Translate this page into:
Review: Merging from traditional to potential novel breast cancer biomarkers
⁎Corresponding author. Aalismailh@gmail.com (Hanan Alismail) alsmailh@ksau-hs.edu.sa (Hanan Alismail)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Breast cancer biomarkers are the main player in decision-making in diagnosis, prognosis, and treatment. Estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) are well-known in breast cancer management. Additionally, the Ki-67 protein is used as a tumor proliferation indicator to asses the cancer aggressiveness. Recently, the field has been rapidly integrating novel biomarkers to develop precise, personalized with high effectiveness in patient care. A group of merging biomarkers, including genomic and transcriptomic signatures, circulating tumor cells (CTCs), cell-free DNA (cfDNA), tumor-infiltrating lymphocytes (TILs), and immune checkpoint proteins such as PD-L1, all showed promising toward revealing tumor behavior, treatment response, and potential metastatic spread. microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are merging as new potential diagnostic tools. All mentioned merging innovative biomarkers showed promising results, yet challenges remain in their validation, standardization, and integration into routine clinical practice. This review will highlight the transition from traditional to novel strategies, developing more effective treatments that improve breast cancer patients’ outcomes and survival.
Keywords
Breast Cancer
Biomarkers
ER
PR
HER2
Ki-67
CTCs
cfDNA
TILs
PD-L1
miRNAs
lncRNAs
Genomics
Prognosis
Therapy
1 Introduction
Breast cancer is recognized to be the most prevalent type of cancer among women; millions have been diagnosed worldwide. Approximately 7.8 million women have been diagnosed with breast cancer in the past five years, according to the World Health Organization (WHO), 2020 (WHO, 2020). 11.7 % of all new cancer cases were reported only in 2020, accounting for 2.3 million women around the world (WHO, 2020). The mortality rate related to breast cancer was reported to be 685,000 deaths in 2020 (GCO, 2020).
Particularly in Saudi Arabia, breast cancer is the most prevalent type of cancer among females, yet the incidence in Saudi Arabia is lower compared to Western countries. Still, there is a big concern due to the steadily rising incidence rate. Breast cancer accounts for 30 % of all of all cancers among women in Saudi Arabia (SHC, 2020). Historically, from 2001 to 2008, 6,922 female breast cancer cases were recorded. The highest percentages occurred in women aged 30–44 and 45–59 years (Alghamdi et al., 2013). Similar to the trend worldwide, in 2020, there were approximately 4,000 new cases of breast cancer diagnosed in Saudi Arabia, accounting for 26 % of newly diagnosed female cancers (Al-daihan & Shafi, 2012). Incidence rate elevation has been correlated with Western lifestyle adoption, decreased physical activity, and changes in dietary patterns (Al-daihan & Shafi, 2012). A high red flag is raised due to the estimation that by 2025, breast cancer incidence in Saudi Arabia may increase up to 350 % (Al-daihan & Shafi, 2012).
Breast cancer is identified as a heterogeneous disorder, which has clinical, physiological, and molecular features. Biomarkers' status is utilized widely in the diagnosis and treatment processes for patients with breast cancer (Duffy et al., 2017). Biomarkers are useful for both patients who develop breast cancer recently and those with recurrence (Duffy et al., 2017). Estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and the cellular proliferation index (kI67) are readily available biomarkers that clinicians depend on. ER, PR, HER2, and kI67 are also essential in determining molecular classification, which is the golden standard for the characterization of breast cancer (Duffy et al., 2017). Routinely, ER, PR and HER2 are examined in breast cancer samples because they are effective and low-priced. In addition, the treatment choices are guided by ER, PR, and HER2 (Howlader et al., 2018).
The classification of breast cancer patients depends on the difference in the survival rate and the treatment. Thus, they are divided into four groups: Luminal A, Luminal B, HER2-amplified, and triple-negative (Meng et al., 2016). Luminal A has the higher survival rate, followed by Luminal B, HER2-amplified, whereas triple-negative appears to have the lowest survival rate among the four groups (Mirabelli & Incoronato, 2013). Along with the different survival rates, there is variation in the treatment for each group. For example, using hormonal therapy is more beneficial for patients with positive hormone receptors, while monoclonal antibodies (trastuzumab) are the main treatment for patients with HER2 overexpression because of their ability to block HER2. Given that the triple-negative group has the lowest survival rate, it becomes the most challenging, and chemotherapies mainly treat it. However, after finishing the treatment, some patients may get completely cured, while others may experience tumor reoccurrence along with marker conversions in terms of their presence or absence (Aurilio et al., 2014).
Guiding clinical decisions for diagnosis, prognosis, and treatment using breast cancer biomarkers is essential. Traditional markers, which are widely recognized in breast cancer management, include estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2); all provide insights into patient care (Duffy et al., 2015; Nicolini et al., 2017). Ki-67, in addition, is a protein reflecting tumor cell proliferation activity and is commonly used to indicate tumor aggressiveness. Recently, in cancer research, there has been an accelerated development toward integrating novel biomarkers supporting more precise, personalized, and effective care for breast cancer patients.
A newly valuable novel generation of biomarkers is emerging into tumor biology’ providing a new pathway toward treatment response into tumor biology. Novel markers have shown promise in predicting tumor biology, treatment response, and potential metastatic spread. These include genomic and transcriptomic signatures, circulating tumor cells (CTCs), cell-free DNA (cfDNA), tumor-infiltrating lymphocytes (TILs), and immune checkpoint proteins such as PD-L1, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) are being recognized as diagnostic tools with potential for cancer detection, monitoring, and therapy prediction. Although innovative biomarkers hold promise toward developing personalized treatment, challenges are still mainly in validating, standardizing, and incorporating them into routine clinical practice.
The diagnosis, prognosis, and therapy of breast cancer are significantly influenced by biomarkers in general. Instead of depending on conventional biomarkers, the discipline is now investigating new ones that provide more accurate and individualized patient care. Fig. 1. Although there have been improvements, no single biomarker has shown enough sensitivity and repeatability for standalone clinical use (Tang & Gui, 2012). Subsequent investigations seek to discover biomarkers for enhancing early identification and recurrence monitoring, as well as for forecasting response to radiation and certain chemotherapies (Nicolini et al., 2017; Tang & Gui, 2012). This review will explore the shift from conventional biomarkers to these new approaches, with the ultimate goal of enhancing treatment results and personalizing for patients with breast cancer.
Illustration of the evolution of medicine towards personalized medicine in the breast cancer research field.
2 Traditional biomarkers in breast cancer
2.1 Estrogen receptor (ER) and progesterone receptor (PR)
Both Estrogen receptor (ER) and progesterone receptor (PR) are essential to predict breast cancer's pathogenesis and treatment response (Cordera & Jordan, 2006). Both are hormone receptors in response to estrogen and progesterone, ER and PR contribute to cancer growth in hormone-sensitive breast tissues by facilitating cancer cell proliferation.
Consequently, these receptors are important targets for hormone therapy since their overexpression is a characteristic of many breast malignancies in certain cancer situations. Aromatase inhibitors and tamoxifen are intended to impede hormone-signaling pathways that are essential for tumor development and survival (Yip & Rhodes, 2014). In addition to being crucial for therapeutic targeting, ER and PR status are also critical for prognostic and predictive cancer care. For clinical significance, at least 1 % of tumor cells must have receptor labeling to be considered ER and PR positive. Research shows that compared to ER-negative cancers, ER-positive tumors often have a higher likelihood of receiving treatment and better survival results. Accordingly, ER status has a significant role in prognosis (Badowska-Kozakiewicz et al., 2015).
In breast cancer assessment, the expression of ER and PR varies significantly in histological subtypes and grades. For instance, higher ER positivity expression rates are reflected in invasive ductal carcinoma (IDC), the most common form of breast cancer, compared to invasive lobular carcinoma (ILC), indicating the receptor expression rate differences across different breast cancer types (Al-timimi & Yousif, 2014; Badowska-Kozakiewicz et al., 2015).
Furthermore, hormonal therapies and guided treatment plans mainly depend on the assessment of hormone receptor expression. Therefore, such assessment of ER and PR status is highly essential in clinical practice for breast cancer patients to optimize treatment selection and improve patient survival rate outcomes (Al-timimi & Yousif, 2014; Yip & Rhodes, 2014). Eventually, personalized treatment is mainly led by hormonal receptor evaluation, allowing clinicians to develop therapeutic strategies with improved efficacy while avoiding unnecessary treatments for patients with hormone-receptor-positive breast cancer.
2.2 Human epidermal growth factor receptor 2 (HER2)
Human Epidermal Growth Factor Receptor 2 (HER2) is overexpressed in approximately 15–30 % of breast cancer cases. Thus, it is considered a crucial prognostic and predictive biomarker for breast cancer (Iqbal & Iqbal, 2014; Shah & Chen, 2010). Unfortunately, overexpression of HER2 is associated with a more aggressive tumor phenotype characterized by susceptible metastasis, poor prognosis, and high recurrence rates (Lv et al., 2016). This concludes that HER2-positive breast cancer is often associated with a more advanced stage at diagnosis, which contributes to these pernicious outcomes.
High amplification of the HER2 gene results in the over-expression of the HER2 protein. Thus, triggers several downstream signaling pathways that promote cell survival, and proliferation, and ultimately will lead to cancer development (Iqbal & Iqbal, 2014). The aggressive behavior of HER2-positive cancers is determined by this dysregulation, highlighting the need to accurately determine HER2 status when selecting a treatment plan.
Assessment of HER2 status is routinely performed using techniques such as immunohistochemistry (IHC) and fluorescent in situ hybridization (FISH). FISH detects HER2 gene amplification, while IHC evaluates the level of HER2 protein expression, both techniques determine the eligibility for HER2-targeted therapies (Shah & Chen, 2010; Lv et al., 2016). Accurate determination of HER2 status influences the choice of therapy and prognosis and is vital for best patient management.
Trastuzumab, a well-known humanized monoclonal antibody that specifically targets HER2, improved HER2-positive breast cancer patients’ clinical outcomes. It works by binding and blocking the HER2 receptor, stopping the receptor's signaling, and leading cancer cells to be destroyed by the immune system (Damodaran & Olson, 2012). Trastuzumab treatment dramatically decreased recurrence rates and increased overall survival among HER2-positive patients, according to clinical trials.
Despite trastuzumab's effectiveness, resistance to the treatment may arise, which is a serious problem when treating HER2-positive breast cancer (Damodaran & Olson, 2012). This resistance may be caused by alterations in the HER2 pathway, activation of alternative growth factor pathways, or changes in the tumor microenvironment. Since the mechanisms are unknown, alternative HER2-targeted medications are badly needed to treat this patient population.
In addition to trastuzumab, other monoclonal antibody therapeutic approaches for HER2-positive breast cancer now include pertuzumab, which targets a distinct HER2 receptor epitope. Tyrosine kinase inhibitors (TKIs), such as lapatinib, work in tandem with trastuzumab to block downstream signaling pathways. By improving treatment effectiveness and delivering cytotoxic drugs directly to HER2-positive cells, antibody-drug conjugates (ADCs) such as trastuzumab emtansine reduce systemic toxicity (Lv et al., 2016). To overcome resistance mechanisms and provide synergistic benefits, a combination of treatments utilizing several modalities is being investigated. This will ultimately improve the results for patients with HER2-positive breast cancer.
2.3 Ki-67
Ki-67 is a vital proliferation marker that is frequently used in the diagnosis and treatment of breast cancer. It offers information on the growth dynamics of tumors (Mannell, 2016). When Ki-67 is present, the malignancy is actively growing during the G1, S, G2, and mitotic stages of the cell cycle. As a stand-alone predictor of outcome, Ki-67 expression is linked to worse clinical outcomes and correlates with both overall survival (OS) and disease-free survival (DFS), with higher expression leading to a lower survival rate and a higher rate of recurrence (Inwald et al., 2013; Azambuja et al., 2007).
Significant differences exist in Ki-67 expression amongst the various molecular subtypes of breast cancer. Luminal A cancers, for example, usually show lower levels of Ki-67, indicating slower rates of proliferation, and have a generally better prognosis. Conversely, triple-negative breast tumors (TNBC), which are notoriously aggressive and result in a dearth of targeted treatments, frequently have higher Ki-67 levels, develop more quickly, and have a worse prognosis (Soliman & Yussif, 2016). As a result, Ki-67 expression helps guide treatment choices and is used to stratify patients according to risk. In both node-negative and node-positive breast cancer patients, a higher level of Ki-67 is directly associated with a higher risk of recurrence and poorer survival outcomes, according to a meta-analysis of 46 studies with a prognostic value of Ki-67 that included 12,155 patients (Azambuja et al., 2007). This emphasizes how crucial Ki-67 is as a standard biomarker that can direct medical judgment.
Despite its importance, Ki-67 is not without difficulties and is measured in a wide range of ways. Diverse sample techniques, scoring schemes, and result interpretation provide difficulties, making its clinical use more difficult (Inwald et al., 2013). The absence of established procedures may lead to inconsistent Ki-67 evaluation results, which would compromise its validity as a biomarker. Furthermore, while there is still discussion on how best to integrate this biomarker into all-encompassing therapy strategies, the role of Ki-67 is being investigated and considered.
Ultimately, Ki-67 remains a potentially important biomarker for the therapy of breast cancer, helping to tailor treatment plans and provide prognostic data (Soliman & Yussif, 2016). As a result, incorporating it into standard clinical practice improves the capacity to identify treatment methods for enhancing patient outcomes in cases of breast cancer and to stratify patients based on risk.
3 Emerging novel biomarkers
3.1 Genomic and transcriptomic signatures
Understanding breast cancer heterogeneity significantly transformed through recent advances in genomic and transcriptomic analyses, resulting in important clinical implications for diagnosis and treatment. Identifying novel molecular subgroups of breast cancer through the integration of both copy number alterations and gene expression profiles from large patient cohorts has enabled researchers to determine genes associated with distinct clinical outcomes. (Curtis et al., 2012). A more nuanced understanding of such integrated approaches allows us to understand how different breast cancer types behave and respond to therapies.
Notably, 117 genes were commonly identified to be altered in breast cancer in a meta-analysis of multiple gene expression studies. The majority of these genes play a crucial role in the development and spread of tumors by regulating the cell cycle and hormone signaling (Abba et al., 2010). All of the aforementioned may be utilized to create prediction models that help with patient survival outcomes, stratifying patients according to risk, and customizing treatment plans.
Significant patterns of mutations resulting from a variety of biological and environmental causes are also displayed by particular mutational signatures. These mutations are associated with a better prognosis and have been connected to immune cell infiltration in luminal breast tumors. According to these results, certain mutations may strengthen the immune system's defenses against the tumor and impact the effectiveness of treatment (Smid et al., 2016). Such correlation reveals the importance of understanding the tumor's genetic makeup and how it interacts with the immune microenvironment.
Accurate breast cancer classification is improved by integrating all genomic, transcriptomic, and proteomic data to create multi-omics signatures. A more thorough profile allows for a more comprehensive understanding of tumor biology, which improves therapeutic response prediction. (Ma et al., 2024). Integrating many omics layers, for example, can ultimately uncover subtle molecular connections underlying the tumor nature to enhance breast cancer diagnosis and prognosis and enable tailored treatment. Fig. 2.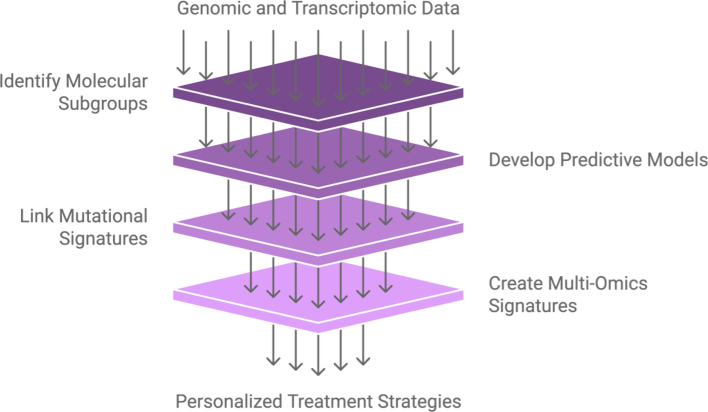
From Genomic data to personalized treatment.
As research continues to evolve in this area, we will likely see an individual’s unique tumor profile with more tailored treatment options matching an individual’s profile. In the long run, this will result in improved patient survival rates and more efficient treatment of breast cancer. By incorporating these cutting-edge analytical methods into clinical practice, oncology will evolve toward precision medicine, where therapies are tailored to the particular genetic and molecular features of each cancer type.
3.2 Circulating tumor cells (CTCs) and cell-free DNA (cfDNA)
Circulating tumor cells (CTCs) and cell-free DNA (cfDNA) have gained novelty as potential biomarkers obtained by liquid biopsies used for breast cancer prognosis and monitoring. Both offer a noninvasive diagnostic approach to monitor tumor dynamics, making the monitoring process much easier, and avoiding the need for invasive tissue biopsies, which can be challenging due to the tumor’s location or the patient’s condition.
Additionally, the ability to repeat samples to identify potential tumor heterogeneity is a benefit of noninvasive liquid biopsies. The genetic composition of tumor cells varies greatly, and this variation may alter as the prognosis worsens or in response to therapy. Clinicians can make better-informed treatment decisions by using liquid biopsies, which provide them with a more thorough picture of the tumor's genetic landscape throughout time (Appierto et al., 2017). For example, before clinical signs appear, physicians may choose to change the treatment plan if the findings of a liquid biopsy show an increase in certain mutations linked to treatment resistance. By enabling prompt treatments based on the changing features of the tumor, the incorporation of blood-based molecular methods may result in more individualized treatment strategies and enhance patient outcomes (Appierto et al., 2017).
Both CTC counts and cfDNA levels have been directly linked to overall survival in several studies, especially in patients with metastatic breast cancer. Both markers are useful indications for determining the course of a disease and the prognosis of a patient since their elevation is associated with a worse prognosis (Rossi et al., 2017; Shaw et al., 2016). The development of prognostic tools utilizing liquid biopsies for early disease recurrence identification or progression monitoring appears to be a viable application of this prediction capacity. With next-generation sequencing technologies (NGS), CTC and matched cfDNA have demonstrated significant mutational heterogeneity across patients with breast cancer. It has been established that some gene mutations, such as those in PIK3CA, TP53, ESR1, and KRAS, are strongly linked to the biological behavior of tumors, especially in metastatic situations where clonal expansion may results in treatment resistance (Shaw et al., 2016). This mutational landscape is important because it can help guide treatment decisions by revealing possible resistance pathways. Remarkably, cfDNA profiles have demonstrated a significant degree of correspondence with the CTC mutation. indicating that cfDNA analysis may be used as an additional method, particularly when CTCs are hard to separate or measure (Shaw et al., 2016). This makes it possible to comprehend tumor dynamics throughout time on a larger scale.
To conclude, a major development in the treatment of breast cancer is the utilization of CTCs and cfDNA in liquid biopsies. These biomarkers have the potential to transform prognosis, monitoring, and treatment approaches by offering real-time insights into tumor biology and dynamics, eventually leading to better patient care. As the field's research progresses, the clinical use of liquid biopsies might eventually become the norm for treating breast cancer.
3.3 Tumor-infiltrating lymphocytes (TILs)
Tumor-infiltrating lymphocytes (TILs) are one of the promising biomarkers, especially with triple-negative and HER2-positive subtypes. The frequency of lymphocyte-predominant breast cancer varies among different subtypes. On average, TILs are present in about 11 % of breast cancers, with this figure increasing to 16 % in HER2-positive cancers and dropping to 6 % in hormone receptor-positive/HER2-negative tumors (Stanton et al., 2016). The significance of TILs as possible markers of the immune landscape of the tumor microenvironment is highlighted by this variability.TILs impact a patient's overall prognosis and are linked to the immune system's reaction to tumor growth. Research has shown that increased TIL levels are associated with improved results and responses to neoadjuvant chemotherapy, highlighting a key area for assessing and promoting treatment effectiveness (Ahn et al., 2015; Stanton et al., 2016).
More specifically, the makeup of TILs can reveal information about the immunological properties of the tumor. About 48 % of cases of breast cancer have CD8 + T-cells, which are important for anti-tumor immunity. In contrast, aggressive subtypes such as triple-negative and HER2-positive breast cancer tend to have more FOXP3 + regulatory T-cells, which are known to suppress immune responses (Stanton et al., 2016).
Despite TILs' potential impact on breast cancer prognosis, there aren't many established techniques for evaluating them. The interpretation of TIL data from various research is made more difficult by the variation in measuring methods (Ahn et al., 2015). Thus, more research is necessary to improve TIL evaluation techniques, pinpoint immune cell subsets linked to improved clinical outcomes, and create treatment plans that boost immune infiltration, especially in patients who are TIL-negative (Dushyanthen et al., 2015). These developments may open the door to tailored immunotherapy strategies for the treatment of breast cancer.
3.4 PD-L1 expression
Expression of PD-L1 (programmed death-ligand 1) has become a key determinant of breast cancer prognosis and therapy, especially for aggressive subtypes like triple-negative breast cancer (TNBC). A complicated interaction between the immune response and the tumor microenvironment is revealed by the correlation between PD-L1 expression and tumor features. Researchers discovered that PD-L1 expression is present in 20–30 % of TNBC patients and that this expression is associated with more aggressive tumor characteristics. High tumor grade and hormone receptor negativity are frequently observed in association with this expression. These traits imply that PD-L1-expressing tumors could behave more aggressively biologically, which might lead to a worse prognosis (Mittendorf et al., 2014; Wimberly et al., 2014).
Additionally, peripheral lymphoid aggregates and tumor-infiltrating lymphocytes (TILs) are favorably correlated with PD-L1 expression. TILs are a sign that the tumor microenvironment is experiencing an aggressive immune response. Since PD-L1 is an essential component of the immune checkpoint pathway, their existence in conjunction with it raises the possibility that the tumor is trying to elude immune surveillance (Cimino-Mathews et al., 2016).PTEN depletion and signaling via the PI3K pathway are two molecular mechanisms that may be involved in the regulation of PD-L1 expression. These pathways are linked to the growth of tumors and may help PD-L1 be upregulated, which would make the tumor more resistant to the immune system (Mittendorf et al., 2014).
Within the clinical community, there is ongoing discussion over the predictive significance of PD-L1 expression. There is no substantial correlation between greater levels of PD-L1 and poorer outcomes, according to some research. However, early clinical studies using PD-1/PD-L1 inhibitors have demonstrated encouraging success in treating metastatic breast cancer, especially in patients with TNBC, suggesting a possible treatment option for this difficult subpopulation (Monneur et al., 2018).
The predictive biomarkers that can identify individuals who are likely to benefit from PD-1/PD-L1 inhibitors require additional investigation. Gaining insight into the immunological characteristics of breast cancer via PD-L1 and TILs may result in more individualized treatment plans, increasing therapeutic results and patient outcomes Fig. 3.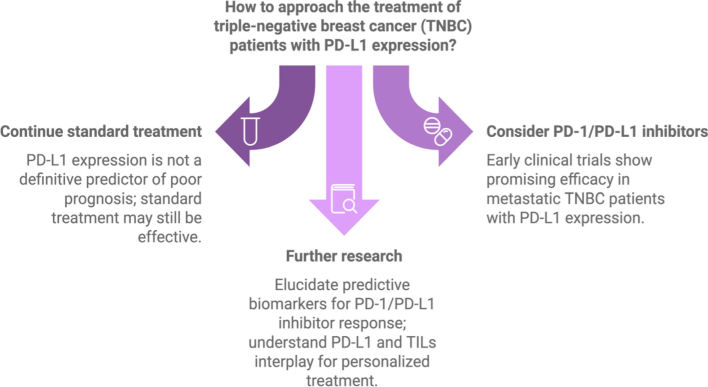
PD-L1 expression guidance towards treatment of triple-negative breast cancer (TNBC).
To sum up, PD-L1 is a biomarker for aggressive disease features and a possible target for immunotherapy in breast cancer, especially TNBC, where therapeutic options are still few. Future research is essential to comprehending how PD-L1 interacts with other biomarkers and how it influences therapy choices.
3.5 MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs)
In the diagnosis of breast cancer, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have been identified as important potential indicators. They are useful tools for improving diagnostic accuracy and enabling early disease identification because of their capacity to control gene expression and their unique expression patterns in different stages of cancer. A more thorough examination of the relationship between these non-coding RNAs and the diagnosis of breast cancer may be summed up as follows:
Normal and malignant breast tissues can exhibit significantly different levels of miRNA and lncRNA expression. Tests for diagnosis can be created using this tissue-specific expression. For instance, it has been repeatedly shown that, in contrast to normal breast cells, several miRNAs, including miR-21, miR-155, and let-7, are either increased or downregulated in breast cancer tissues. A molecular signature that helps in the diagnosis of breast cancer may be obtained by quantifying their levels using methods such as quantitative reverse transcription PCR (qRT-PCR) (Malih et al., 2016).
There are several subtypes of breast cancer, each with unique biological traits, making it a diverse illness. Treatment options depend on the ability of miRNA and lncRNA profiles to distinguish between these subtypes. For instance, some miRNA signatures can differentiate between triple-negative, HER2-positive, luminal A, and luminal B breast tumors (Lo et al., 2016). Clinicians can better customize treatment plans and improve patient outcomes by recognizing these subgroups upon diagnosis.
Because miRNAs are stable in physiological fluids, they are good candidates for liquid biopsy applications, which can offer a non-invasive early cancer detection method. The existence of tumors has been associated with elevated levels of circulating miRNAs, which makes it possible to track the course of the disease and its response to treatment. Studies have shown, for example, that some miRNAs, when found in blood samples, correspond with the early stages of breast cancer, allowing for an earlier detection (Amorim et al., 2016). High-risk people may benefit most from this non-invasive technique as it allows for routine monitoring without the dangers of conventional biopsy techniques.
Although miRNA and lncRNA expression levels are mostly associated with diagnosis, they can also offer prognostic data that aid in clinical decision-making. For instance, a worse prognosis and more aggressive illness have been linked to elevated levels of certain miRNAs. This information might be extremely important when deciding on the kind and urgency of therapy needed after a diagnosis. To avoid harm to patients with good prognoses, prognostic biomarkers can help physicians choose more aggressive treatment plans for individuals with high-risk profiles (Panoutsopoulou et al., 2018).
Diagnostic accuracy can be improved by combining miRNA and lncRNA profiles with more conventional diagnostic techniques like imaging and histopathology analysis. Combining the results of a liquid biopsy with those of a mammogram, for example, may help discover malignancies that might otherwise go undetected by imaging alone. Furthermore, combining genetic information from miRNAs and lncRNAs with pathological and clinical data can help create a more thorough knowledge of the illness and support the creation of individualized diagnostic strategies.
4 Challenges and future directions
Rapid advancements in biomarker research for breast cancer are opening up new possibilities for diagnosis, prognosis, and therapy selection. Nevertheless, a number of obstacles still need to be overcome before these innovative biomarkers may be used in clinical settings Fig. 4. This is a thorough examination of the difficulties and potential paths in the field: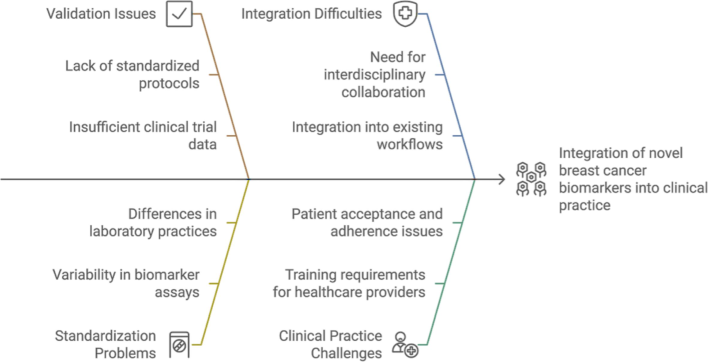
Challenges in integrating novel breast cancer biomarkers into clinical practice.
4.1 Validation and standardization
Large-Scale Studies Are Necessary: Although several new biomarkers have promise in early research, large-scale validation in a variety of clinical contexts and populations is desperately needed. This will make it easier to guarantee that the biomarkers are trustworthy and efficient across a range of demographic groupings.
Standardization of Testing Procedures: The absence of established procedures for biomarker testing may cause inconsistent outcomes and impede the clinical uptake of these tests. For biomarker evaluations to be more consistent and reproducible, standardized procedures for sample collection, processing, and analysis must be established.
Regulatory Approval: Before being regularly employed in clinical settings, biomarkers must pass a stringent screening process to get regulatory approval. Comprehensive clinical trials are one way to demonstrate therapeutic value and relevance.
4.2 Integration into clinical practice
Multidisciplinary Collaboration: Oncologists, pathologists, molecular biologists, and bioinformaticians must work together to integrate new biomarkers into standard clinical practice. To provide thorough recommendations for the use of biomarkers in clinical decision-making, this cooperative approach is required. STC Tumor Board Platform is sponsoring an effort in Saudi Arabia that provides cancer care throughout the country and facilitates direct connections between patients and doctors. It facilitates peer-to-peer consultation, smooth coordination between doctors, and cooperative conversations regarding diagnosis and treatment strategies.
Creation of Guidelines and Procedures: To interpret biomarker data and convert them into practical treatment solutions, precise guidelines and procedures are required. This entails setting positive result criteria, outlining the clinical significance of distinct biomarker profiles, and making suggestions for further treatment choices in light of biomarker results.
Education and Training: To evaluate new biomarkers and their implications for patient treatment, healthcare practitioners need to get education and training. This is essential to guaranteeing the efficient use of biomarkers in therapeutic contexts.
4.3 Personalized medicine
Customizing Treatments: Personalized medicine, in which therapies are adapted to the unique features of each patient's tumor, is the ultimate objective of using biomarkers in the treatment of breast cancer. This entails using a mix of new and established indicators to guide therapy choices.
Maximizing Efficacy and Reducing Side Effects: Physicians can choose treatments that are more likely to be successful while avoiding those that could result in needless side effects by knowing the distinct molecular profile of each patient's cancer. For example, focusing on particular mutations or pathways found by genetic testing may result in more successful therapy for some individuals.
Longitudinal Monitoring: By integrating biomarkers into regular monitoring, information on the course of a disease and the effectiveness of treatment may be obtained. Clinicians can make well-informed judgments about therapy modifications or the need for alternative therapies depending on the patient's changing cancer profile by evaluating biomarkers at various time periods.
4.4 Emerging technologies and research
Developments in Genomic Technologies: The discovery of new biomarkers has been completely transformed by the quick development of genomic technologies, such as next-generation sequencing (NGS). More investigation into these indicators' functional implications will advance knowledge and strengthen their use in therapeutic contexts.
Combining Multiple Omics Methods: Integrating information from proteomics, metabolomics, transcriptomics, and genomes can yield a more thorough knowledge of the biology of breast cancer. The discovery of novel biomarkers and therapeutic targets might result from this all-encompassing strategy.
The use of artificial intelligence (AI) and machine learning: can assist analyze complicated biomarker data, allowing for the identification of trends and improved patient outcome prediction. By facilitating the incorporation of biomarkers into clinical workflows, these technologies can enhance the process of making decisions.
5 Conclusion
In breast cancer, the development of biomarkers—from conventional markers like HER2, progesterone receptor, and estrogen receptor (ER) to cutting-edge choices like genomic signatures, circulating tumor cells (CTCs), and microRNAs—represents a substantial move toward more individualized and efficient therapies. To fully realize the promise of these developments, it is imperative to address the issues of validation, standardization, and integration into clinical practice. The field may get closer to personalized treatment for breast cancer by encouraging cooperation among medical experts, funding research and technology, and creating clear guidelines. This will eventually improve patient outcomes and quality of life.
CRediT authorship contribution statement
Hanan Alismail: Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Breast cancer biomarker discovery in the functional genomic age: a systematic review of 42 gene expression signatures. Biomark. Insights. 2010;5:103-118.
- [Google Scholar]
- Current issues and clinical evidence in tumor-infiltrating lymphocytes in breast cancer. J. Pathol. Translat. Med.. 2015;49:355-363.
- [Google Scholar]
- Breast cancer in Saudi Arabia: A review. Asian Pac. J. Cancer Prev.. 2012;13(4):955-959.
- [Google Scholar]
- The incidence rate of female breast cancer in Saudi Arabia: an observational descriptive epidemiological analysis of data from Saudi Cancer Registry 2001–2008. Breast Cancer (Dove Medical Press). 2013;5:103-109.
- [Google Scholar]
- Immunohistochemical determination of estrogen and progesterone receptors in breast cancer: pathological correlation and prognostic indicators. Inter. J. Cancer Res.. 2014;10(2):75-82.
- [Google Scholar]
- How to study and overcome tumor heterogeneity with circulating biomarkers: the breast cancer case. Semin. Cancer Biol.. 2017;44:106-116.
- [Google Scholar]
- A meta-analysis of estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 discordance between primary breast cancer and metastases. Eur. J. Cancer. 2014;50(2):277-289.
- [Google Scholar]
- Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br. J. Cancer. 2007;96(10):1504-1513.
- [Google Scholar]
- Badowska-Kozakiewicz, A., Patera, J., Sobol, M. and Przybylski, J., 2015. The role of estrogen and progesterone receptors in breast cancer – immunohistochemical evaluation of estrogen and progesterone receptor expression in invasive breast cancer in women. Contemporary Oncology (Poznan), 19, Pp. 220–225.Cimino-.
- Steroid receptors and their role in the biology and control of breast cancer growth. Semin. Oncol.. 2006;33(6):631-641.
- [Google Scholar]
- The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature. 2012;486(7403):346-352.
- [Google Scholar]
- Targeting the human epidermal growth factor receptor 2 pathway in breast cancer. Hosp. Pract.. 2012;40(2):15-17.
- [Google Scholar]
- Biomarkers in breast cancer: Where are we and where are we going? Adv. Clin. Chem.. 2015;71:1-23.
- [Google Scholar]
- Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM) Eur. J. Cancer. 2017;75:284-298.
- [CrossRef] [Google Scholar]
- Relevance of tumor-infiltrating lymphocytes in breast cancer. BMC Med.. 2015;13:202.
- [Google Scholar]
- Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol. Biomark. Prev.. 2018;27(6):619-626.
- [Google Scholar]
- Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res. Treat.. 2013;139(2):539-552.
- [Google Scholar]
- Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol. Biol. Inter.. 2014;2014:1-12.
- [Google Scholar]
- Molecular mechanisms and translational therapies for human epidermal receptor 2 positive breast cancer. Int. J. Mol. Sci.. 2016;17(10):1-14.
- [Google Scholar]
- A review on trends in development and translation of omics signatures in cancer. Comput. Struct. Biotechnol. J.. 2024;23:954-971.
- [Google Scholar]
- A brief review on long noncoding RNAs: a new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumor Biol.. 2016;37:1479-1485.
- [Google Scholar]
- PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum. Pathol.. 2016;47(1):52-63.
- [Google Scholar]
- Receptor conversion in metastatic breast cancer: a prognosticator of survival. Oncotarget. 2016;7(44):71887-71903.
- [Google Scholar]
- Usefulness of traditional serum biomarkers for management of breast cancer patients. BioMed Res. Inter.. 2013;2013:685641
- [Google Scholar]
- PD-L1 expression and PD-1/PD-L1 inhibitors in breast cancer. Bull. Cancer. 2018;105(3):263-274.
- [Google Scholar]
- Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin. Cancer Biol.. 2017;52(Pt 1):56-73.
- [Google Scholar]
- miRNA and long non-coding RNA: molecular function and clinical value in breast and ovarian cancers. Expert Rev. Mol. Diagn.. 2018;18:963-979.
- [Google Scholar]
- Cell-free DNA and circulating tumor cells: comprehensive liquid biopsy analysis in advanced breast cancer. Clin. Cancer Res.. 2017;24:560-568.
- [Google Scholar]
- Saudi Health Council, 2020. Cancer Incidence Report Saudi Arabia 2020 [Internet]. [cited 2024 Sep 13]. Available from: https://nhic.gov.sa/eServices/Documents/2020.pdf.
- Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat. Commun.. 2016;7:12934.
- [Google Scholar]
- Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol. Med.. 2016;13(4):496-504.
- [Google Scholar]
- Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: a systematic review. JAMA Oncol.. 2016;2(10):1354-1360.
- [Google Scholar]
- Biomarkers in the diagnosis of primary and recurrent breast cancer. Biomarker Medicine. 2012;6(5):567-585.
- [Google Scholar]
- PD-L1 expression correlates with tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy in breast cancer. Cancer Immunol. Res.. 2014;3(3):326-332.
- [Google Scholar]
- World Health Organization, 2020. Breast cancer: Prevention and control [Internet]. [cited 2024 Sep 13]. Available from: https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/.
- Estrogen and progesterone receptors in breast cancer. Future Oncol.. 2014;10(14):2293-2301.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103551.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







