Translate this page into:
Taxon-specific zeta-crystallin of camel eye lens: A comparative in silico studies
⁎Corresponding author. amalik@ksu.edu.sa (Ajamaluddin Malik)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The eye lens is a specialized organ of the visual system, which is transparent due to its complex geometry and unique protein composition. Nature has designed it to focus light on the retina throughout the life span. Several factors (post-translational modifications, thermal and solar radiations) causes aggregation of the lens proteins and result in early-onset of cataract. A cataract is one of the major causes of blindness worldwide. It's interesting how camel eyes maintain lens transparency in a harsh desert climate? The camel eye lenses contain a novel protein zeta (ζ)-crystallin in a bulk amount that is also present in two other animals (guinea pig and Japanese frog) but is adopted for milder habitats. The camel lens ζ-crystallin is a poorly characterized protein. This study has investigated the physicochemical and structural properties of camel, guinea pig, and Japanese frog’s ζ-crystallin. The docking results showed a strong affinity between NADPH and ζ-crystallin. RMSD, RMSF, radius of gyration, and SASA analysis in the ligand-free and bound states showed distinct prope6rties in these ζ-crystallins. Moreover, the hydrophobicity of camel and Japanese frog ζ-crystallin is lower than ζ-crystallin of other mammalian sources. The unique physicochemical and structural properties of taxon-specific ζ-crystallin are likely to maintain lens transparency.
Keywords
Zeta-crystallin
Structure modeling
Eye lens proteins
Camel
Protein solubility
- NADPH
-
Nicotinamide adenine dinucleotide phosphate reduced
- RMSD
-
Root-mean-square deviation
- RMSF
-
root-mean-square fluctuation
- Rg
-
radius of gyration
- SASA
-
solvent accessible surface area
Abbreviations
1 Introduction
The lens is a fundamental part of the visual pathway, located between the exterior and the retina, that is designed as a specialized tissue for the single known purpose of allowing clarity of vision throughout life. The lens is composed of a high concentration of water-soluble proteins, called crystallins, which constitute approximately 90% of all proteins in the lens (Jaenicke 1996). This high concentration of crystallins (∼500 mg/ml) increases the refractive index and maintains the transparency of the eye. The adult eye lens is devoid of nuclei and cellular organelles to minimize light scattering (Ivanov et al., 2005, Gong et al., 2007). As a result, the lens proteins synthesized during embryonic stages can persist and function throughout the life span of an organism, withstanding high levels of UV light, temperature fluctuations, and dehydration. These factors may initiate misfolding and aggregation of crystallins, resulting in increased light scattering and lens opacification, leading to cataracts (Sharma and Santhoshkumar 2009, Al-Shabib et al., 2019a,b). Camels being desert-dwelling animals, have eye lenses that are exposed to very high ambient temperatures, dehydration, and high levels of ultraviolet radiation.
Three ubiquitous proteins (α-,β-andγ-) crystallins are present in all lenses. Additionally, a fourth taxon-specific crystallin, ζ-crystallin (EC 1.6.5.5), has been identified in camels (Camelus dromedarius), guinea pigs (Cavia porcellus), and Japanese frogs (Hyla japonica). This ζ-crystallin possesses quinone oxidoreductase activity (Rao et al., 1992, Duhaiman et al., 1995, Fujii et al., 2001) and constitutes approximately 10% of the total lens proteins of C. dromedarius (Garland et al., 1991). In humans and other animal lenses, ζ- crystallin is present at an enzyme level (Rao et al., 1997). However, C. dromedarius, C. porcellus, and H. japonica habitats are markedly different. Interestingly, why quinone oxidoreductase was recruited as crystallin in diverse evolutionary animals and the role of ζ-crystallin in the lens is not yet well understood.
In this study, we performed in silico studies to determine the physicochemical, phylogenetic, structural properties, and ligand binding of ζ-crystallin in C. dromedarius, C. porcellus, and H. japonica.
2 Materials and methods
2.1 ζ- crystallin sequences
The sequence of C. dromedarius ζ-crystallin fetched from European Nucleotide Archive (EMBL accession no: GADU01072870.1). The accession numbers of Homo sapiens, Cavia porcellus, and Hyla japonica ζ-crystallin were BAD96870.1, NP_001166407.1, and BAB41213.1, respectively.
2.2 Multiple sequence alignment and phylogenetic analysis relationships
The amino acid sequences were compared with those obtained from the protein database using PSI-BLAST algorithm. Next, homologous sequences of ζ- crystallin were aligned using the MAFFT sequence alignment technique (Katoh and Standley 2013). Next, the alignment output was color-coded using the Jalview workbench (Waterhouse et al., 2009). Finally, evolutionary divergence among the homologous sequences and the phylogenetic tree were constructed using the BLOcks SUbstitution Matrix (Henikoff and Henikoff 1992).
2.3 Sequence analysis
The physicochemical properties of C. dromedarius ζ-crystallin and its sequence homologs such as molecular weight, aliphatic index, theoretical pI, grand average hydropathy (GRAVY), and instability index (II) were calculated using the ProtParam tool (Gill and von Hippel 1989).
2.4 Homologous modeling
The three-dimensional structures of ζ-crystallin belonging to C. dromedarius, C. porcellus, and H. japonica were predicted using the Swiss-model server (Biasini et al., 2014). The crystal structure of H. sapiens ζ-crystallin (PDB id: 1YB5) at 1.85 Å resolution was used as a template. Homology modeling was carried out based on the following criteria: (1) Sequence identity > 60%; (2) 100% coverage; and (3) Template structure with a bound ligand. All the structures were analyzed and processed using PyMOL (PyMOL, 2006).
The three-dimensional structure models of C. dromedarius, C. porcellus, and H. japonica ζ-crystallin were validated using PROCHECK (Morris et al., 1992), ProSA-Web (Sippl 1995, Wiederstein and Sippl 2007) and QMEAN (Benkert et al., 2009) servers.
2.5 Molecular docking
The modeled 3D structures of ζ-crystallin belonging to all species in the trial were used for docking studies. NADPH (ZINC85551904) ligand was taken from the Zinc15 database and converted to a PDB format. AutoDock Vina (Trott and Olson 2010) was used to determine the binding site between ligands and proteins. Initially, potential binding sites were detected based on blind docking where the boxes were sufficiently large enough to cover the entire protein. PyMOL and Discovery Studio Visualizer (Biovia 2015) were used to visualize the structures.
2.6 Molecular dynamic simulations
The GROMACS 5.1.2 version was used to carry out the MD simulation for 50 ns at 300 K under a pressure of 1 bar using a GROMOS96 43a1 force-field. Computational tools such as PyMOL, Visual Molecular Dynamics (VMD) (Humphrey et al., 1996), and QtGrace were used to visualize, evaluate, and analyze MD trajectories. The structural coordinates of the ζ-crystallin of all species and NADPH were pre-processed in SPDBV. The topology and force-field parameters for NADPH were generated from the PRODRG server and merged into the parent.gro file of species’ ζ-crystallin to make the complex. Energy minimization was carried out using the steepest descent to remove bad connections to prevent structural hindrances in the solvated systems. The final MD ran at 50 ns for both systems, and the resulting trajectories were saved for further analysis using inbuilt utilities of GROMACS such as gmx rms, gmx rmsf, gmx gyrate, and gmx sasa (Naqvi et al., 2020).
3 Result and discussion
3.1 Multiple sequence alignment of ζ-crystallin
The amino acid sequence of ζ-crystallin of C. dromedarius was 100% identical to the two-hump domesticated Bactrian camel (C. ferus) (accession no: XP_006178374.1). Sequence alignment of C. dromedarius ζ-crystallin with H. sapiens, C. porcellus, and H. japonica showed 87, 83, and 64% identity, respectively (Table 1). The sequence homology was 79–97% between C. dromedaries ζ-crystallin and other mammalian sources. Average sequence homology of the thirteen mammalian ζ-crystallins was 87%. Whereas the sequence homology of amphibian (H. japonica) ζ-crystallin was 64%. The multiple sequence alignment of ζ-crystallin from various sources is presented in Fig. 1. The output showed a high degree of conservancy.
ζ-CrystaIIin
(NCBI Ref. Seq)
Amino acid residues
Identity
pl
MW (Da)
Aliphatic index
hydropathicity (GRAVY)
Instability index
Camelus dromedarius
GADU01072870*
329
100
7.1
35176
96.05
-0.033
30.93
Homo sapiens
BAD96870.1
329
87
8.56
35193
99.91
0.056
27.61
Lama guanicoe
Q28452.1
330
97
7.67
35187
96.03
-0.002
29.54
Pan troglodytes
XP_001167460.1
329
87
8.82
35232
99.6
0.033
26.51
Macaca mulatta
XP_0011001 74.1
329
86
8.9
35242
99.6
0.028
27.56
Pongo abelii
NP_001126904.1
329
86
8.33
35083
98.72
0.039
27.49
Sus scrofa
NP_001038049.1
330
86
8.24
35222
103.73
0.119
27.82
Bos taurus
NP_776450.2
318
87
7.74
34183
97.52
0.007
27.03
Ailuropoda melanoleuca
XP_002916952.1
330
85
8.57
35440
100.76
0.046
24.18
Canis lupus familiaris
NP_001239326.1
330
84
8.64
35521
104.33
0.057
27.99
Cavia porcellus
NP_001166407.1
329
83
7.69
35202
94.53
0.026
25.29
Oryctolagus cuniculus
XP_002715963.1
329
84
9
35242
103.13
0.074
31.6
Mus musculus
AAH43076.1
331
79
8.47
35368
94.56
-0.022
31.89
Hylajaponica
BAB41213.1
329
64
6.76
35565
88.05
-0.067
22.28

Amino acid sequence alignment of 13 mammalian and one amphibian ζ- crystallins. The multiple sequence alignment was generated with the MAFFT Multiple Sequence Alignment on a Jalview workbench. According to conservancy, residues are color-coded. Ligand (NADP) binding residues are labeled by (*).
3.2 Phylogenetic analysis of ζ-crystallin
The phylogenetic relationships among ζ-crystallins from various sources and evolutionary divergence are shown in Fig. 2. The C. dromedarius ζ-crystallin grouped with that of Lama guanicoe and Sus scrofa. H. sapiens and the other mammals showed evolutionary divergence. Due to low similarity, H. japonica (ζ-crystallin) was separated from other mammalian species.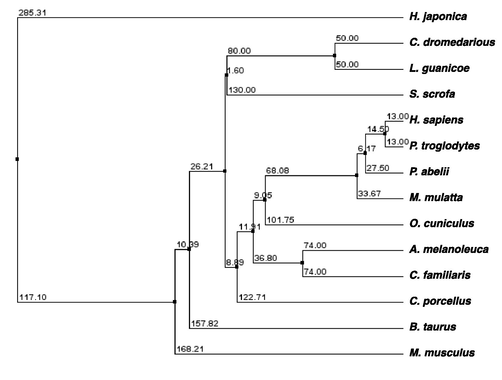
The phylogenetic tree of camel ζ-crystallin and homologous genes. The phylogenetic tree was generated with the BLOSUM62 from MAFFT Multiple Sequence Alignment.
3.3 Physicochemical properties of ζ-crystallin
The physicochemical properties of C. dromedarius ζ-crystallin and its sequence homologs are shown in Table 1. The calculated pI of C. dromedarius, C. porcellus, and H. japonica ζ- crystallin was close to neutral pH (7.1, 7.69, and 6.76, respectively). The ζ-crystallin was expressed at very high levels in the lens of C. dromedarius, C. porcellus, and H. japonica constituting approximately 10% of the total lens soluble proteins in these animals (Garland et al., 1991). Except for Bos taurus (bovine) ζ-crystallin, the pI of all other mammalian ζ- crystallin is in the alkaline region (pI > 8.0), suggesting that the solubility of the proteins is minimal at this pI. Interestingly, ζ-crystallin made up most of the soluble lens proteins in C. dromedarius, C. porcellus, and H. japonica, and their isoelectric points were close to the physiological pH. It is noteworthy that maintaining such a high degree of solubility is required throughout their lifespan. It may be possible that α-crystallin acts as a molecular chaperone to assist ζ-crystallin in maintaining its solubility. The pH inside the lenses of these animals is not known. It is also possible that the pH inside the lenses is different in these animals or some other factors may enhance their solubility.
The Grand Average of Hydropathy (GRAVY) value of a protein indicates its solubility (Khan et al., 2018). A positive value is indicative of a hydrophobic nature, while a negative value points to the hydrophilic nature of the protein (Kyte and Doolittle 1982). The ζ-crystallin of C. dromedaries, L. guanicoe, M. musculus, and H. japonica was relatively more hydrophilic than the other species (Table 1). C. porcellus ζ-crystallin was more hydrophobic than that of C. dromedarius, M. musculus, and H. japonica. In earlier studies, it was observed that when C. porcellus ζ-crystallin was expressed in E.coli, it was largely recovered as insoluble inclusion bodies. But, when C. porcellus ζ-crystallin was co-expressed with the GroEL/ES chaperone system, the solubility increased markedly (Goenka and Rao 2001). In contrast, M. musculus and H. japonica ζ-crystallin were expressed in E.coli without any assistance from a chaperone system (Fujii et al., 2001, Simpanya et al., 2010). Similarly, C. dromedarius recombinant ζ-crystallin was produced in E.coli without a chaperone system in our lab (Malik et al., 2016, Malik et al., 2017). Moreover, the yield of pure recombinant M. musculus ζ-crystallin was five-fold higher than recombinant C. porcellus ζ-crystallin (Simpanya et al., 2010), which could be due to the hydrophilic nature of the M. musculus ζ-crystallin.
The instability index provides an estimate of the in vitro stability of the test protein. An instability index value < 40 indicates that the protein is stable (Guruprasad et al., 1990). The instability index of all three ζ-crystallins examined were < 40. The instability index of the M. musculus ζ-crystallin (31.89) was the highest, while H. japonica ζ-crystallin was the lowest (22.28). The aliphatic index (AI) is used to indicate the thermostability of globular proteins (Ikai 1980). All (ζ-crystallins had a high aliphatic index, which varied between 88.05 and 104.33.
3.4 Structural studies of ζ-crystallin
The physiological role of ζ-crystallin in lenses is not well understood. Truncated mutants of ζ-crystallin cause cataracts in C. Porcellus. The structure of ζ-crystallin from C. dromedarius, C. porcellus, and H. japonica ζ-crystallin is not known, but a high-resolution structure of homologous H. sapiens ζ-crystallin (PDB: 1YB5) is available. A Swiss model server generated a 3D model structure of the (ζ-crystallin of the three species under study. Ligand (NADPH) bound human ζ-crystallin was superimposed on the ζ-crystallin of C. dromedarius, C. porcellus, and H. japonica (supplementary Fig. S1). The overall topology of ζ-crystallin from these animals was similar to human ζ-crystallin. The sequence identity of H. sapiens ζ-crystallin with C. dromedarius, C. porcellus, and H. japonica was 87, 83, and 64%, respectively.
Different parameters of the modeled structures were evaluated using PROCHECK, ProSA-Web, and QMEAN servers. The PROCHECK analyses evaluate the stereo-chemical quality of the protein structure. The PROCHECK analysis was generated using a Ramachandran plot. As shown in supplementary Fig. S2, this plot ensured the quality of the model structure of C. dromedarius, C. porcellus, and H. japonica ζ-crystallins with 92.5, 91.8 and 91.6% of the residues in the ‘most favored region’, respectively. According to the Ramachandran plot, 7.1, 7.8, and 8% of the residues were in the ‘additional allowed region’. Furthermore, only 0.4% of the residues were in the disallowed regions in all model structures.
The QMEAN Z-score evaluates the degree of nativeness in the model structure by comparing it to the template structure solved by X-ray crystallography or NMR spectroscopy. The QMEAN Z-scores range between 0 and 1. The QMEAN analysis of all three models is shown in Supplementary Fig S3. The three plots showed that the Z-score QMEAN of C. dromedarius, C. porcellus, and H. japonica were 0.48, 0.76, and 0.4, respectively. The red marker in the QMEAN graph of C. dromedarius and H. japonica ζ-crystallin structure is located within the dark zone while in the graph of C. porcellus it is located on the boundary of the dark zone, indicating an overall good quality of all modeled structures.
The ProSA analysis was used to analyze the overall quality of the three structure models of C. dromedarius, C. porcellus, and H. japonica ζ-crystallin by calculating Z-score based on the plot of experimentally determined protein structure (Supplementary Fig. S4). The Z-score for C. dromedarius, C. porcellus, and H. japonica ζ-crystallin structures were −8.71, −8.44, and −9.09, respectively. The Z-score in all three modeled structures displayed in the plot was in the range of all experimentally determined protein chains in the current PDB. If the ProSA Z-score is located outside a characteristic range of native conformation of proteins, then the model or experimental structures are erroneous (Sippl 1995).
3.5 Molecular docking
The docking results showed that NADPH has good docking scores with ζ-crystallin of the three species. The ligand, NADPH, showed strong binding affinities of −9.3, −10.8, and −9.8 kcal/mol towards C. dromedarius, C. porcellus, and H. japonica ζ-crystallin, respectively (Table 2). As shown in Fig. 3A, the C. dromedarius ζ-crystallin-NADPH complex interacted with the aid of seven hydrogen bonds within a distance range of 1.98–2.98 Å and six hydrophobic interactions within a distance range of 3.04–5.10 Å. The interaction between C. porcellus ζ-crystallin and NADPH) was relatively stronger than between the NADPH complexes with C. dromedarius and H. japonica ζ-crystallin. It occurred due to NADPH forming nine hydrogen bonds with distances between 1.93 and 3.39 Å and thirteen hydrophobic bonds at distances between 2.99 and 5.37 Å with the side chains of C. porcellus ζ-crystallin(Fig. 3B). The interaction between NADPH and H. japonica ζ-crystallin was relatively stronger than C. dromedarius ζ-crystallin as it formed eleven hydrogen bonds (with distances between 2.69 and 3.38 Å) and ten hydrophobic interactions (with distances between 3.33 and 5.34 Å) (Fig. 3C).
S.No.
Ligand
Receptor
Binding energy (kcal/mol)
Hydrogen interaction (Bond distance-Å)
Hydrophobic interaction (Bond distance-Å)
1
NADPH
ZINC85551904
C. dromedarius ζ-crystallin model
−9.3
Tyr53 (2.60), Ser158 (2.34), Gly160 (2.94, 2.98) Val161 (2.83), Ser318 (1.98), Gly323 (2.01)
Met224 (5.07), Leu225 (5.10), Lys249 (3.61) Leu315 (3.04), Ala321 (4.23), Lys324 (3.61)
2
NADPH
ZINC85551904
C. porcellus
ζ-crystallin model−10.8
Ser158 (3.11), Gly160 (3.16), Val161 (3.39), Gly162 (2.94), Gly181 (3.17), Tyr205 (2.81), Asn229 (2.35), Val269 (1.93), Thr321 (3.34)
Val50 (4.64), Ile131 (5.37), His155 (3.80), Gly156 (3.40), Gly159 (3.49), His200 (3.72, 3.76), Arg201 (2.99, 4.12), Leu225 (3.69, 3.96) Arg249 (4.36), Gly320 (3.20)
3
NADPH
ZINC85551904
H. japonica
ζ-crystallin model−9.8
Asn48 (2.69), Tyr53 (3.09), Ser158 (2.70), Val161 (3.32), Gly181 (3.38), His200 (3.25), Arg201 (3.20), Gln249 (2.89, 3.02), Ala321 (3.15), Lys324 (3.16)
Pro49 (4.18, 4.54), Gly159 (3.33), Gly160 (3.69), Val161 (5.09), His200 (3.99), Arg201 (5.11), Leu225 (4.68), Cys228 (4.91), Leu315 (5.34)
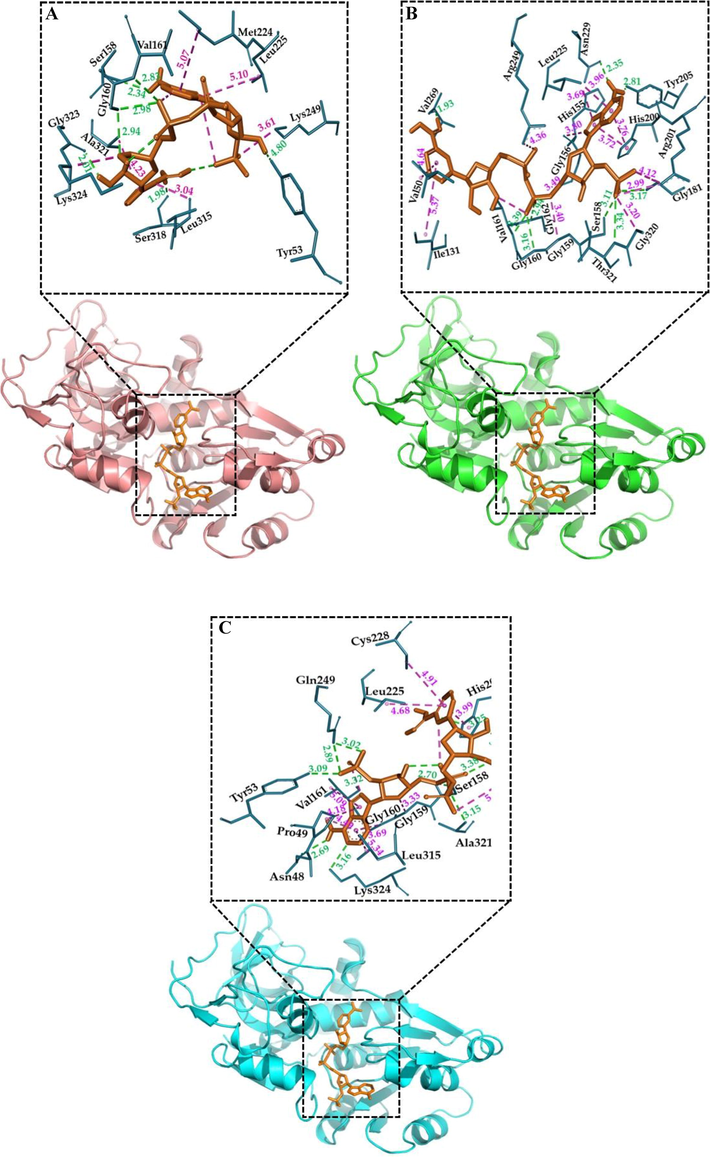
Interaction between ζ-crystallin and NADPH through molecular docking. (A) animated structure of C. dromedarius ζ-crystallin binding with NADPH in the central calyx as the best position of binding, (B) animated structure of C. porcellus ζ-crystallin binding with NADPH in the central calyx, (C) animated structure of H. japonica ζ-crystallin binding with NADPH in the central calyx. The H-bonding is shown in a dashed green line and hydrophobic interactions in a dashed pink line.
3.6 Molecular dynamic study of ζ-crystallin and ligand
MD simulations have been used extensively to understand the conformational changes, stability, and dynamics of the binding mechanisms of ligands to a protein under explicit solvent conditions (Al-Shabib et al., 2019a,b, Khan et al., 2019). Here, the stability of the complexes (C. dromedarius, C. porcellus, and H. japonica ζ-crystallin-NADPH docked complexes) were evaluated by performing molecular dynamic simulations for 50 ns at 300 K. Binding of a small molecule in the binding pocket of a protein can lead to large conformational changes (Ali et al., 2014, Al-Shabib et al., 2020). Fig. 4 shows the root mean square deviations (RMSDs) of C. dromedarius, C. porcellus, and H. japonica ζ-crystallin with and without the ligand. The RMSD analysis provides valuable information on the conformational changes in structure during the simulation. We have determined the RMSD across Cα-atoms of all three ζ-crystallins in the absence and presence of NADPH. As shown in Fig. 4A, it is apparent that the RMSD of C. dromedarius ζ-crystallin without the ligand increased initially and stabilized within 2 ns of simulation. Conversely, the RMSD of the C. dromedarius ζ-crystallin-NADPH complex stabilized within 15 ns and remained constant during the 15–50 ns of simulation time, indicating a stable complex formation. The average RMSD values of C. dromedarius ζ-crystallin in the absence and presence of NADPH were estimated to be 0.34 and 0.49 nm, respectively, within the acceptable limit of 0.2 nm. The average RMSD values for C. porcellus ζ-crystallin in the absence and presence of NADPH were found 0.52 nm and 0.39 nm, respectively (Fig. 4B). Similarly, the average RMSD values for H. japonica ζ-crystallin alone or complexed with NADPH were found to be 0.44 nm and 0.42 nm, respectively (Fig. 4C). Here, in all three simulations, the RMSD values fluctuated by<0.2 nm, confirming the stability of the ζ-crystallin-NADPH complex during the 50 ns of simulation. After adding NADPH, the RMSD values of C. dromedarius, C. porcellus, and H. japonica ζ-crystallin complexes were stabilized in approximately 15, 2, and 5 ns, respectively. Stabilization of the ζ-crystallin-NADPH complexes depended on the complexes' binding energies, and results indicated that C. dromedarius ζ-crystallin-NADPH > C. porcellus ζ-crystallin-NADPH > H. japonica ζ-crystallin -NADPH (Table 2). Overall, the RMSD plots of C. dromedarius, C. porcellus, and H. japonica ζ-crystallins showed equilibration throughout the 50 ns MD simulation, suggesting stable NADPH complexes' formation (Fig. 4).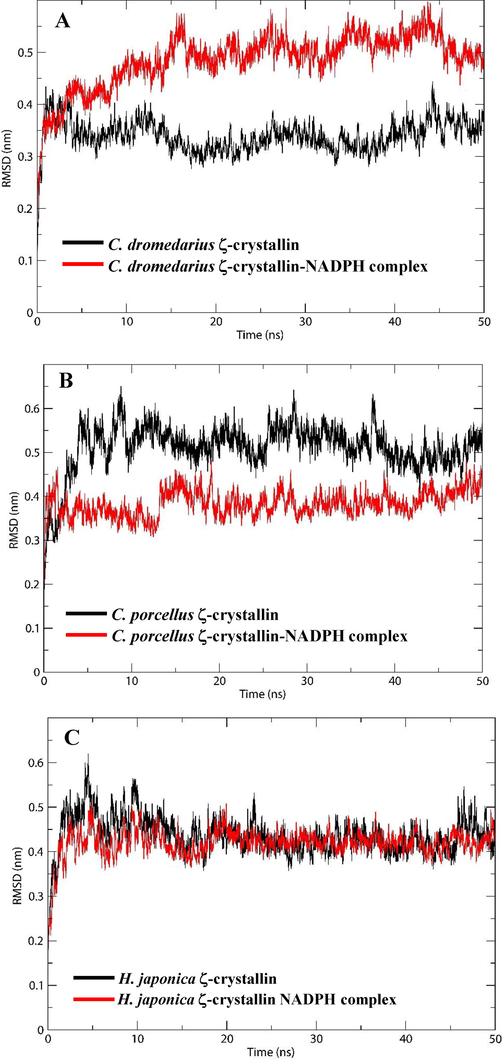
Characterization of ζ-crystallin-NADPH interaction parameters as a function of MD simulation time. (A) RMSD of C. dromedarius ζ-crystallin alone and complexed with NADPH, (B) RMSD of C. porcellus ζ-crystallin alone and complexed with NADPH, (C) RMSD of H. japonica ζ-crystallin alone and complexed with NADPH.
Root mean square fluctuation (RMSF) measures the flexibility in the protein 3D structure due to side-chain movements. This study estimated the flexibility and rigidity of the ζ-crystalline side chains in the absence and presence of NADPH by measuring the RMSF during a 50 ns simulation. In Fig. S5A, it is observed that the average RMSF values of C. dromedarius ζ-crystallins alone and complexed with NADPH were 0.16 and 0.19 nm, respectively. The average RMSF of C. porcellus ζ-crystallins in the absence or presence of NADPH was 0.18 and 0.19 nm, respectively (Fig. S5B). Similarly, the average RMSF of H. japonica ζ-crystallins with and without NADPH was also 0.18 and 0.19 nm, respectively (Fig. S5C). The fluctuating regions on the ζ-crystallins of C. dromedarius, C. porcellus, and H. japonica spanned across the protein molecule regardless of the presence of NADPH. These residual fluctuations were found to be minimized upon binding of the NADPH throughout the simulation. However, several random residual fluctuations were also observed when ζ-crystallin bound to NADPH (Fig. S5).
The radius of gyration (Rg) measures the overall compactness of a protein–ligand complex during MD simulation. Moreover, Rg analyses also provide information on the protein folding pattern, stability, and conformational changes in the absence and presence of ligands. Here, we have assessed the Rg of C. dromedarius, C. porcellus, and H. japonica ζ-crystallins alone and as complexes with NADPH as a function of simulation time (Fig. S6). The average Rg values of C. dromedarius ζ-crystallins alone or complexed with NADPH were determined to be 3.01 and 2.78 nm, respectively. A 0.23 nm decrease in C. dromedarius ζ-crystallin-NADPH complex indicates that the binding of NADPH increased the compactness of C. dromedarius ζ-crystallins (supplementary Fig. S6A). NADPH binding with C. porcellus and H. japonica ζ-crystallins lead to a slight increase in the Rg of both ζ-crystallins indicating that the binding of NADPH resulted in a partial loss of compactness in both ζ-crystallins (Table 3 and Fig. S6B & C).
Complex
Average RMSD
(nm)
Average RMSF
(nm)
Average Rg
(nm)
Average SASA (nm2)
Volume (nm3)
Density (g 1-1)
C.dromedarius ζ-crystallin
0.34
0.16
3.01
281.98
1941.93
1013.63
C.dromedarius ζ-crystallin + NADPH
0.49
0.19
2.78
259.14
1374.69
1001.96
C. porcellus ζ-crystallin
0.52
0.18
2.86
250.73
2108.33
1013.38
C. porcellus ζ-crystallin + NADPH
0.39
0.19
2.88
266.15
1540.19
999.30
H. japonica ζ-crystallin
0.44
0.18
2.89
255.80
2069.98
1013.95
H. japonica ζ-crystallin + NADPH
0.42
0.19
2.96
259.04
2069.49
1014.24
Measurement of the solvent-accessible surface area (SASA) of a protein shows the changes in solvent exposure of amino acid residues after ligand binding (Khan et al., 2019, Al-Shabib et al., 2020). This study assessed how the SASA was affected by the ligand binding of ζ-crystallins obtained from the three species under study at different simulation times (Fig. S7). We observed that the mean SASA of C. dromedarius ζ-crystallins alone and in its complex form was 281.98 and 259.14 nm2, respectively (Fig. S7A). A decrease in the SASA of the C. dromedarius ζ-crystallin-NADPH complex indicated that the solvent surface exposure area decreased with NADPH binding. The mean SASA values for C. porcellus alone and when complexed with NADPH were found to be 250.73 and 266.15 nm2, respectively (Table 3). An increment in C. porcellus (-crystallins) SASA was observed with NADPH binding (Fig. S7B). This increment in the SASA can be presumed to result from the exposure of some of the internal residues in C. porcellus ζ-crystallins to solvents due to conformational changes arising from complexing with NADPH. The solvent surface exposure area of H. japonica ζ-crystallins increased when bound to NADPH, indicating that minor conformational changes occurred after NADPH binding (Table 3 and Fig. S7C). After the simulation, the volume and density of the (-crystallins alone and when complexed with NADPH were also calculated (Table 3).
4 Conclusions
ζ-Crystallin (quinone oxidoreductase) is a metabolic enzyme that is relatively abundant in the eye lenses of C. dromedarius, C. porcellus, and H. japonica. Our study showed that the overall folding topology of C. dromedarius, C. porcellus, and H. japonica ζ-crystallins were remarkably similar to that of H. sapiens ζ-crystallin. The pI of the ζ-crystallins obtained from C. dromedarius, C. porcellus, and H. japonica was close to the physiological pH. It is interesting to note that, there was a high concentration of ζ-crystallins in a soluble form in the lenses of these species. The hydrophobicity of C. porcellus ζ-crystallin was relatively higher compared to that of C. dromedaries and H. japonica, and it was correlated with its protein expression pattern in E. coli. It is well-known that proteins with a lower hydrophobic index are easy to express and remain soluble in E.coli. It raises the question of why nature has recruited an aggregation-prone protein in the lens of C. porcellus that has to remain soluble permanently without a turnover. Our in silico data indicated that differences in physicochemical properties of ζ-crystallin may lead to differences in protein folding and stability in the lenses of C. dromedarius, C. porcellus, and H. japonica ζ-crystallin may facilitate their adaptation to different environmental conditions.
Acknowledgements
The authors are grateful to the Researchers Supporting Project Number (RSP-2021/360), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
To the best of our knowledge, no conflict of interest, financial or others, exists. All authors are fully aware of this submission.
References
- A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci.. 2014;15(5):456-476.
- [Google Scholar]
- A quercetin-based flavanoid (rutin) reverses amyloid fibrillation in beta-lactoglobulin at pH2.0 and 358K. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2019;214:40-48.
- [Google Scholar]
- Allura red rapidly induces amyloid-like fibril formation in hen egg white lysozyme at physiological pH. Int. J. Biol. Macromol.. 2019;127:297-305.
- [Google Scholar]
- Molecular interaction of tea catechin with bovine beta-lactoglobulin: a spectroscopic and in silico studies. Saudi Pharm. J.. 2020;28(3):238-245.
- [Google Scholar]
- QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37(Web Server issue):W510-W514.
- [Google Scholar]
- SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res.. 2014;42(Web Server issue):W252-W258.
- [Google Scholar]
- Discovery studio modeling environment. San Diego: Dassault Systèmes; 2015.
- Purification and characterization of zeta-crystallin from the camel lens. Biochem. Biophys. Res. Commun.. 1995;215(2):632-640.
- [Google Scholar]
- Taxon-specific zeta -crystallin in Japanese tree frog (Hyla japonica) lens. J. Biol. Chem.. 2001;276(30):28134-28139.
- [Google Scholar]
- zeta-Crystallin is a major protein in the lens of Camelus dromedarius. Arch. Biochem. Biophys.. 1991;285(1):134-136.
- [Google Scholar]
- Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem.. 1989;182(2):319-326.
- [Google Scholar]
- Expression of recombinant zeta-crystallin in Escherichia coli with the help of GroEL/ES and its purification. Protein Expr. Purif.. 2001;21(2):260-267.
- [Google Scholar]
- Connexins in lens development and cataractogenesis. J. Membr. Biol.. 2007;218(1–3):9-12.
- [Google Scholar]
- Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng.. 1990;4(2):155-161.
- [Google Scholar]
- Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A.. 1992;89(22):10915-10919.
- [Google Scholar]
- Thermostability and aliphatic index of globular proteins. J. Biochem.. 1980;88(6):1895-1898.
- [Google Scholar]
- Microarray analysis of fiber cell maturation in the lens. FEBS Lett.. 2005;579(5):1213-1219.
- [Google Scholar]
- Stability and folding of ultrastable proteins: eye lens crystallins and enzymes from thermophiles. FASEB J.. 1996;10(1):84-92.
- [Google Scholar]
- MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol.. 2013;30(4):772-780.
- [Google Scholar]
- An intermittent amyloid phase found in gemini (G5 and G6) surfactant induced beta-sheet to alpha-helix transition in concanavalin A protein. J. Mol. Liq.. 2018;269:796-804.
- [Google Scholar]
- Effect of cetyltrimethylammonium bromide (CTAB) on the conformation of a hen egg white lysozyme: a spectroscopic and molecular docking study. Spectrochim. Acta A Mol. Biomol. Spectrosc.. 2019;219:313-318.
- [Google Scholar]
- A simple method for displaying the hydropathic character of a protein. J. Mol. Biol.. 1982;157(1):105-132.
- [Google Scholar]
- Expression, purification and properties of redox-sensitive eye lens zeta-crystallin of Arabian camel. Protein Pept. Lett.. 2016;23(6):573-580.
- [Google Scholar]
- Spectral and thermal properties of novel eye lens zeta-crystallin. Int. J. Biol. Macromol.. 2017;102:1052-1058.
- [Google Scholar]
- Stereochemical quality of protein structure coordinates. Proteins. 1992;12(4):345-364.
- [Google Scholar]
- Evaluation of pyrazolopyrimidine derivatives as microtubule affinity regulating kinase 4 inhibitors: Towards therapeutic management of Alzheimer's disease. J. Biomol. Struct. Dyn.. 2020;38(13):3892-3907.
- [Google Scholar]
- PyMOL “ v. S. N. Y., NY, USA, 2006.”.
- Identification and characterization of the enzymatic activity of zeta-crystallin from guinea pig lens. A novel NADPH:quinone oxidoreductase. J. Biol. Chem.. 1992;267(1):96-102.
- [Google Scholar]
- Guinea pig and bovine zeta-crystallins have distinct functional characteristics highlighting replacements in otherwise similar structures. Biochemistry. 1997;36(18):5353-5362.
- [Google Scholar]
- Lens aging: effects of crystallins. Biochim. Biophys. Acta, Proteins Proteomics. 2009;1790(10):1095-1108.
- [Google Scholar]
- Expression and purification of his-tagged recombinant mouse zeta-crystallin. Protein Expr. Purif.. 2010;69(2):147-152.
- [Google Scholar]
- Knowledge-based potentials for proteins. Curr. Opin. Struct. Biol.. 1995;5(2):229-235.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189-1191.
- [Google Scholar]
- ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res.. 2007;35(Web Server issue):W407-W410.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.101973.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







