Translate this page into:
A symmetric DNA encryption process with a biotechnical hardware
⁎Corresponding author. esrasatir@duzce.edu.tr (Esra Şatir),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
The growing rate of internet/network technologies day by day dramatically increases the formation of data in the world. As the flow of information increases on the network, time security threats are also increasing for users. In order to protect data, cryptography and steganography have been used from the past to the present. The goal of cryptography is to transfer the message between sender and receiver in a way that is incomprehensible to the observer. Nowadays, DNA cryptography is a shining branch in the field of cryptography. The primary purpose here is to employ DNA as a carrier and to employ modern biological techniques as application tools. In this study, a DNA cryptography technique was proposed by integrating DNA encoding and DNA operators into the Feistel network structure. Here, DNA itself was used as a carrier instead of traditional digital media such as image, text or video, while its biological tools were being used as implementation tools. Besides, the developed simulation software and the synthesized DNA sequence were digitally and biologically integrated into specifically created biotechnical hardware. Experimental results demonstrated that the proposed study has efficient outcomes for cryptographic requirements; capacity of nearly 100%, brute force attack nearly 12 × 106 years for only one block, key space that is 280 for only one block, and entropy analyses close to 2. Besides, the implementation of the proposed method has been verified by vitro experiments.
Keywords
DNA computing
DNA encoding
DNA encryption
Text encryption
Security analysis
1 Introduction
The growing rate of network technologies day by day dramatically increases the formation of data in the world. As the flow of information increases on the network, time security threats are also increasing for users. In order to protect the data, cryptography and steganography methods have been used from the past to the present. While cryptography codes the data, steganography hides it (Kaundal and Verma, 2014).
Cryptography is one of the most remarkable solutions for security issues. In cryptography, data is transferred between the sender and recipient over an untrusted medium. Cryptographic algorithms are classified by depending on the use of keys during encryption and decryption. These categories are symmetric cryptography and asymmetric cryptography. In the first one, encryption and decryption processes are implemented by using the same key. In the second one, encryption and decryption processes are implemented by using different keys. Here, each party has a pair of keys called private key and public key. The private key is always kept secret, while the public key can be shared (Namasudra et al., 2020).
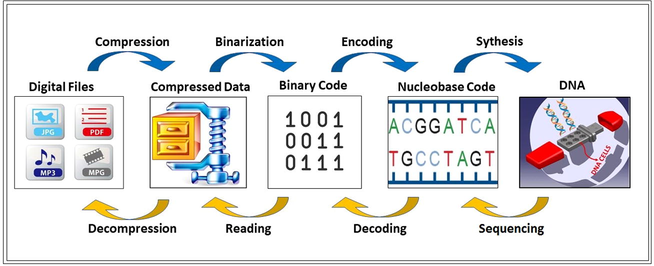
Recently, DNA computing has been applied to cryptography. In DNA cryptography, DNA is an information carrier instead of any other medium like text, image, video, etc. Thus, this approach takes advantage of biological technology to achieve encryption. In other words, DNA is used as the carrier medium, while modern biological techniques are being used as application tools. However, it has disadvantages like expensive experimental equipment, complicated operations, and complex biotechnology. Hence, it still cannot be widely applied in the field of cryptography. In order to overcome these problems, some operations of DNA computing are employed to confuse information (Wang et al., 2015). Nowadays, the DNA concept is popular in the field of information security since it has a complex structure. Here, unlike the traditional computer computation concepts like 0 and 1, data are encrypted and stored by using DNA bases, Adenine (A), Cytosine (C), Guanine (G), and Thymine (T). So, the sender can choose any combination of these bases during the encryption process by improving the security of the data (Namasudra et al., 2020).
The motive of this area is to employ helpful metaphors from natural biological models to generate efficient security alternatives in the field of cryptography. In this paper, depending on the Central Molecular Biology principle (CMB) for encryption and decryption of systems, a novel encryption approach by combining DNA carrier medium, DNA encoding, and DNA XOR operation with Feistel network structure has been derived. Besides, the proposed DNA encryption process was both digitally and biologically integrated into designed biotechnical hardware. In order to provide confusion of the data, DNA encoding and DNA XOR have been integrated into Feistel Network. Another significant finding about DNA is that the capacity of electrical and optical media was exceeded by DNA. A gram of DNA may contain 1021 bases or about 108 terabytes of data (Gehani et al., 2003). Finally, we aim to increase the number of hidden bits (capacity) by employing DNA as the carrier medium instead of traditional multimedia carriers (image, text, video and etc.) in cryptography.
The proposed study has been organized as six sections. In Section 2, similar studies in the literature have been mentioned. In Section 3, the used methodologies have been mentioned, and the proposed approach has been explained step by step by providing short samples. In Section 4, the simulation and design of biotechnical hardware have been explained. The results of the performed experiments have been shared and discussed in Section 5. A general outcome has been expressed in Section 6. Finally, the benefitted supports have been mentioned in Section 7.
2 Related work
The concept of DNA computing was firstly discussed in 1994 by Adleman with the goal of solving the Hamiltonian path problem. In 1995, Lipton solved the scheduling problem for 2-bit numbers, which is in another NP (non-deterministic Polynomial) complexity class, by using DNA molecules in a test tube (Kaundal and Verma, 2014). In 2003 by Chen, DNA cryptography algorithms based on molecular computing and single-used cipher were designed. Encryption/decryption steps were carried out on 2-dimensional images (Chen, 2003). In 2005, Tanaka et al. proposed a DNA cryptography algorithm based on asymmetric encryption. In the study, public keys were synthesized by helping non-identical mixtures. After key generation, the message was encoded with the first public key, and it was tied with the second public key into a DNA sequence. Asymmetric encryption is more secure than symmetric encryption, but it is slower than symmetric encryption when it is compared with symmetric encryption in terms of speed (Tanaka et al., 2005). In 2011, Kumar and Singh proposed a new method based on writing secret data in DNA sequences. In the proposed algorithm, they encrypted the word “HELLO” as plaintext by 350-bit of a single-used key. First, the word “HELLO” was converted to ASCII code equivalent. Then, they had the equivalent of binary pattern in 35 bits and 35 × 10 = 350 bits One Time Pad sequence. By using this produced nucleotide sequence and ssDNA key, they sent and received the data through the encryption/decryption perfectly (Kumar and Vijayaraghavan, 2011). A method based on RSA encryption algorithm that is encrypted via DNA sequences was proposed by Monika and Upadhyaya In 2015. The concerning method was adapted to SSL (Secure Socket Layer) technology in order to provide a higher level of security during the communication (Monika and Upadhyaya, 2015).
In 2016, Mousa proposed a DNA-Genetic Encryption Technique called D-GET. The primary purpose here is to make the technique more secure and less predictable. In this technique, the binary form of any digital data is converted to DNA sequencing, reshaped, encrypted, crossovered, mutated, and then reshaped. These main steps of D-GET are repeated three times or more. Experimental results demonstrated that his proposed technique had multilayer protection stages against different attacks and a higher level of security based on the multi-stages and genetic operations (Mousa, 2016).
In 2016 Goyat and Jain proposed a DNA-based cryptographic algorithm to secure the entire communication and storage of cloud servers for implementation and design. Their DNA-based cryptographic technique was basically developed using the substitution and other basic operator’s implementation. The experimental performance of their study in terms of time and space complexity provided effective and low resource-consuming techniques for data security in cloud-based systems. (Goyat and Jain, 2016).
In 2017 Ahmed and Mohammed developed a novel hybrid security algorithm called RC4-DNA-Alg. This algorithm uses both the symmetric stream cipher RC4 and DNA-indexing algorithms to provide secure data hiding with high complexity in the scope of steganography framework. The performance evaluation of their proposed scheme was measured by considering these three parameters; conditional entropy, randomness tests, and encryption time. Their result showed outperformance in security and distorted in hybrid cipher when compared to the native RC4 (Ahmed and Mohammed, 2017).
In 2018 Thangavel and Varalakshmi proposed a DNA cryptosystem to secure the original data within the DNA. They called their method Enhanced ElGamal cryptosystem. Their method is an asymmetric cryptosystem that has the purpose of solving key management issues in the cloud. They aim to overcome this problem by transferring the key file securely between the Data Owner and the Data User. They mentioned that Enhanced ElGamal cryptosystem provided better user authentication results. Besides, they added that it had better performance against security attacks (Thangavel and Varalakshmi, 2018).
In 2018 Narendren et al. proposed DNA based algorithm which acts as an additional layer to the RSA algorithm. In their method, a round function was employed that is based on the process of protein synthesis from DNA. They mentioned that the security analysis of their proposed scheme has proven to give good results (Narendren. et al., 2018). In 2018, Sharma and Sohal presented a novel cryptographic technique for cloud systems. Their approach uses client-side data encryption for encrypting the data before uploading it onto the cloud. Besides, it can also be considered a multifold symmetric-key cryptography technique that is based upon DNA cryptography. They compared their study with the existing symmetric-key algorithms; DNA, AES, DES, and Blowfish. They mentioned that their experimental results illustrated better outcomes when compared with the traditional algorithms in terms of ciphertext size, encryption time, and throughput (Sohal and Sharma, 2018).
In 2019 Basu et al. developed a cryptography algorithm inspired by the Central Dogma of Molecular Biology (CDMB). They implemented encryption and decryption stages via Genetic Coding simulation, which implies conversion from binary to DNA bases, Transcription, which means conversion from DNA to mRNA, and Translation which means conversion from mRNA to protein in a reversible way. Here, input blocks have the length of 16-bits. The final form of the blocks was concatenated to form the final ciphertext in the form of protein bases. Besides, they trained A Bidirectional Associative Memory Neural Network (BAMNN) for key generation. They mentioned that their method showed efficient encryption and decryption times when compared with the other existing systems in the literature (Basu et al., 2019).
In 2020 Tahir et al. developed a new model based on a genetic algorithm (GA) called CryptoGA to cope with data integrity and privacy issues on cloud data. In their method, GA was used to generate the keys for encryption and decryption, which are integrated with a cryptographic algorithm to ensure the privacy and integrity of cloud data. In experiments, execution time, throughput, key size, and avalanche effect were considered for evaluation and comparison. They expressed that experimental results analysis ensured the integrity and preserved the privacy of the user’s data against unauthorized parties. Moreover, they mentioned that CryptoGA was robust and provided a better performance on selected parameters when compared with the other state-of-the-art cryptographic algorithms like DES, 3DES, RSA, Blowfish, and AES (Tahir et al., 2021).
In 2020, Indrasena et al. proposed a bio-inspired cryptographic DNA system. Their method consists of three phases: encryption, key generation, and decryption. In their method, transcription and translation and some of the inverse procedures were used from reproducing the normal procedures in the genetic encoding for encryption and decryption of the data. Moreover, Central Dogma of Molecular Biology (CMDB) was employed. They compared their method with the traditional cryptographic techniques and reported 67% increased processing time for the encryption process and decryption process, respectively (Indrasena Reddy et al., 2020).
In 2021, Thabit et al. designed a cryptography algorithm that has two layers to improve cloud computing security. In their study, the first layer was inspired by Shannon’s diffusion and confusion theory. The second layer was inspired by the structures of genetic coding for cryptographic purpose. They employed simulation of natural processes of genetic cryptography (translation from binary to DNA bases), transcription (regeneration from DNA to mRNA), and translation (regeneration from mRNA to protein). They obtained remarkable experimental results which can be used to secure applications on cloud computing in terms of size and execution time. (Thabit et al., 2021).
In this study, a DNA cryptography technique has been proposed by integrating DNA encoding and DNA operators into the Feistel network structure. Here, DNA itself was used as a carrier instead of traditional digital media such as image, text, or video, while modern biological tools were being used as implementation tools. Besides, the developed simulation software and the synthesized DNA sequence were both digitally and biologically integrated into specifically created biotechnical hardware. Since storing data in DNA environment was also aimed, the amount of data (bits) that can be embedded in a DNA base (A, C, G, and T) becomes an important problem here. When the studies in the literature were examined, it was seen that the number of bits that can be encoded per nucleotide did not exceed two. In the proposed study, this rate has been increased with the original compression algorithm that is carried out before the encryption process. When the biologically synthesized DNA sequence was compared with the simulated DNA sequence obtained by the decoding process in the simulation, 100% match success was obtained. The simulation software was also integrated into the hardware that can execute in a plug-and-play manner. These are the main features that make the proposed study versatile in terms of DNA-based data storage.
3 The proposed method
In this section, first, the main principle of DNA cryptography has been mentioned by briefly expressing the biological structure of DNA. Then the proposed algorithm has been explained with the details of each substep by providing the concrete samples.
3.1 DNA cryptography
DNA is a hereditary material in all living organisms and carries genetic information. DNA includes an array that is non-parallel two biopolymers wrapped around each other to create the double helix structure. Each DNA sequence includes four types of nucleotides: Adenine (A), Guanine (G), Cytosine (C), and Thymine (T) (Monika and Upadhyaya, 2015). In DNA structure, Adenine matches with Thymine, Guanine matches with Cytosine, which is called Watson-Crick complement. Every two sequences in this structure, are anti-parallel. Here, if a sequence starts with 3, it ends with 5, and if the other sequence starts with 5, it ends with 3 (Kaundal and Verma, 2014).
In DNA cryptography, DNA pairs are used as information carriers. When compared with the other methods, the large processing power of DNA molecules renders DNA cryptography more advanced. As a result, it is expected that DNA chip (hardware) technology will replace the existing silicon chips in the future. Thus, processing data on computers will increase tremendously. Additionally, there is a need for a more secure algorithm since traditional concepts of cryptographic algorithms such as DES, RSA can be broken. The advantages of DNA cryptography are DNA's exceptional storage capacity, low power consumption, and remarkable processing performance (Monika and Upadhyaya, 2015).
First, any kind of digital data is converted to binary to perform DNA encoding. Then by DNA encoding, the binary form of data is converted to DNA bases. Here, encryption is performed by benefitting DNA operators like DNA XOR, DNA shifting, etc. Then the simulated DNA sequence is synthesized. The synthesized DNA is then sequenced to obtain the original data in the beginning. Here, decryption is performed to obtain the DNA bases as the result of DNA encoding stage. Then DNA decoding is performed to form the binary form of the data. Finally, this binary form is read and converted to the digital file in the beginning.
3.2 Preliminaries
In this subsection, a brief description of the used symbols in equations has been provided.
Input file array in ASCII. In fact, this file can be in any form since it is going to be converted to binary. However, in the simulation, it is preferred to be ASCII
Input file vector in Base 2
: String array whose words consist of three bits.
w: word of G whose length is 3 bits
: Compressed DNA sequence
Bit Array of D via DNA Encoding
B’ whose elements are in the length of 12 bits.
w': word of G’ whose length is 12 bits
Left half of w’
Right half of w’
Next left half of w’
Next right half of w’
s: Output of S-box
Encrypted DNA sequence
Binary form of E via DNA encoding
B″ whose elements are in the length of 12 bits.
Compression process: In the proposed study, the compression method was formed by using the analytical plane and the well-known DNA base-bit transformation (Patent Pending ID number of 2017/00459). Thus, the complex relationship between input and output was contributed while the compression capacity was being increased by nearly 180%.
3.3 Compression process
Step 1: In this step, a text file is received as input data and it is converted to binaries as mentioned in Eq. (1):
For example:
Text: ESRA SATIR DUZCE UNIVERSITESI. [30 characters]
Text to Binary: (01000101, 01010011, 01010010, 01000001, 00100000, 01010011, 01000001, 01010100, 01001001, 01010010, 00100000, 01000100, 01010101, 01011010, 01000011, 01000101, 00100000, 01010101, 01001110, 01001001, 01010110, 01000101, 01010010, 01010011, 01001001, 01010100, 01000101, 01010011, 01001001, 00101110) (30 × 8 = 240 bits)
Step 2: In this step, the resultant data are divided into groups, each containing 3 bits (if not, it is concatenated with a concerning number of 0 s). Here, by obeying the mentioned compression process (please refer to subsection 3.2.) DNA bases are expressed as vectors. Clusters are set with the help of the analytical plane. The base vectors of DNA are located within each cluster on X-axis, while the coordinates in Y-axis are being expressed as two bits. First, the coordinates of each triple bit are found, and then the corresponding base is assigned. Afterward, the corresponding cluster of the base is determined, and each binary cluster is subjected to the bit-base transformation. The mathematical process of this step is as follows as shown in Eq.2:
G = (010, 001, 010, 101, 001, 101, 010, 010, 010, 000, 010, 010, 000, 001, 010, 011, 010, 000, 010, 101, 010, 001, 001, 001, 010, 100, 100, 010, 000, 001, 000, 100, 010, 101, 010, 101, 101, 001, 000, 011, 010, 001, 010, 010, 000, 001, 010, 101, 010, 011, 100, 100, 100, 101, 010, 110, 010, 001, 010, 101, 001, 001, 010, 011, 010, 010, 010, 101, 010, 001, 000, 101, 010, 100, 110, 100, 100, 100, 101, 110)
In G array, we have a total of 240 bits. The concerning DNA bases, Cluster Equivalents, and Primer bases have been estimated by considering the mentioned compression procedure (please refer to subsection 3.2.).
DNA Bases:
TGTAGATTTTTTTGTGTTTATGGGTCCTTGTCTATAAGTGTGTTTTTATGCCCATCTGTAGGTGTTTATGTATCCCCCAC (80 bases)
Cluster Equivalents:
1,0,1,0,0,0,1,1,1,0,1,1,0,0,1,1,1,0,1,0,1,0,0,0,1,0,0,1,0,0,0,0,1,0,1,0,0,0,0,1,1,0,1,1,0,0,1,0,1,1,0,0,0,0,1,1,1,1,1,0,0,0,1,1,1,1,1,0,1,0,0,0,1,0,1,0,0,0,0,1 (80 bits)
Primers:
GGACGCACGGGAGTAAGGATGCAGCAACCGACCGGAGGAT (40 bases)
Step 3: In this step, the compressed DNA sequence is obtained by concatenating DNA bases and primers as given in Eq. (3):
D = (GGACGCACGGGAGTAAGGATGCAGCAACCGACCGGAGGAT//TGTAGATTTTTTTGTGTTTATGGGTCCTTGTCTATAAGTGTGTTTTTATGCCCATCTGTAGGTGTTTATGTATCCCCCAC) (40 + 80 = 120 bases)
3.4 Encryption process
In the proposed study, DNA encoding (Kumar and Singh, 2011) and DNA XOR (Mondal and Mandal, 2017) are integrated into the Feistel network. The Feistel network has been depicted in Fig. 1. This integration has been performed by using traditional DES S-boxes. In Feistel networks, the design of F function is crucial since it measures the originality and complexity of the approach. Here, a novel F function has been developed by benefitting DNA XOR and DNA shifting, as depicted in Fig. 2(b). Fig. 2(a) depicts the usage of DES S-box. As a result, the proposed scheme is an example of symmetric encryption.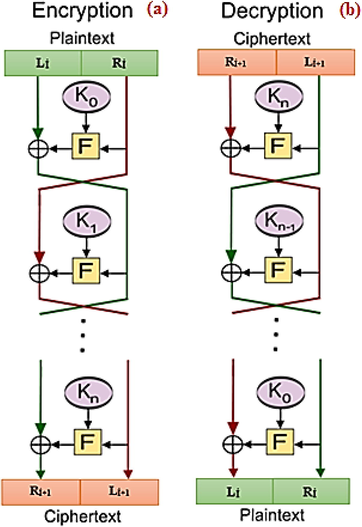
Block diagram of the Feistel Network Coding (a) Encryption process (b) Decryption process (Knudsen and Robshaw, 2011).
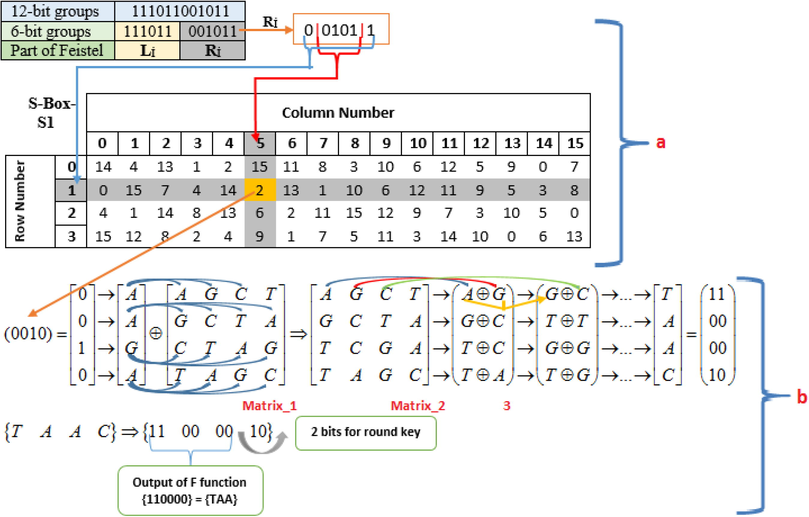
(a) The use of DES S_Box_S1 (b) The designed F function.
Mathematically, the formulation of Feistel network encryption can be shown in Eqs. (4) and (5).
Step 1: In this step, the obtained base sequence after compression is converted to binary via DNA Base-Bit transformation, expressed as follows by Eq. (6):
Step 2: In this step, the obtained bit sequence is divided into groups of 12 bits blocks via Eq. (7).
Here, each group is again divided into two equal parts in itself. These 6-bits groups are called left, Li and right Ri. Thus, Li and Ri are ready to submit to Feistel network structure as described in Table 1.
D
TGTAGATTTTTTTGTGTTTATGGGTCCTTGT…
B′
111011001011111111111111101110111111001110101011010111111011…
G′
111,011,001,011
111,111,111,111
101,110,111,111
…
6 bits groups
111,011
001,011
111,111
11,111
101,110
111,111
…
…
Part of Feistel
Lİ
Rİ
Lİ
Rİ
Lİ
Rİ
Lİ
Rİ
Step 3: In Feistel networks, F function makes the structure original according to the designer. In the proposed study, a novel F function has been formed by using DES S-Boxes, DNA XOR, and DNA shifting operators. Pseudo codes of this process have been provided below for ease of understanding:
-
Truncate w’ into two equal pieces (Ri, Li)
-
Exchange left and right pieces
-
(Li) ↔ (Ri)
-
For x = 1 to 8
-
{
-
s = S_Box_Sx (Ri)
-
For y = 1 to 5
-
e’ = DNA_F_Function(s) (Refer to Figure 2.b)
-
}
In Fig. 2(a), Ri from Table 1 enters S-Box_S1. Hence the output becomes in the length of 4 bits. The bases corresponding to the obtained 4 bits are subjected to DNA XOR with Matrix_1 in Fig. 2(b). As a result, Matrix_2 is obtained. Matrix_2 is subjected to DNA XOR operation again, but this time as depicted in a way marked as 3 in Fig. 2(b). The result still consists of 4 bases and, accordingly, 8 bits. Also, this is the output of our F function. Here, 2 LSB bits of each 8 bit are the round keys, as shown in Fig. 2(b). The remaining 6 bits, namely 3 bases, are ready to enter XOR process with Li in the Feistel network. In total, 8 loops are made for 8 S-Boxes (S1,…,S8), and Matrix_1 is shifted 5 times for each S-Box loop. Thus, Li+1 and Ri+1 are formed after 40 loops. Eventually, the encoded DNA chain is obtained. In Fig. 2(a) and (b), only one round of the developed F function has been presented. Pseudo codes of this process have been provided below for ease of understanding:
Due to Feistel structure, notice that
Li + 1 = Ri
Ri + 1 = e’ XOR Li
e = Li + 1 // Ri + 1
E = {e: e = Li + 1 // Ri + 1})
In Fig. 3, a flowchart of the encryption process has been provided by separating and marking each step.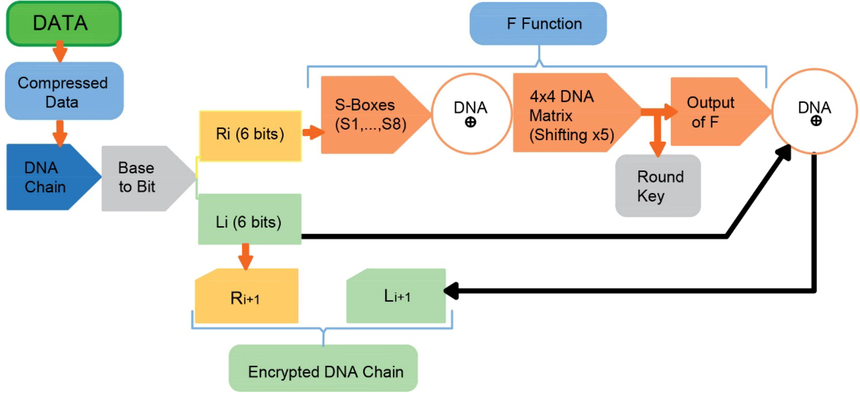
Flowchart of the encryption process.
3.5 Decryption process
Owing to the symmetric structure of encryption process, the decoding process can be easily obtained by inverting the encryption process. The plain data can be obtained back by adapting the encrypted DNA sequence to the Feistel network structure, as seen in Fig. 1(b).
The decoding phase of the Feistel network can be mathematically shown as follows via Eqs. (8) and (9):
Step 1: In this step, the encrypted DNA chain is divided into groups of 6 bases for presenting the Feistel network, as depicted in Fig. 1(b). These obtained 6-base groups are expressed as Li+1 and Ri+1. They are in the form of two half in order to be adapted to the Feistel structure again, but this time, inversely. This process is shown via Eq. (10):
Step 2: In this step, B″ is separated into words, each of which length is 12 bits. We call the resulting array T. In the decryption process, Li+1 is the entry of F function. It takes Li, directly and here F function runs in reverse order of the encryption process. The 6-bit groups formed in Ri+1 and the result of F function enters DNA XOR processing. Hence, the obtained result is Ri. This process is shown by Eq. (11):
Pseudo codes of this process have been provided below for ease of understanding:
-
Truncate w″ into two equal pieces (Ri′, Li′)
-
Notice that: Li′ = Li + 1 and Ri′ = Ri + 1 since the decryption process is the reverse function of encryption.
-
For y = 1 to 5
-
{
-
e′ = reverse (DNA_F_Function (Li′)
-
For x = 1 to 8
-
s = reverse (S_Box_Sx (e′))
-
}
Step 3: Due to the structure of Feistel, Li+1 becomes the new Li. The resultant Li and Ri groups are subjected to Bit-Base transformation, and thus, the plain DNA sequence is obtained.
Ri + 1 = s XOR Li′
Li + 1 = Ri′
[B′] = Li + 1 // Ri + 1
Notice that D corresponds to DNA encoding of B. Here, after the decompression process that is the reverse function of compression process, B is obtained. Since B is the binary form of input data I, the initial input file has been obtained. In Fig. 4, flowchart of the decryption process has been provided by separating and marking each step.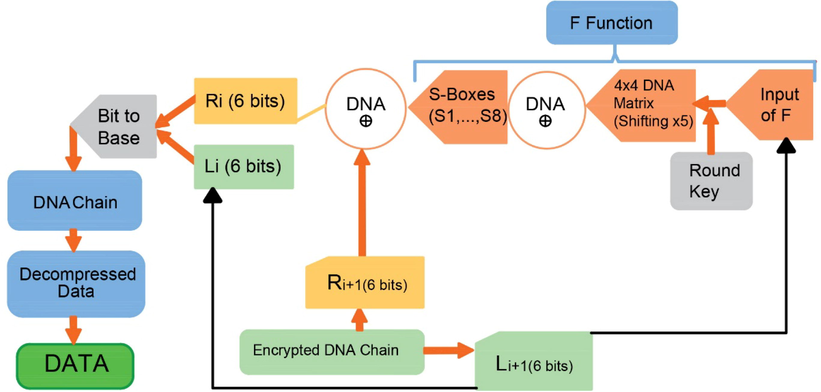
Flowchart of the decryption process.
4 Simulation and the design of biotechnical hardware
Simulation of the proposed method has been carried out by using C# language. Software testing has been performed by a computer with i7, 2.2 GHz processor, 6 GB RAM, and Win7 64bit operating system. The simulation output, including encryption and decryption phases, has been presented in Fig. 5.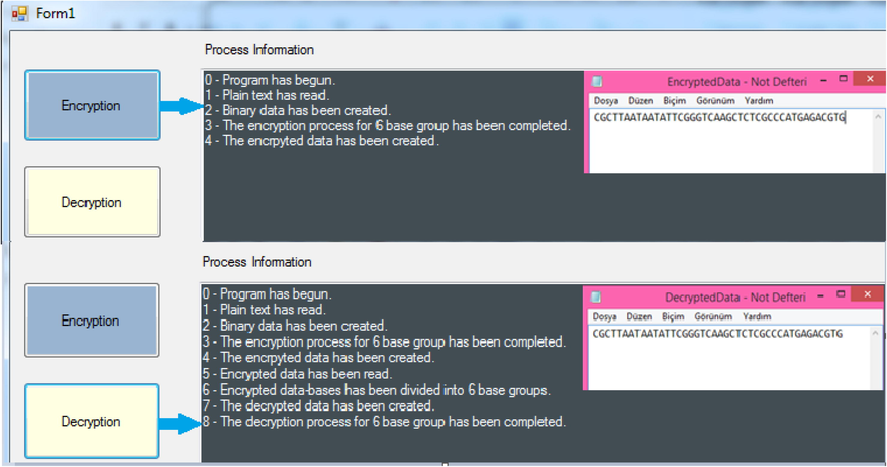
Encryption and decryption phases interface of the simulation software.
In Fig. 5, flow of the simulation software can be followed in “Process Information” part. The concerning output after encryption and decryption processes can be obtained via the given note pad file on the right side of the interface.
Biotechnical hardware has a unique design that is drawn in CATIA V5 program. The created design was manufactured with the help of a 3D printer. For a detailed view, Fig. 6 needs to be considered. DNA cells in Fig. 6 in biotechnical hardware provide the possibility for storing the synthesized DNA. On the other hand, the simulation software that produces the concerning digital DNA sequence resulting from encryption or decryption is ready to be run on any Windows-based system.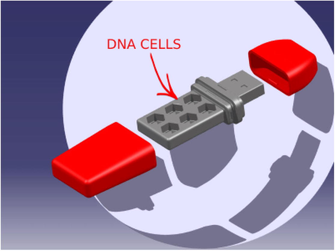
The internal structure of the developed biotechnical hardware.
When the general specifications of the studies in the literature were examined, it was seen that the processes of DNA computing were gradually increasing. However, it was noted that this process always linearly depended on the speed of the biological process. Furthermore, most of the performed studies stayed limited by the simulation. In the proposed study, this lack is addressed. First of all, a novel DNA cryptography algorithm that is suitable with the nature of DNA was improved. Then, the limitation of the simulation was solved by integrating the simulation software into the produced plug-and-play biotechnical hardware. The internal structure of the developed biotechnical hardware is illustrated in Fig. 6. Here, the integrated DNA cells were designed to store the synthesized DNA sequence in the form of DNA drops. These DNA drops are needed to be diluted by a special solution before sequencing. Hence, we designed each DNA cell to store 1 mg of DNA. It is widely known that 1 g of DNA can store 108 Terabytes of data. Since we have 6 DNA cells in the designed biotechnical hardware, we are able to store 6 × 105 TBytes of data.
5 Experimental results
In this section, the performed biological experiments, performance, and security analyses and their results have been presented by providing concrete samples as much as possible.
5.1 In vitro experiments
In the proposed study, 9588 bp of encrypted DNA sequence, which is obtained via the simulation software, has been synthesized with the support of Düzce University Scientific Research Projects Coordination ship. After synthesizing DNA chain, it is an essential requirement to obtain the original DNA sequence by re-reading it. This process is called DNA sequencing. Fig. 7 shows the sequence result of the synthesized DNA chain. Here, after sequencing of 9588 bp DNA chain, it has been seen that the synthesized and the sequenced DNA chains match without loss of information. The synthesized 9588 bp DNA chain has been encapsulated in a plasmid.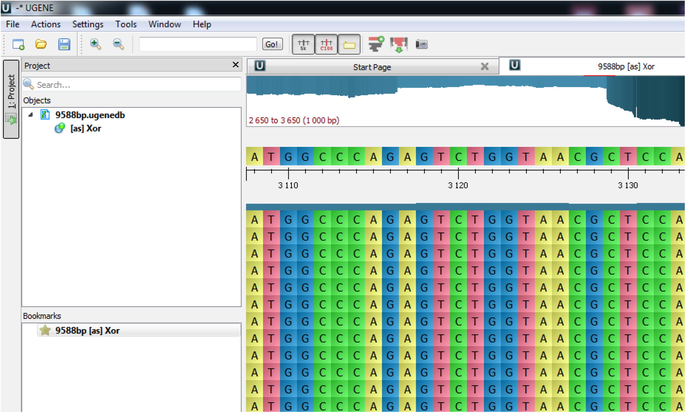
DNA chain sequence results.
5.2 Requirement analysis
There are a set of requirements that must be fulfilled by each DNA encryption algorithm. (UbaidurRahman et al., 2015). In the proposed study, these given requirements have been considered for comparison. In Table 2, comparison results of the existing DNA encryption algorithms in terms of these fulfillments have been presented. x- Indication of minimum level of supporting. √- Indication of Acceptable Level of Supporting. * - Partial fulfilment.
Authors
DNA Encoding of Complete character set fulfillment
Dynamic Encoding Table Generation
Unique sequence for encoding of every character of plaintext to DNA sequence for every session
Robustness of encoding
Biological Process Simulation
Dynamicity of the encryption process
G. Cui et al. (Cui et al., 2009)
x
x
x
x
*
*
Q. Zhang et al. (Zhang et al., 2009)
x
x
x
x
*
x
S. Sadeg et al. (Sadeg et al., 2010)
x
x
x
x
√
x
S.T. Amin et al. (Amin et al., 2006)
x
x
x
x
√
x
O. Tornea & M.E. Borda (Tornea and Borda, 2009)
x
x
x
x
*
x
M. Sabry et al. (Sabry et al., 2012)
x
x
x
x
*
*
X. Wang & Q. Zhang (Wang and Zhang, 2009)
x
x
x
x
x
x
A. Akanksha et al. (Akanksha et al., 2012)
√
x
x
x
x
x
N. H. UbaidurRahman et al. (UbaidurRahman et al., 2015)
√
√
√
√
√
√
S. Goyat & S. Jain (Goyat and Jain, 2016)
*
x
x
*
√
*
H. M. Mousa (Mousa, 2016)
*
*
*
√
√
*
R. Ahmed & I. Mohammed (Ahmed and Mohammed, 2017)
*
x
x
*
√
x
M. Thangavel &P. Varalakshmi (Thangavel and Varalakshmi, 2018)
x
√
√
*
*
Narendren et al. (Narendren. et al., 2018)
x
x
*
√
x
S. Basu et al. (Basu et al., 2019)
*
√
*
*
√
√
Tahir et al. (Tahir et al., 2021)
*
√
*
√
√
√
F. Thabit et al. (Thabit et al., 2021)
√
√
√
√
√
√
The Proposed Study
√
x
√
√
√
√
In the proposed study, it is clear that the majority of the predicted requirements have been accommodated. Since the proposed study converts plaintext to binary, it is possible to encode the entire character set via DNA. However, dynamic encoding is not possible. Since DNA encoding and DNA operators integrated Feistel Network are used, and different round keys are generated in every loop in only one session, a unique sequence for encoding every character of plaintext to DNA sequence has been fulfilled. This also renders the encryption process dynamic. Because, for encoding only 12 bits block, at least 40 loops are performed. In every loop, we have a round key whose length is 2 bits. In total, the key length is at least 80 bits for encryption of 12 bits blocks, and for every block, keys are also changed. These are the main features that support the robustness and dynamicity of encryption. The proposed algorithm has been simulated, and laboratory experiments have also been performed, as explained in subsection 5.1, which address the biological process simulation issue.
5.3 Capacity analysis
This section compares the capacity rates of the proposed study and the reached existing studies in the literature. Capacity is the density of information transport that the ciphertext has. In DNA encryption, it is expressed by the rate of data that can be embedded per nucleotide. Table 3 shows the capacity comparisons of the existing studies and the proposed study.
Authors
Length of Plaintext Input (bits)
Length of Ciphertext Output (bits)
Capacity (Input/Output)
S.T. Amin et al. (Amin et al., 2006)
90,300
88,410,189
0,001
O. Tornea & M.E. Borda (Tornea and Borda, 2009)
10
148
0,067
M. Sabry et al. (Sabry et al., 2012)
4
20
0,2
A. Akanksha et al. (Akanksha et al., 2012)
1
4
0,25
N.H. UbaidurRahman et al. (UbaidurRahman et al., 2015)
4
16
0,25
X. Wang & Q. Zhang (Wang and Zhang, 2009)
3
9
0,33
M. Sohal & S. Sharma (Sohal and Sharma, 2018)
40,000
46,640
0,85
The proposed study
40,000
40,320
0,99
In Table 3, lengths of plaintexts, ciphertexts, and their ratios, namely capacity values, have been presented. Lengths of plaintexts and ciphertexts have been provided in bits. As presented in Table 3, the proposed method has a more efficient capacity rate than the existing studies in the literature.
5.4 Brute force attack analysis
Evaluation of the brute force algorithm is simple, but it has a massive number of steps for processing to decrypt the password. For this purpose, the attacker begins to try every single possible combination for password candidates and see whether it results in a decrypted file (Gautam and Jain, 2015).
In this study, the key length is at least 80 bits since the input has at least one block with a length of 12 bits. The key is obtained from 40 loops, each of which contains a round key having 2 bits. It is nearly impossible to break it via a brute force attack that is performed by employing today’s technology. When the brute force attack is performed for the proposed study,
-
The average CPU (Central Processing Unit) speed for an average computer is 3 × 109 Hz
-
Consider that one year is about 3 × 107 s,
-
The formula in terms of year is as follows given in Eq. (12):
-
Accordingly, the time to perform the brute force attack is calculated as follows for only one block:
Besides, the proposed method has a biological process due to the nature of DNA. This situation adds an extra layer of security that is performed biologically. Therefore, classical attacking techniques stay pointless to break the proposed scheme.
5.5 Key space
Key space represents the possible number of keys used for encryption algorithm. A higher key space makes the algorithm more secure against brute force attacks since it makes the key achieving calculations computationally infeasible within a given interval of time (Roy et al., 2020). Namely, the key space regarding an encryption scheme signifies the total number of possible keys (Roy et al., 2020). We have at least 40 loops for only one block of input data in the encryption phase in the proposed scheme. In each loop, a round key in the length of two bits is produced. Therefore, we have four combinations to find the round key that is produced in each loop (22 = 4). Since there are 40 loops in the proposed scheme, the total length of the key is 40 × 2 = 80 bits. Here, we have 280 possible combinations to extract the hidden key in the proposed scheme. Moreover, this combination number depends on the length of plaintext. As the length of plaintext increases, the length of the key increases, too.
5.6 Information entropy
Information entropy is the most significant feature for measuring randomness of a source. The information entropy H(m) of a message source m can be estimated by the following formula in Eq. (13):
Here, L is the total number of symbols in m, and p(mi) is the probability of occurrence of the symbol mi. Assuming there are 256 symbols with the same probability entropy value, H(m) = 8 (Bakhshandeh and Eslami, 2013). In the proposed DNA cryptography scheme, the source message consists of four symbols; A, G, C, T. It has seen that for the plain and cipher texts which have the lengths between 100 and 100.000 bases, (via incrementing the lengths 10× by 10×) the estimated entrophy values were very close to 2. This is a quite efficient ratio when we consider that there were 4 probabilities.
6 Conclusion
In this paper, depending on the Central molecular biology principle for encryption and decryption of systems, a novel encryption approach by combining DNA carrier medium, DNA encoding, and DNA XOR operation with Feistel network structure has been derived. Besides, the proposed DNA encryption process was both digitally and biologically integrated into designed biotechnical hardware.
The performed experiments showed that DNA cryptology has a high potential to be a new method in data security. Experimental results demonstrated that the proposed study has efficient outcomes in terms of capacity, brute force attack, key space, and entropy analyses. Besides, the implementation of the proposed method has been verified by vitro experiments.
However, in the scope of this study, there are still some obstacles. First of all, this study bases on the symmetric encryption process. For a more secure scheme, asymmetric encryption-based structures have to be examined. In DNA structure, random access to data is another problem since this technology is a brand new field. Here, we need extra techniques to speed up the whole process when we consider the processing time and the speed of the synthesized DNA instead of the silicon medium. As mentioned above, dynamic encoding of each character is another requirement that increases the robustness of an algorithm. However, the design has to be performed without occupying the memory and causing any speed loss in this process. All of these issues are planned to be tackled in future studies.
Funding
The experimental stages of the proposed study were supported by Düzce University Scientific Research Projects Coordinationship (project number 2016.06.01.502) in the project called “Data Storage in the DNA Chain.” Furthermore, the compression algorithm in the proposed study is in the national patent process with the Patent Pending ID number of 2017/00459.
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial or non-financial interest in the subject matter or materials discussed in this manuscript.
References
- Developing a New Hybrid Cipher Algorithm using DNA and RC4. Int. J. Adv. Comput. Sci. Appl.. 2017;8:71.
- [CrossRef] [Google Scholar]
- Implementation of DNA algorithm for secure voice communication. Int. J. Sci. Eng. Res.. 2012;3:1-5.
- [Google Scholar]
- A DNA-based implementation of yaea encryption algorithm. Proc. 2nd IASTED Int. Conf. Comput. Intell. CI. 2006;2006:116-120.
- [Google Scholar]
- An authenticated image encryption scheme based on chaotic maps and memory cellular automata. Opt. Lasers Eng.. 2013;51:665-673.
- [CrossRef] [Google Scholar]
- Bio-inspired cryptosystem with DNA cryptography and neural networks. J. Syst. Archit.. 2019;94:24-31.
- [CrossRef] [Google Scholar]
- A DNA-based, biomolecular cryptography design. Proc. - IEEE Int. Symp. Circuits Syst.. 2003;3:822-825.
- [CrossRef] [Google Scholar]
- DNA computing and its application to information security field. 5th Int. Conf. Nat. Comput. ICNC. 2009;2009 6:148-152.
- [CrossRef] [Google Scholar]
- Gautam, T., Jain, A., 2015. Analysis of brute force attack using TG — Dataset. https://doi.org/10.1109/IntelliSys.2015.7361263
- Gehani, A., LaBean, T., Reif, J., 2003. DNA-based Cryptography. pp. 167–188. https://doi.org/10.1007/978-3-540-24635-0_12
- A secure cryptographic cloud communication using DNA cryptographic technique. In: in: 2016 International Conference on Inventive Computation Technologies (ICICT). 2016. p. :1-8.
- [CrossRef] [Google Scholar]
- A secured cryptographic system based on DNA and a hybrid key generation approach. Biosystems. 2020;197:104207
- [CrossRef] [Google Scholar]
- The block cipher companion. Springer Science & Business Media; 2011.
- Secret data writing using DNA sequences. In: in: 2011 International Conference on Emerging Trends in Networks and Computer Communications (ETNCC). IEEE. 2011. p. :402-405.
- [CrossRef] [Google Scholar]
- Kumar, S., Vijayaraghavan, R., 2011. Solid state drive (SSD) FAQ.
- DNA-Genetic Encryption Technique. Int. J. Comput. Netw. Inf. Secur.. 2016;8:1-9.
- [CrossRef] [Google Scholar]
- A light weight secure image encryption scheme based on chaos & DNA computing. J. King Saud Univ. - Comput. Inf. Sci.. 2017;29:499-504.
- [CrossRef] [Google Scholar]
- Secure Communication Using DNA Cryptography with Secure Socket Layer (SSL) Protocol in Wireless Sensor Networks. Procedia Comput. Sci.. 2015;70:808-813.
- [CrossRef] [Google Scholar]
- Towards DNA based data security in the cloud computing environment. Comput. Commun.. 2020;151:539-547.
- [CrossRef] [Google Scholar]
- Narendren., Yathish, Y.B., Mohan, C., 2018. A Cryptosystem using Two Layers of Security-DNA and RSA Cryptography Roy, S., Rawat, U., Sareen, H.A., Nayak, S. K., 2020. IECA: an efficient IoT friendly image encryption technique using programmable cellular automata. J. Ambient Intell. Human Comput. 11, 5083–5102. https://doi.org/10.1007/s12652-020-01813-6.
- IEVCA: An efficient image encryption technique for IoT applications using 2-D Von-Neumann cellular automata. Multimed Tools Appl.. 2021;80(21-23):31529-31567.
- [CrossRef] [Google Scholar]
- Three Reversible Data Encoding Algorithms based on DNA and Amino Acids’ Structure. Int. J. Comput. Appl.. 2012;54:24-30.
- [CrossRef] [Google Scholar]
- Sadeg, S., Gougache, M., Mansouri, N., Drias, H., 2010. An encryption algorithm inspired from DNA. 2010 Int. Conf. Mach. Web Intell. ICMWI 2010 - Proc. 344–349. https://doi.org/10.1109/ICMWI.2010.5648076
- BDNA-A DNA inspired symmetric key cryptographic technique to secure cloud computing. J. King Saud Univ. – Comput. Inf. Sci.. 2018;34(1):1417-1425.
- [Google Scholar]
- CryptoGA: a cryptosystem based on genetic algorithm for cloud data security. Cluster Comput.. 2021;24(2):739-752.
- [CrossRef] [Google Scholar]
- Public-key system using DNA as a one-way function for key distribution. BioSystems. 2005;81(1):25-29.
- [CrossRef] [Google Scholar]
- A new data security algorithm for the cloud computing based on genetics techniques and logical-mathematical functions. Int. J. Intell. Networks. 2021;2:18-33.
- [CrossRef] [Google Scholar]
- Enhanced DNA and ElGamal cryptosystem for secure data storage and retrieval in cloud. Cluster Comput.. 2018;21(2):1411-1437.
- [CrossRef] [Google Scholar]
- Tornea, O., Borda, M.E., 2009. DNA Cryptographic Algorithms. pp. 223–226. https://doi.org/10.1007/978-3-642-04292-8_49.
- A novel DNA computing based encryption and decryption algorithm. Procedia Comput. Sci.. 2015;46:463-475.
- [CrossRef] [Google Scholar]
- Wang, X., Zhang, Q., 2009. DNA computing-based cryptography. BIC-TA 2009 - Proceedings, 2009 4th Int. Conf. Bio-Inspired Comput. Theor. Appl. 67–69. https://doi.org/10.1109/BICTA.2009.5338153
- Security analysis on a color image encryption based on DNA encoding and chaos map. Comput. Electr. Eng.. 2015;46:433-446.
- [CrossRef] [Google Scholar]
- Zhang, Q., Guo, L., Xue, X., Wei, X., 2009. An image encryption algorithm based on DNA sequence addition operation. BIC-TA 2009 - Proceedings, 2009 4th Int. Conf. Bio-Inspired Comput. Theor. Appl. 75–79. https://doi.org/10.1109/BICTA.2009.5338151







