 1). Additionally, a secretory immunoglobulin A-binding protein, EsiB, and five hypothetical proteins also were detected. Annotated by, *: PGAP v5.1, **: RASTtk, ***: BLASTp.
Product
Length (bp)
Locus Tag
Integrase domain-containing protein
876
FED29_021010*
RepA, replication initiation
534
**, ***
RepB, plasmid replication
243
FED29_021020*
ParA-like, ATPase chromosome-partitioning
630
FED29_021030*
ParB, chromosome partitioning
198
FED29_021025*
RelB/StbD antitoxin to RelE/StbE (replicon stabilization)
243
FED29_021050*
RelB/DinJ antitoxin (Type II toxin-antitoxin)
261
FED29_021055*
RelE/ParE toxin (Type II toxin-antitoxin)
222
FED29_021035*
EsiB, immunoglobulin A-binding protein
624
FED29_021480*
3.3 Heavy metal and antibiotic resistance of A. veronii CTe-01
A. veronii CTe-01 demonstrated resistance to various toxic metal (loid) salts, including K2TeO3, HgCl2, AgNO3, K2CrO4, CdCl2, ZnSO4·7H2O, and NiSO4·6H2O In silico analysis revealed potential heavy metal (loid) resistance genes on the chromosome (Table 2). Antibiotic MIC results showed intrinsic resistance to penicillin, ampicillin, ampicillin/sulbactam, amoxicillin/clavulanic acid, erythromycin, and cefazolin (Table 3). Both strains tested were resistant to clindamycin, oxacillin, linezolid, daptomycin, and synercid; showed intermediate resistance to piperacillin/tazobactam and rifampicin; and were susceptible to cefepime, ceftazidime, gentamycin, levofloxacin, moxifloxacin, ertapenem, nitrofurantoin, ticarcillin/clavulanic acid, trimethoprim/sulfamethoxazole, and tobramycin. S: sensitive, I: intermediate, R: resistant.
Heavy metal
A. veronii CTe-01
E. coli BW25113
Genomic analysis
MIC (µg/mL)
Gene detected
Function
K2TeO3
150
1
terABD
TeR
HgCl2
20
3
merA
HgR
AgNO3
50
7
cusAB
AgR
K2CrO4
60
40
chrA
CrR
CdCl2
100
75
czcD
Cd transport
ZnSO4·7H2O
250
130
czcD
Zn transport
NiSO4·6H2O
450
250
nikACE2
Ni transport
CoCl2·6H2O
150
250
czcD
CoR
CuSO4
250
550
cutCE, corC,scsABCD, cpxAR
Cu homeostasis
Antibiotic
MIC (µg/mL)
A. veronii CTe-01
E. coli BW25113
Penicillin
> 8
R
< 2
S
Ampicillin
>8
R
≤ 8
S
Ampicillin/Sulbactam
≥ 8/4
R
≤ 8/4
S
Amoxicillin/Clavulanic acid
≤4/2
R
≤ 4/4
S
Erythromycin
> 4
R
2
I
Cefazolin
≥ 4
R
≤ 2
S
Ceftriaxone
≤ 8
S
≤ 8
S
Cefuroxime
≤ 4
S
≤ 4
S
Ciprofloxacin
≤ 1
S
≤ 1
S
Aztreonam
≤ 8
S
≤ 8
S
Amikacin
≤ 8
S
≤ 8
S
Imipenem
≤ 1
S
≤ 1
S
Piperacillin
16
I
≤ 16
S
3.4 Improved genomic annotation and identification of resistance-associated genes, pathogenicity factors, and phage-related sequences on A. veronii CTe-01 genome
PGAP annotations of the A. veronii CTe-01 draft genome revealed significant improvements: contigs decreased from 272 to 200, pseudogenes from 162 to 145, rRNAs increased from 4 to 14, and tRNAs from 40 to 107.
Analysis of the raw data revealed several resistance genes, including β-lactamases like ampH/ampS, blaOXA12, blaTEM, and cphA4, among others. Additionally, type III secretion systems (acr, asc, exs) and motility genes, key pathogenicity factors, were predicted (Table 4). A search for heavy metal resistance genes identified candidates for Cu, Co, Zn, Cd, As, Ni, Hg, and Te (Table 5). The draft genome of A. veronii CTe-01 showed four regions with phage-associated genes related to Aliivibrio fischeri, Bacillus cereus, Acinetobacter baumannii, and E. coli (Table 6). Predicted phage and prophage proteins, included recombinase/integrase sequences and proteins related to phage DNA synthesis and structure, were found. Components of the psp operon, such as pspABC, were also detected using RASTtk (Table 7). *: not annotate with PGAP.
Gene
Product/Description
Database
Acr, aop, asc
associated to Type III secretion system (T3SS)
VFDB_CORE
acr1, acr2
Acr1, gatekeeper; Acr2 chaperone
acrGHRV
AcrG, AcrH, AcrR, chaperones; AcrV, V-tip protein
aopN
AopN, gatekeeper
ascBY
AscB, AscY chaperones
ascCD
AscC outer membrane, AscD inner membrane rings
ascRSTU
AscR, AscS, AscT, AscU, export apparatus
ascEHIJKLNOQVX
AscEHIJKLNOQVX proteins
ampHS
β-lactamases
RESFINDER/KMERFINDER
blaOXA-12
Associated to cefazolin inactivation
CARD
blaOXA-72
β-lactamase OXA-72, carbapenem-hydrolyzing
MEGARES
blaTEM-1
Antibiotic inactivation
CARD/MEGARES
blaTEM-10,
β-lactamase TEM-10
RESFINDER/KMERFINDER
blaTEM-101
β-lactamase TEM-101
ARGANNOT
cphA4
β-lactamase, carbapenem resistance
KMERFINDER
exeG
ExeG, pseudopilin
VFDB_CORE
exsABCD
ExsABCD proteins
fimACD
FimACD proteins
fliGM
FliG, FliM, flagellar motor switches
tapTW
TapT, TapW, pilus ATPases
Resistance subsystem
Role
Locus Tag
Arsenic-antimonite
ArsA, arsenite/antimonite pump-driving ATPase
FED29_015035
Arsenic
ArsBCDR proteins
FED29_015040, FED29_015045,
FED29_015030, FED29_000690
Cobalt-magnesium-zinc homeostasis
CorA, magnesium/cobalt transporter; ZntB, zinc transporter
FED29_018305
Cobalt-zinc-cadmium
CzcD, cobalt/zinc/cadmium resistance
FED29_006865
Cu, Pb, Cd, Zn, Hg homeostasis
Copper-translocating P-type ATPase; lead-cadmium-zinc-mercury transporting ATPase
FED29_006880, FED29_018810
FED29_013500
Copper
CutACE, Cu resistance proteins
FED29_016755, FED29_009335, FED29_008035
ScsABCD, suppression copper sensitivity proteins
FED29_015795, FED29_015800, FED29_015805, FED29_015810
Copper-silver homeostasis
CusAB, copper/silver efflux RND transporters
FED29_006475
FED29_006480
Mercury
MerA, mercuric reductase
FED29_001585
MerR family, transcriptional regulator
FED29_011840
Nickel homeostasis
HypA, HypB, [NiFe] hydrogenase nickel
FED29_003565, FED29_003560
NikA2, substrate-binding protein; NikC2, permease; NikE2, ATP-binding protein
FED29_003535, FED29_003545, FED29_003555
Chromium
ChrA, chromate transport
FED29_019065
Tellurite
TerABC, tellurite resistance
FED29_020300, FED29_020305, FED29_020310
Fluoroquinolones
LiuR, regulator; DNA gyrase subunit B
FED29_008890, FED29_012790
Multidrug
MacA, macrolide-specific efflux protein
FED29_002090
Multi antimicrobial extrusion protein
FED29_007815
Characteristic
Region (N°)
1
2
3
4
Region position (NODE)
32
49
51
62
Start and end position
804–35 093
1970–16 680
275–11 348
32–6624
Completeness (score)
Questionable (90)
Incomplete (60)
Incomplete (50)
Questionable (70)
Phage hit proteins
29
10
9
6
Specific keywords
integrase, tail, portal
transposase, tail,terminase
transposase, tail
transposase
Different phage species similar
27
9
6
3
Most common phage
Douglas 12A4(NC_021068)
Shanette
(NC_028983.1)vB_AbaM_ME3
(NC_041884.1)SH2026Stx1
(NC_049919.1)
Host of most common phage
A. fischeri
B. cereus
A. baumannii
E. coli
GC %
59.25
45.91
52.10
54.91
Subsystem
Role
Locus Tag
Integrase
Phage antirepressor protein
FED29_017795
Rha family transcriptional regulator
FED29_017950
DNA replication protein O
FED29_017920
Phage immunity repressor protein C
FED29_009230
Phage integrase
FED29_017780
Tyrosine-type recombinase/integrase
FED29_004490
Phage regulatory protein/antirepressor Ant
FED29_017900
Phage DNA synthesis
Abi family protein
FED29_012710
Adenine DNA methyltransferase
FED29_017835
Peptidylprolyl isomerase
FED29_014880
DNA invertase
FED29_015945
GpQ, capsid protein
FED29_010635
Prohead core protein
*
Ogr protein
FED29_010640
AAA family ATPase
FED29_004150
AlpA, transcriptional regulator
FED29_012715
Site-specific recombinase
FED29_010770
Phage shock psp operon
PspABC, phage shock proteins
FED29_015785
FED29_010340
FED29_010345
4 Discussion
4.1 Characterization of A. veronii CTe-01
A. veronii CTe-01 was isolated from wastewater containing domestic, hospital, and industrial effluents, rich in various microorganisms and chemicals (Tataje-Lavanda et al., 2019). This bacterium can utilize various carbohydrate sources and shows diverse enzymatic activities, which could contribute to its survival in the complex environment. Its β-hemolytic property may be related to aerolysin (Aer), encoded by the aer gene, which was the only gene detected among those investigated. This property may contribute to degrade animal or organic compounds in the environment, similar to processes in domestic wastewater (Tataje-Lavanda et al., 2019). β-hemolysis from aerolysin is a common feature in Aeromonas isolates from fish, clinical, and food samples (Janda and Abbott, 2010). In contrast, other strains have additional hemolytic genes like act, hly, and ast (Sun et al., 2021).
4.2 Characterization of plasmid pCTe-01
The plasmid pCTe-01 is about 9 kb in size. Analysis of the intact plasmid and fragments from BamHI, BsTNI, and PstI digestion matched in silico predictions using SnapGene v1.1.3. The MboI enzyme did not cut the DNA, likely due to absent recognition sites or methylation (Fig. 1). These results confirm the plasmid size and Illumina sequence, though the plasmid sequence remains incomplete.
In silico analysis predicted that pCTe-01 replicates using repA and repB genes, which encode for DNA replication initiation and transcriptional regulation. These genes, part of the repABC operon, are found in plasmids with low copy number, as seen in pCTe-01. They belong to several incompatibility groups (Pérez-Oseguera and Cevallos, 2013). Additionally, also predicted it use ParAB partitioning proteins for segregation, and type II toxin-antitoxins (TA) for stabilization (Kamruzzaman et al., 2021), but not antibiotic resistance genes (Table 1). The absence of resistance genes on pCTe-01 suggests that A. veronii CTe-01 carries these genes on its chromosome, and the observed resistance genes in this bacterium may have been acquired through phages or other mobile elements integrated into the genome (Table 6), rather than plasmids, as no mobilization genes were detected. The plasmid pCTe-01 also encodes EsiB, a protein that interacts with secretory immunoglobulin A, potentially aiding the bacterium in evading the neutrophil response during infections in fish and aquatic animals (Pastorello et al., 2013). Additionally, also contains a gene for an integrase domain-containing protein (Table 1), which phages use to integrate into the bacterial genome, suggesting exposure to MGEs similar to those in A. veronii C198 (Hatrongjit et al., 2020). Several hypothetical proteins were also identified and warrant further investigation.
4.3 Heavy metals and antibiotic MICs
A. veronii CTe-01 showed resistance to various heavy metals (loids) and the in silico analysis suggest genes related (Table 2). This is the first demonstration of tellurite resistance and the terABD operon in A. veronii, suggesting a possible link. However, similar traits and genes have been reported in A. caviae (Arenas et al., 2014). The bacterium forms black colonies, indicating tellurite reduction, but the mechanism in this species is unknown. In A. caviae, the dihydrolipoamide dehydrogenase enzyme (LpdA) reduces tellurite to elemental tellurium (Arenas et al., 2014). The copA and czcA genes, involved in copper and cadmium-zinc resistance, respectively, were not found in A. veronii CTe-01. However, they have been detected in 25 % and 61 % of 36 Aeromonas spp. isolates from shellfish Ruditapes philippinarum, respectively (Dahanayake et al., 2019). Similar genes for Cu, Pb, Cr, Hg, and Cd resistance have been observed in other Aeromonas species, supporting the idea of natural bioremediation of heavy metals (De Silva et al., 2018).
A. veronii CTe-01 showed resistance to β-lactams like penicillin, ampicillin, and cefazolin, as well as to the macrolide erythromycin, while remaining sensitive to several other antibiotics (Table 3). These resistances may be related to the presence of β-lactamases and extended-spectrum β-lactamases (Table 4), which are common in these bacteria (De Silva et al., 2018). Additionally, Aeromonads are known for resistance to tetracyclines, quinolones, and cephalosporins (Zhou et al., 2019). The presence of multiple antibiotic resistance genes in environmental bacteria represents a global public health concern, particularly with the rising prevalence of resistances.
4.4 Genome analysis of A. veronii CTe-01: Identification of antibiotic and heavy metal resistance, virulence genes, potential phages, and prophages through in silico analysis
The resistome analysis suggest that resistance to penicillin and ampicillin (Table 3) may be associated with extended-spectrum β-lactamase genes such as blaTEM-10, blaTEM-101, blaOXA-12, and others (Table 4). Some of these genes, such as blacphA3 and blaOXA-12, have been observed in A. veronii C198 (Hatrongjit et al., 2020), and blaOXA-12 and cphA3, in A. veronii XhG1.2 (Das et al., 2021). These genes, known as mobile, are widely disseminated among enteric bacteria. They can be acquired horizontally through MGEs (Piotrowska and Popowska, 2015). The resistance to cefazolin displayed could be related to blaOXA-12 (Table 4), an antibiotic-inactivation gene detected in Aeromonas (Hatrongjit et al., 2020). The resistance to erythromycin is likely associated with the presence of the macrolide-specific efflux protein MacA (Table 5).
The heavy metal resistances observed in A. veronii CTe-01 could be associated with various genes (Table 2, Table 5), similar to other members of its genus (De Silva et al., 2018). However, further investigation is needed to explore its potential as a bioremediator.
Several genes related to the asc family type III secretion system (T3SS) have been identified on the chromosome of A. veronii CTe-01 (Table 4). This virulence system is also found in other bacteria such as Salmonella enterica, Shigella spp., Citrobacter rodentium, and pathogenic E. coli (Sanchez-Garrido et al., 2022). In A. salmonicida, the T3SS is recognized as the primary virulence system (Frey and Origgi, 2016). A comparison between the T3SS components of A. veronii CTe-01 and A. salmonicida reveals significant similarity, indicating that these genes might contribute to the pathogenic traits and resistance profile in A. veronii. However, no previous reports were found on the presence of T3SS or the RelBE toxin-antitoxin system in this species. Other pathogenicity factors detected were the fim and fli genes, related to fimbria and the flagellar motor (Table 4), which may contribute to virulence, by aiding cell surface adherence (Fernández-Bravo and Figueras, 2020).
Phage and prophage-related sequences on A. veronii CTe-01 gDNA (Table 6) suggest these MGEs originate from water-associated species, such as A. fischeri, B. cereus, A. baumannii, and E. coli. Additionally, several components, including integrase, phage DNA synthesis elements, and the phage shock pspABC operon, have been identified (Table 7). This operon is critical in phage stress response, PspA protein helps maintain membrane integrity where damage is perceived by PspBC, as reported in Yersinia enterocolitica (Flores-Kim and Darwin, 2016).
The specific compounds produced by A. veronii during heavy metal activity −such as volatile mercury, reduced tellurium, and nanodots- have not yet been identified. Consequently, the functional roles of genes and proteins remain undemonstrated. Future research will focus on completing the A. veronii CTe-01 genome using PacBio or nanopore technologies, exploring its evolutionary relationships with other bacteria and plasmids, analyzing its transcriptome in the presence of specific heavy metals, and identifying the resulting compounds for potential applications.
5 Conclusions
The findings suggest that A. veronii CTe-01 may serve as a genetic reservoir for antibiotic and heavy metal resistance genes, as well as virulence factors. These genetic elements could potentially be transferred horizontally to other bacteria in aquatic environments, contributing to the emergence of new bacterial strains with altered characteristics and ecological roles.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Luis Tataje-Lavanda: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Phillip Ormeño-Vásquez: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Conceptualization. Ricardo Choque-Guevara: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Rosa Altamirano-Díaz: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Conceptualization. Manolo Fernández-Díaz: Writing – review & editing, Writing – original draft, Methodology, Investigation. Juan C. Tantaleán: Writing – review & editing, Writing – original draft, Validation, Resources, Methodology, Investigation, Formal analysis, Conceptualization.
Acknowledgements
To the Laboratory BioSLab EIRL and FARVET SAC for their support in conducting the microbiological assays and DNA sequencing, respectively.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Complete genome sequence data of multidrug-resistant Aeromonas veronii strain MS-18-37. Data Brief. 2019;23:10-13.
- [CrossRef] [Google Scholar]
- Horizontal gene exchange in environmental microbiota. Front. Microbiol.. 2011;2:1-19.
- [CrossRef] [Google Scholar]
- On the mechanism underlying tellurite reduction by Aeromonas caviae ST dihydrolipoamide dehydrogenase. Biochimie. 2014;102:174-182.
- [CrossRef] [Google Scholar]
- PHASTER: a better, faster version of the PHAST phage search tool. Nucl. Acids Res.. 2016;44:W16-W21.
- [CrossRef] [Google Scholar]
- The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1-15.
- [CrossRef] [Google Scholar]
- Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120.
- [CrossRef] [Google Scholar]
- VFDB 2016: Hierarchical and refined dataset for big data analysis - 10 years on. Nucl. Acids Res.. 2016;44:D694-D697.
- [CrossRef] [Google Scholar]
- Antibiotic and heavy metal resistance genes in Aeromonas spp. isolated from marketed Manila Clam (Ruditapes philippinarum) in Korea. J Appl Microbiol. 2019;127:941-952.
- [CrossRef] [Google Scholar]
- Prevalence of virulence and antimicrobial resistance genes in Aeromonas species isolated from marketed cockles (Tegillarca granosa) in Korea. Lett Appl Microbiol. 2020;71:94-101.
- [CrossRef] [Google Scholar]
- Genome sequencing and annotation of multi-virulent Aeromonas veronii XhG1.2 isolated from diseased Xiphophorus hellerii. Genomics. 2021;113:991-998.
- [CrossRef] [Google Scholar]
- Frozen white-leg shrimp (Litopenaeus vannamei) in Korean markets as a source of Aeromonas spp. harboring antibiotic and heavy metal resistance genes. Microb. Drug Resist.. 2018;24:1587-1598.
- [CrossRef] [Google Scholar]
- Complete sequences of IncU plasmids harboring quinolone resistance genes qnrS2 and aac(6′)-Ib-cr in Aeromonas spp. from ornamental fish. Antimicrob Agents Chemother. 2016;60:653-657.
- [CrossRef] [Google Scholar]
- MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020;48:D561-D569.
- [CrossRef] [Google Scholar]
- Draft genome of Raoultella planticola, a high lead resistance bacterium from industrial wastewater. AMB Express. 2023;13
- [CrossRef] [Google Scholar]
- Draft genome analysis for Enterobacter kobei, a promising lead bioremediation bacterium. Front Bioeng Biotechnol. 2023;11
- [CrossRef] [Google Scholar]
- An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020
- [CrossRef] [Google Scholar]
- Interactions between the Cytoplasmic Domains of PspB and PspC Silence the Yersinia enterocolitica Phage Shock Protein Response. J Bacteriol. 2016;198:3367-3378.
- [CrossRef] [Google Scholar]
- Type III secretion system of Aeromonas salmonicida undermining the host’s immune response. Front Mar Sci 2016
- [CrossRef] [Google Scholar]
- Galardini, M., Biondi, E.G., Bazzicalupo, M., Mengoni, A., 2008. CONTIGuator : a bacterial genomes assembler for structural insights on draft genomes Abstract : Genome 2–6.
- ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212-220.
- [CrossRef] [Google Scholar]
- Omics technology draws a comprehensive heavy metal resistance strategy in bacteria. World J Microbiol Biotechnol 2024
- [CrossRef] [Google Scholar]
- Genomic analysis of Aeromonas veronii C198, a novel Mcr-3.41-harboring isolate from a patient with septicemia in Thailand. Pathogens. 2020;9:1-13.
- [CrossRef] [Google Scholar]
- Isolation, Identification, and Characterization of Aeromonas veronii from Chinese Soft-Shelled Turtle (Trionyx sinensis) Microorganisms. 2023;11
- [CrossRef] [Google Scholar]
- The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:35-73.
- [CrossRef] [Google Scholar]
- Biological functions of type ii toxin-antitoxin systems in bacteria. Microorganisms 2021
- [CrossRef] [Google Scholar]
- The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348-3357.
- [CrossRef] [Google Scholar]
- Biochemical and molecular characterization of tetracycline-resistant Aeromonas veronii isolates from catfish. Appl Environ Microbiol. 2006;72:6461-6466.
- [CrossRef] [Google Scholar]
- EsiB, a Novel Pathogenic Escherichia Coli Secretory Immunoglobulin A-Binding Protein Impairing Neutrophil Activation.. 2013;mBio 4:1-11.
- [CrossRef]
- RepA and RepB exert plasmid incompatibility repressing the transcription of the repABC operon. Plasmid. 2013;70:362-376.
- [CrossRef] [Google Scholar]
- The type III secretion system effector network hypothesis. Trends Microbiol. 2022;30:524-533.
- [CrossRef] [Google Scholar]
- Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc Natl Acad Sci U S A. 2007;104:9481-9486.
- [CrossRef] [Google Scholar]
- Aeromonas hydrophila and Aeromonas veronii predominate among potentially pathogenic ciprofloxacin- and tetracycline-resistant Aeromonas isolates from Lake Erie. Appl Environ Microbiol. 2014;80:841-848.
- [CrossRef] [Google Scholar]
- Taxonomy, virulence determinants and antimicrobial susceptibility of Aeromonas spp. isolated from bacteremia in southeastern China. Antimicrob Resist Infect Control. 2021;10:1-9.
- [CrossRef] [Google Scholar]
- Draft Genome Sequence of Heavy Metal-Resistant Aeromonas veronii CTe-01, Isolated from a Peruvian Wastewater Treatment Plant. Microbiol Resour Announc. 2019;8:1-3.
- [CrossRef] [Google Scholar]
- Bacterial flora associated with diseased freshwater ornamental fish. Journal of Veterinary Research (poland). 2017;61:445-449.
- [CrossRef] [Google Scholar]
- Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother.. 2012;67:2640-2644.
- [CrossRef] [Google Scholar]
- Taxonomy, virulence genes and antimicrobial resistance of Aeromonas isolated from extra-intestinal and intestinal infections. BMC Infect. Dis.. 2019;19:1-9.
- [CrossRef] [Google Scholar]







