Translate this page into:
Combining of molecular 16S rRNA gene and metabolic fingerprinting through biolog system for the identification of streptomycetes in Saudi Arabia
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Many of the conventional identification methods for the group of streptomycetes deal with attempts to reveal their metabolic activity as well as their genetic makeup. The current study dealt with the possibility of combining one of the most prevalent molecular identification methods of bacteria, which is 16S rRNA gene sequencing along with the use of some basic standard methods and linking them with the Biolog system for bacterial identification. The study was conducted on the isolated Streptomyces (isolate EA3) with an ability of cellulose utilization, a well-known activity of the genus Streptomyces in nature. Identification of the isolate was achieved by standard phenotypic methods. The isolate was metabolically fingerprinted using the Biolog MicroPlate. Molecular identification was also performed using analysis of 16 s rRNA. The characteristic band of about 1487 bp was obtained. The sequencing analysis was performed and revealed that the isolate is very close to Streptomyces cinereoruber. The obtained sequence, 405 bp, was deposited in GenBank under accession, MT907291.1. The study concludes on the importance of linking the molecular and metabolic fingerprints of this genus of bacteria because of its enormous role in the natural processes and its numerous benefits.
Keywords
16srRNA
Biolog MicroPlate
Streptomyces
Molecular
Fingerprinting
Identification
1 Introduction
Streptomycetes, especially the genus Streptomyces, have significant roles in many biological processes (Ait Barka et al., 2016). This group of microorganisms is characterized by its multiple metabolic activities that reflect its nature. Many studies deal with attempts to identify this group from different environments and study its activities particularly the soil habitats. There are many methods of identification from standard methods to molecular. It is important to reach a more correct definition, at least to the genus level, using a combination of standard and molecular methods.
The cellulolytic activity of the genus Streptomyces is one of the most important activities in nature (Ventorino et al., 2016). This establishes the basic principle in many studies for the isolation and identification of this important group of bacteria. Here in this study the activity of cellulose decomposition also was chosen since it is one of the primary activities of this group of bacteria.
Many studies on actinobacteria involving morphological experiments and one molecular method are sufficient for identification to the genus level, and to determine the species further molecular and biochemical investigations are required (Salamoni et al., 2010).
Molecular identification of Streptomyces by targeting the 16S ribosomal RNA gene is usually the regular and basic practice for identification of this genus (Kämpfer, 2006). Employing a combination of genotypic and phenotypic identification methods is the most reliable approach (Anderson and Wellington 2001; Kämpfer and Glaeser, 2012). Some studies combined the use of Biolog method and 16S rRNA gene in bacterial identification (Wragg et al., 2014).
The current study aimed to isolate some streptomycetes based on their cellulolytic activity of Avicel microcrystalline cellulose. This substrate was used specifically due to its selective nature for efficient decomposers of insoluble cellulose. Regular phenotypic studies for identification were considered besides determining the metabolic fingerprint using the Biolog-MicroPlate. Eventually, the molecular identification targeting the 16S ribosomal RNA gene was performed.
2 Materials and methods
2.1 Isolation and detection of cellulase activity
Compost samples from Baljurashi (Al-Baha, KSA) was the source of isolation. The age of this compost was about 45 days old at the stage of maturation. This compost was previously prepared with frequent methods and composed initially from common household plant remnants without any microbial enhancement. The serial dilution technique was used for isolation in agar medium contained cellulose as the only carbon source. The culturing medium composition was (g/L): Microcrystalline cellulose (Avicel PH-101, Sigma), 20; NaNO3, 2.0; KCl, 0.5; MgSO4·7H2O, 0.5; K2HPO4 (anhydrous basis), 1.0; Agar, 20; FeSO4·7H2O, 0.01; and water, 1L. The final pH of this medium was adjusted to 7.0 before sterilization, inoculation, and incubation for five days at 45 °C. Cellulase activity was detected by iodine solution according to Kasana et al. (2008). Clear zones of cellulose hydrolysis surrounding the isolated colonies after the addition of iodine solution were recorded.
2.2 Phenotypic identification
Morphological studies and analysis of cell hydrolysate all were performed following what was mentioned in Kämpfer (2006).
2.2.1 Morphological studies
These studies included morphological characters on agar media and scanning electron microscopy.
2.2.2 Analysis of cell hydrolysate
Total cell hydrolysate was analyzed to determine the wall chemotype as the presence of sugars and amino acids pattern.
2.3 Metabolic fingerprinting
Following the manufacturer's directions, the metabolic fingerprinting was created using the 96 well Biolog plate third generation for aerobic bacteria and was read using semi-automated MicroStation Biolog software.
2.4 Molecular identification
2.4.1 DNA extraction and PCR for 16S rRNA gene
DNA has been extracted and purified following the kit’s manual (51306, QIAGEN). The forward and reverse universal bacterial primers for PCR were 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′), respectively, as described by Monciardini et al. (2002). The PCR mixture has prepared in a 50 μl final volume. The mixture contained; dNTPs, 200 mM; primer, 0.5 μM; 10X PCR buffer, 5 μl (15 mM MgCl2, 100 mM Tris–HCl, 500 mM KCl; pH 8.3), DNA template, 1 μl; and DNA Taq polymerase (Sigma), 1 U. Applied Biosystems ABI 9800 Thermal Cycler was used for amplification. The reaction standard conditions of denaturation, annealing, elongation, and final extension were followed. The amplified product was purified using a high pure PCR product purification kit (Sigma). The purified product was recognized by horizontal 2% agarose gel electrophoresis. A 100-bp DNA Ladder (Sigma) was used as a size marker.
2.4.2 Sequencing of 16s rRNA gene and phylogenetic analysis
An ABI-310 DNA-sequencer (Applied Biosystems) was used for sequencing using the same PCR primers. Sequencing was done following the kit’s instructions (4337449, Applied Biosystems). The final assembled sequence was used in GenBank to search for similar sequences then deposited. Phylogenetic analysis was done using the MEGA X program (Kumar et al., 2018).
3 Results
3.1 Isolation and detection of the cellulase activity of the isolate EA3
The isolate EA3 was obtained from the used compost. This isolate was grown on the cellulose substrate Avicel as a sole source of carbon. Fig. 1 shows the cellulase activity of the isolate EA3 by the appearance of zones of clearance.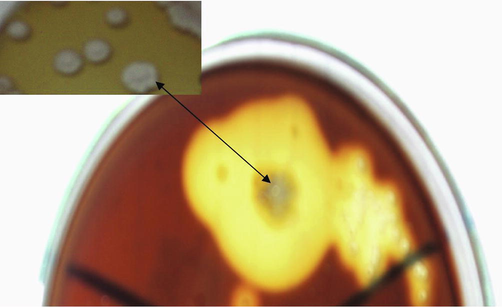
Cellulase activity of the isolate EA3 on Avicel agar flooded with iodine solution.
3.2 Phenotypic identification of the isolate EA3
Growth of the isolate was good on the different used agar media. The spore chain is flexible with a smooth surface of spores (Fig. 2). The total cell hydrolysate contained LL-diaminopimelic acid and glycine, while any sugars were not detected. Based on these characteristics, the isolate EA3 belongs largely to the genus Streptomyces.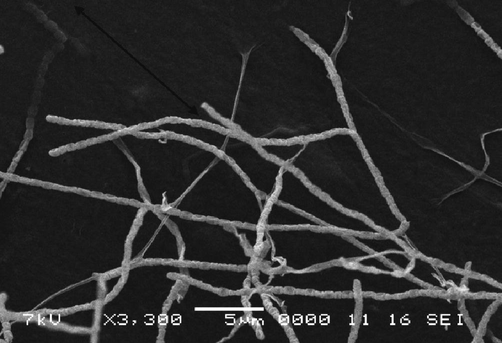
Scanning electron micrograph of the isolate EA3.
3.3 Metabolic fingerprinting of the isolate EA3
The metabolic fingerprinting of the isolate EA3 was created by the Biolog plate shown in Fig. 3. Data in (Table 1) were the recorded results after reading the plate and represented a metabolic fingerprint for this isolate. Among the recorded results is the ability of the isolate to utilize D-Cellobiose, an intermediate during cellulose decomposition. (+): Positive result. (−): Negative result. (?): Positive or Negative result (borderline).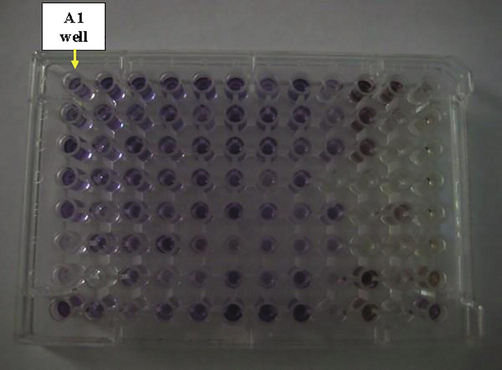
The Biolog plate inoculated with the isolate EA3.
1
2
3
4
5
6
7
8
9
10
11
12
A
Negative Control
Dextrin
D-Maltose
D-Trehalose
D-Cellobiose
Gentiobiose
Sucrose
D-Turanose
Stachyose
Positive Control
pH 6
pH 5
–
+
+
+
+
+
–
+
–
+
+
–
B
D-Raffinose
α-D-Lactose
D-Melibiose
β-Methyl-D-Glucoside
D-Salicin
N-Acetyl-D-Glucosamine
N-Acetyl-β-D-Mannosamine
N-Acetyl-D-Galactosamine
N-Acetyl Neuraminic Acid
1% NaCl
4% NaCl
8% NaCl
–
+
?
+
–
+
+
+
–
+
–
–
C
α-D-Glucose
D-Mannose
D-Fructose
D-Galactose
3-Methyl Glucose
D-Fucose
L-Fucose
L-Rhamnose
Inosine
1% Sodium Lactate
Fusidic Acid
D-Serine
+
–
+
?
+
+
+
?
+
+
–
–
D
D-Sorbitol
D-Mannitol
D-Arabitol
myo-Inositol
Glycerol
D-Glucose- 6-PO4
D-Fructose- 6-PO4
D-Aspartic Acid
D-Serine
Troleandomycin
Rifamycin SV
Minocycline
?
+
+
+
+
?
–
+
–
–
–
–
E
Gelatin
Glycyl-L-Proline
L-Alanine
L-Arginine
L-Aspartic Acid
L-Glutamic Acid
L-Histidine
L-Pyroglutamic Acid
L-Serine
Lincomycin
Guanidine HCl
Niaproof 4
+
–
+
?
+
+
?
–
+
–
+
–
F
Pectin
D-Galacturonic Acid
L-Galactonic Acid Lactone
D-Gluconic Acid
D-Glucuronic Acid
Glucuronamide
Mucic Acid
Quinic Acid
D-Saccharic Acid
Vancomycin
Tetrazolium Violet
Tetrazolium Blue
–
?
–
+
–
–
–
–
–
–
–
–
G
p-Hydroxy- Phenylacetic Acid
Methyl Pyruvate
D-Lactic Acid Methyl Ester
L-Lactic Acid
Citric Acid
α-Keto-Glutaric Acid
D-Malic Acid
L-Malic Acid
Bromo-Succinic Acid
Nalidixic Acid
Lithium Chloride
Potassium Tellurit
–
–
–
–
–
+
–
+
–
+
–
–
H
Tween 40
γ-Amino-Butryric Acid
α-Hydroxy-Butyric Acid
β-Hydroxy-D,L-Butyric Acid
α-Keto-Butyric Acid
Acetoacetic Acid
Propionic Acid
Acetic Acid
Formic Acid
Aztreonam
Sodium Butyrate
Sodium Bromate
+
–
–
–
+
+
+
+
–
+
–
+
3.4 Molecular identification of the isolate EA3
Molecular identification of the isolate EA3 was achieved through analysis of the 16S rRNA gene. The purity of the obtained PCR product was checked by electrophoresis and the characteristic fragment of about 1487 bp was obtained. The product is subjected to sequencing with the same forward and reverse primers and has assembled into a continuous sequence 405 bp in length. This partial sequence of the isolate EA3 was aligned with the partial sequences of the neighboring sequences in GenBank then deposited under accession MT907291.1. The Neighbor-Joining method has been used to construct the phylogenetic tree (Fig. 4). Based on the phylogenetic tree, the isolate EA3 was found to belong to the genus Streptomyces. Fig. 5 shows the partial sequence of the isolate EA3 clustered with the partial sequences of the nearby Streptomyces cinereoruber strains. Among these strains, the isolate EA3 was most closely related to that of Streptomyces cinereoruber subspecies cinereoruber strain LS SPJ (accession, MK808032.1) and showed 99 % sequence identity with it. Fig. 6 shows the differences in nitrogenous bases between the isolate EA3 and the aligned sequence of the closely related Streptomyces cinereoruber subspecies cinereoruber strain LS SPJ. The computed base composition of the isolate EA3 sequence is represented in Table 2 and shows the high GC content of this sequence of about 62.47 %, a characteristic feature of this group of bacteria.
Neighbor-joining of the partial 16S rRNA gene sequence of the isolate EA3 (No.1) with the partial sequences of the nearby streptomycetes in the GenBank.
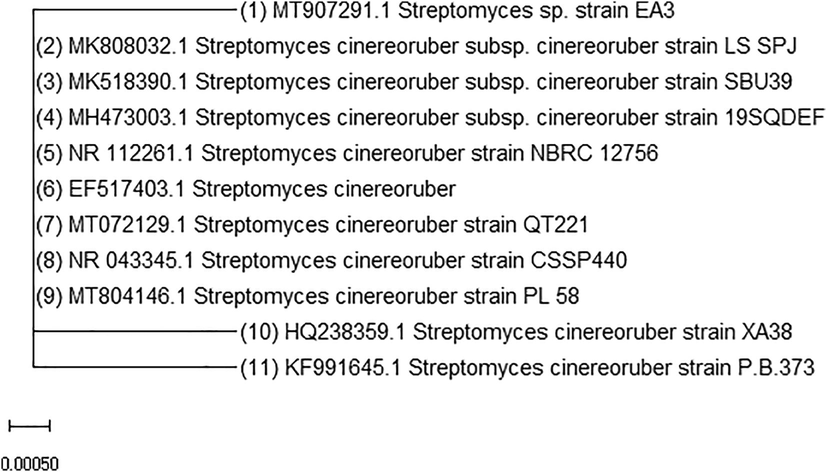
Neighbor-joining of the partial 16S rRNA gene sequence of the isolate EA3 (No.1) with the partial sequences of the nearby Streptomyces cinereoruber strains in the GenBank.

The differences in nitrogenous bases (shown in red) in the aligned sequence the isolate EA3 (Query) and the nearest strain in the GenBank Streptomyces cinereoruber subsp. cinereoruber strain LS SPJ (Subject).
Total (bp)
T(U)(%)
C(%)
A(%)
G(%)
GC Content (%)
AT Content (%)
405
16.79
26.66
20.74
35.80
62.46
37.53
4 Discussion
This study was planned to isolate some streptomycetes from compost since the genus Streptomyces is predominant in it (Low et al., 2015). Moreover, studies on microcrystalline cellulose (Avicel) utilization by streptomycetes was most prevailing (Ray, 2015). The target of using such substrate was primarily to aid in selection of the efficient organism and excluding many of others with no hydrolytic activity on insoluble cellulose simultaneously. The isolate EA3 was obtained as intended on Avicel cellulose as a sole source of carbon.
Based on classical phenotypic studies, the isolate EA3 was found to belong to the genus Streptomyces. The Biolog Plate assay for characterization of enzymatic potential and identification of bacteria was formerly approved as a metabolic fingerprint (Winding and Hendriksen, 1997). Also, Biolog Microplates methods were identical for the rapid identification of bacteria (Al-Dhabaan and Bakhali, 2017). So, the metabolic fingerprinting of the isolate EA3 was created by the Biolog plate, noting that so far there is no Biolog plate database available for identification of this group of bacteria.
Identification by molecular methods is essential in many studies conducted to discover the different environments and activities of bacteria (Bahamdain et al., 2020). One of the basic molecular methods until present to define actinomycetes is the use of sequencing of 16S rRNA gene (Ait Barka et al., 2016). So, this approach was used here in this study to confirm the identification of the genus and assist in the identification of the species. Usually, the size of the PCR amplicon using the universal primers is in the range of about 1500 bp. Here, the fragment was of about 1487 bp. Using the same primers, the assembled partial sequence 405 bp in length has a high GC content typical of streptomycetes. Also, the isolate EA3 was found to more likely to belong to Streptomyces cinereoruber. Thus, correlations between genotypic and phenotypic traits are useful to best describe species within the Streptomyces genus and hence its applications.
Many recent studies referred to the great potential and applications of streptomycetes. In all studies, the identification methods usually represent the basic practice. Among these studies are the following: Shen et al. (2021), Buzón-Durán et al. (2020) and Al-Dhabi et al. (2019). Some bacterial identification studies employed the Biolog system and the 16S rRNA gene sequencing as compared to conventional methods. Such studies have approved the considerable high accurate identification of these methods compared to the conventional methods (Morgan et al., 2009). Similar studies combined the use of Biolog method and 16S rRNA gene with promising results in bacterial identification notably to the genus level. However, the results of identification to the genus level by 16S rRNA gene sequencing was significantly better (Wragg et al., 2014).
5 Conclusion
This study demonstrated the importance of linking genetic and phenotypic datasets of the genus Streptomyces, especially in important biological processes. The isolate in this study with its unique activity in decomposing crystalline cellulose has potential in multiple applications. Ultimately, the study recommends establishing databases of metabolic fingerprints for the different types of this important genus of bacteria and linking them to basic molecular fingerprints to facilitate and support their identification.
Conflict of interest
The author declares that there is no conflict of interest.
References
- The Actinobacteria: taxonomy, physiology and their natural products. Microbiol. Mol. Biol. Rev.. 2016;80:1-43.
- [Google Scholar]
- Analysis of the bacterial strains using Biolog plates in the contaminated soil from Riyadh community. Saudi J. Biol. Sci.. 2017;24(4):901-906.
- [Google Scholar]
- Composting of vegetable waste using microbial consortium and biocontrol efficacy of Streptomyces Sp. Al-Dhabi 30 isolated from the Saudi Arabian environment for sustainable agriculture. Sustainability. 2019;11(23):6845.
- [Google Scholar]
- The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol.. 2001;51(3):797-814.
- [Google Scholar]
- Molecular identification and phylogenetic analysis of some rare actinomycetes obtained from Al-Lith hot spring area of Saudi Arabia. Biosc. Biotech. Res. Comm.. 2020;13(3):1037-1049.
- [Google Scholar]
- Applications of Streptomyces spp. enhanced compost in sustainable agriculture. Biology of composts. Springer 2020:257-291.
- [Google Scholar]
- The family Streptomycetaceae, part I: taxonomy. In: Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., eds. The Prokaryotes. New York, NY: Springer New York; 2006. p. :538-604.
- [CrossRef] [Google Scholar]
- Prokaryotic taxonomy in the sequencing era–the polyphasic approach revisited. Environ. microbiol.. 2012;14(2):291-317.
- [Google Scholar]
- A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. microbiol.. 2008;57(5):503-507.
- [Google Scholar]
- Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K., 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35(6), 1547.
- Taxonomic diversity and antimicrobial activities of actinomycetes from manure composts. Res. J. Microbiol.. 2015;10(11):513-522.
- [Google Scholar]
- New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol.. 2002;42(3):419-429.
- [Google Scholar]
- Comparison of the Biolog OmniLog Identification System and 16S ribosomal RNA gene sequencing for accuracy in identification of atypical bacteria of clinical origin. J. Microbiol. Methods. 2009;79(3):336-343.
- [Google Scholar]
- Microbial avicelase: An overview. Bull. Environ. Pharmacol. Life Sci.. 2015;4(4):3-13.
- [Google Scholar]
- Preliminary characterization of some Streptomyces species isolated from a composting process and their antimicrobial potential. World J. Microbiol. Biotechnol.. 2010;26(10):1847-1856.
- [Google Scholar]
- Identification and application of Streptomyces microflavus G33 in compost to suppress tomato bacterial wilt disease. Appl. Soil Ecol.. 2021;157:103724.
- [CrossRef] [Google Scholar]
- Lignocellulose-adapted endo-cellulase producing Streptomyces strains for bioconversion of cellulose-based materials. Front. Microbiol.. 2016;7:2061.
- [Google Scholar]
- Biolog substrate utilisation assay for metabolic fingerprints of soil bacteria: incubation effects. In: Insam H., Rangger A., eds. Microbial communities. Berlin, Heidelberg: Springer Berlin Heidelberg; 1997. p. :195-205.
- [Google Scholar]
- Comparison of Biolog GEN III MicroStation semi-automated bacterial identification system with matrix-assisted laser desorption ionization-time of flight mass spectrometry and 16S ribosomal RNA gene sequencing for the identification of bacteria of veterinary interest. J. Microbiol. Methods. 2014;105:16-21.
- [Google Scholar]







