Translate this page into:
Biosynthesis of copper nanoparticles using Bacillus flexus and estimation of their potential for decolorization of azo dyes and textile wastewater treatment
⁎Corresponding authors. sabir.hussain@gcuf.edu.pk (Sabir Hussain), melshikh@ksu.edu.sa (Mohamed S. Elshikh)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract

Abstract
In many underdeveloped countries, textile industries discharge their effluents without any treatment. The untreated textile wastewater consists of both azo dyes and heavy metals which not only change the physicochemical and biological properties of soil and water but also affect human health. In recent years, copper-based nanomaterials have attained worldwide attention because of their unique properties and potential for decolorization of azo dyes and wastewater treatment. The present study demonstrates the bacterial synthesis of copper nanoparticles (Cu-NPs) using Bacillus flexus strain isolated from a textile wastewater followed by their application for photocatalytic degradation of various azo dyes and treatment of actual wastewater. The FT-IR analysis confirmed the presence of various functional groups including proteins on Cu-NPs which improved their stability. The scanning electron microscopy (SEM) images revealed the spherical shape of Cu-NPs with a size of range between 17–34 nm. Similarly, the XRD analysis of biosynthesized Cu-NPs showed various diffraction peaks at 44.5°, 51.5°, 74.75 which confirmed the crystalline nature of nanoparticles. While studying the photocatalytic decolorization of azo dyes by Cu-NPs, it was observed that 88.164 ± 0.19 %, 88.452 ± 1.89 %, 90.433 ± 1.81 %, 64.770 ± 1.02 %, 46.774 ± 1.61 %, and 67.274 ± 2.89 % of reactive black-5, congo red, malachite green, methylene blue, reactive red-2 and blue direct, respectively, were decolorized after 4 h of solar irradiation at 50 ppm concentration. Additionally, the biosynthesized nanoparticles also resulted in reduction of various parameters like EC, pH, TDS, COD, color intensity, sulphates, and phosphates in the textiles wastewater. The reduction of COD, sulfates, and phosphates was about 39.659 %, 43.157 %, and 49.493 %. The results of current work suggest that biosynthesized copper nanoparticles might serve as a potential green solution for the decolorization of various dyes including wastewater treatment.
Keywords
Bacterial isolation
Biosynthesis
Copper nanoparticles
Photocatalysis
Dye degradation
Wastewater treatment
- BOD
-
Biological oxygen demand
- COD
-
Chemical oxygen demand
- EC
-
Electrical conductivity
- Cu-NPs
-
Copper nanoparticles
- UV-vis
-
Ultraviolet visible spectroscopy
- SEM
-
Scanning electron microscopy
- FTIR
-
Fourier transform infrared spectroscopy
- XRD
-
X-ray diffraction
Abbreviations
1 Introduction
Water is one of the most important natural resource on earth not only for human beings but also for all other living organisms. All essential phenomena of the biosphere depend on the availability of water. Along with other human requirements, a huge amount of water is also used for industrial processes. Among different industries, the textile industries utilize an enormous amount of water for different activities such as dyeing and washing and similarly this untreated water becomes part of the environment as wastewater (Imran et al., 2015). It is reported that, after dyeing one kilogram (Kg) of fabric, about 40–65 L of wastewater is produced (Balarak et al., 2019). Approximately 20 % of applied quantity of different dyes is lost during the process of washing and dyeing and discharged along with wastewater (Fathima et al., 2018). The major problem with untreated textile wastewater is its color due to the presence of these dyes in large quantities. It is reported that dyes have many carcinogenic and mutagenic properties. Moreover, several environmental problems such as elevation of various parameters like pH, BOD, suspended bodies, and COD of aquatic bodies are associated with untreated textile effluents (Imran et al., 2015, Subramanian e al., 2022; Bilal et al., 2023). Thus, dyes loaded textile wastewaters limits downstream usage such as drinking, washing, irrigation, and recreation (Imran et al., 2015).
Many physical (adsorption, membrane filtration, coagulation and filtration method), chemical (oxidation like chlorination, ozonation, bleaching and advanced oxidation method with heterogenous and homogenous catalysts) and biological (employing bacteria, fungi, yeast, algae, and plants) methods have been used for the treatment of dyes loaded textile wastewater (Singh et al., 2017). Above mentioned physiochemical methods are associated with many drawbacks such as they are expensive and make use of dangerous chemicals. Furthermore, they are also a source of secondary pollutants such as production of sludge (Bharagava et al., 2020). In contrast, biological methods are environmentally-friendly and even they have ability to completely mineralize many azo dyes. Many reports have described the role of microorganisms in the removal of azo dyes from textile effluents (Hussain et al., 2013, Thangaraj et al., 2021). Microorganisms involved in decolorization of azo dyes produce many enzymes such as azo reductase, laccase, peroxidase, MG reductase, and aminopyrine N-demethylase (Souza et al., 2022). However, the rate of biodegradation process depends on many factors including survival activity, and adaptability of microbes with chemical properties of azo dyes and other xenobiotics (Manjarrez Paba et al., 2021). Although biological methods are eco-friendly but the old methods may not be much effective to fulfill the strict standards of water quality and to remove emergent contaminants.
During the past few years, use of nanomaterials has attained worldwide attention for the degradation of various dyes. They are preferred because of production of biodegradable end products such as aromatic amines which can be further biodegraded easily by microbes (Nazar et al., 2018). Various studies have highlighted the catalytic potential of different metal nanoparticles including silver, titanium, zinc and nickel nanoparticles for the decolorization of different dyes (Edison et al., 2016, Saravanan et al., 2017, Kiran et al., 2020, Mustafa et al., 2023). Among other metal nanoparticles, copper nanoparticles are of excessive interest because of their low cost, availability, and unique properties such as catalytic, antibacterial, biocidal, optical, magnetic and heat transfer abilities (Punniyakotti et al., 2020, Ghaffar et al., 2021). Furthermore, in comparison to silver-nanoparticles, copper nanoparticles (Cu-NPs) have more chemical and physical stability and ease of mixing with polymers (Palani and Elangovan, 2022). The Cu-NPs find their applications in many fields like health (antibacterial, anticancer, dentistry, etc.), food and agriculture (antifungal, nano herbicides, nano-fertilizers, improving texture, etc.), industry (nano composites, cosmetics, nano-pigments, paper coating) and environment (nano remediation, wastewater treatment, biodegradation of polymer, environmental catalysts, etc.) (Ball et al., 2019). For example, Cu-NPs have the ability for dechlorination of dichloromethane (a ground water contaminant) with NaBH4 by using them as catalyst (Huang et al., 2012). They showed 90 % reduction of dichloromethane within 1 h using NaBH4 and no degradation was observed without NaBH4 (Huang et al., 2012).
Different physical and chemical methods like precipitation-pyrolysis method, sonochemistry, cathodic vacuum arc and solvothermal reactions have been reported for physicochemical synthesis of copper-based nanoparticles (Botsa et al., 2019). All such methods make use of the chemicals which make them eco-hazardous and avoid their applications in other fields of clinical, medicine and biology (Akintelu et al., 2020). So, the researchers are more interested toward biological method for nanoparticles synthesis because of the above-mentioned problems that are associated with physicochemical methods. Numerous bacterial species are reported to synthesize copper nanoparticles like Salmonella typhimurium (Ghorbani et al., 2015), Shewanella oneidensis (Kimber et al., 2018), Halomonas elongate (Rad et al., 2018), Streptomyces sp. (Hassan et al., 2019), Escherichia coli (Zaynitdinova et al., 2020), Pseudomonas stutzeri (Zaynitdinova et al., 2020), Pseudomonas fluorescens (Bayat et al., 2021). Similarly, various studies have highlighted the capability of copper nanoparticles in facilitating the photocatalytic degradation of different dyes (Fathima et al., 2018, Noman et al., 2020). For wastewater treatment, Anjum et al. (2019) described the nanotechnology as one of the most advanced process. In this backdrop, following research investigates an innovative and sustainable approach for synthesizing Cu-NPs. The potential of bacterial strain was harnessed with an aim to not only reduce the environmental impact but also enhance its cost effectiveness. For in-depth understanding of Cu-NPs, various cutting-edge techniques such as, UV–Vis spectroscopy, FTIR, SEM, and XRD were used. This study was not only focused on green synthesis but also holds an immense promise in catalytic biodegradation of various azo-dyes. Furthermore, the potential of nanoparticles for the treatment of textile waste water was also explored, thus contributing to environmental conservation. This is the research that promises to not only captivate but also contribute to a greener and more sustainable future.
2 Materials and methods
2.1 Materials
All the reagents or chemicals namely nutrient agar, nutrient broth, and copper sulfate pentahydrate, used for experimental analysis, were procured from Sigma Aldrich or UNI-Chem and used without any purification techniques. The dyes used in this study namely congo red (CR), reactive black-5 (RB-5), methylene blue (MB), reactive red-2 (RR-2), and blue direct (BD) were also purchased from Sigma Aldrich. Moreover, the glassware used during experimental analysis was washed with distilled water and autoclaved.
2.2 Collection of samples for the isolation of bacterial strain
Wastewater samples were collected from Five-star Textile Industry Faisalabad. Bacterial strains were isolated using serial dilution method. Dilutions (10−4 to 10−6) were spread on plates of nutrient agar amended with the solution of copper sulfate (1 mM). These plates were incubated for 48 h in incubator at 30 °C. The bacterial colonies having ability to tolerate stress were purified by repeated streaking method on freshly prepared plate and labelled. Among different isolates, strain FB-1 was used for the biosynthesis of nanoparticles.
2.3 Biosynthesis of copper nanoparticles
For the biosynthesis of copper nanoparticles (Cu-NPs), the colony of bacterial strain was inoculated in nutrient broth medium along with two controls (The first control containing only the uninoculated medium and the second control containing the uninoculated medium along with the precursor salt), and incubated under shaking condition at 120 rpm for 48 h at 28 °C. The culture was centrifuged for 10 min at 7000 rpm to remove the pellet for extracellular synthesis of Cu-NPs. The supernatant was then added with 10 mM copper sulfate pentahydrate solution and incubated again under shaking condition for 24 h at 120 rpm. Equal concentration of precursor salt solution was also added to the second control. The change in color from bluish-to-bluish green in comparison to control confirmed the biosynthesis of Cu-NPs. The reaction mixture was again subjected to centrifugation for about 10 min at 7000 rpm at 4 °C to remove the supernatant. The pellet was washed with double distilled water to remove the impurities and placed in an oven to heat dry the nanoparticles. They were then ground into fine powder.
2.4 Molecular characterization and phylogenetic study
The selected bacterial isolate having potential of Cu-NPs synthesis was identified on molecular basis. The 16 S rRNA gene was amplified using forward and reverse primers f-D1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and r-D1 (5′-AAGGAGGTGATCCAGCC-3′) (Weisburg et al., 1991). The amplified gene products of 16 S rRNA were sequenced commercially by Macrogen, South Korea. The retrieved sequence of 16 S rRNA gene was trimmed analyzed by Sequence Scanner software package and trimmed on both sides to cut the bad sequences. The BLASTn was used for identification of strain. Then the phylogenetic tree was constructed by using NJ (neighbour-joining) method to describe the identity of selected strain.
2.5 Characterization of biosynthesized copper nanoparticles
The bacterial synthesis of Cu-NPs was confirmed using Ultraviolet Visible Spectroscopy (Shimadzu, Kyoto, Japan). It determined the maximum absorption spectra of samples. UV–vis spectroscopy analyzed the reaction mixture at a wavelength ranging from 300 to 800 nm (Noman et al., 2020).
Scanning Electron Microscopy (SEM) was employed to describe the shape and surface texture of biosynthesized Cu-NPs (Instrument: JSM-6490). SEM was conducted by following the method of Tiwari et al. (2016). For SEM analysis, about 10 μL of homogenous cell free colloidal solution was placed on cover slip made up of glass and dried at 100 °C. After that, the glass cover slip was cooled and fixed on aluminum stub using adhesive carbon tape.
The crystalline nature of biosynthesized nanoparticles was determined using X-ray diffraction analysis. To determine the XRD pattern, the drop was coated on glass substrate and placed in X-ray diffractometer apparatus (JCPDS Card #=04-0836) which was operated at 40 mA and 45 KV voltage of current with Cu Kα rays. The crystalline nature of biosynthesized nanoparticles was analyzed, and Debye-Scherrer’s formula, was used for their average size Whereas,
D = “Crystalline size”, K = “Sharrer’s constant” and its value is “0.9”, λ = Wavelength of “x-ray source”, β = “Width at 1/2 maximum of reflection peaks” and, θ = “Peak position”.
Functional groups and associated proteins that participate in stability of biosynthesized Cu-NPs were determined by Fourier-Transform Infrared Spectroscopy (FT-IR) (FTIR-Bruker TENSOR- 27). The dried powder of biosynthesized Cu-NPs was taken to FTIR measurement and the FTIR spectra were attained. It is useful method to ascertain the various type of chemical bonds in a molecule by making an IR absorption spectrum that functioned as molecular fingerprint. The analysis of FT-IR was conducted in a range of 4000–350 cm−1.
2.6 Catalytic degradation of azo dyes
2.6.1 Optimization of concentration of Cu-NPs for RB-5 decolorization
Various concentrations of Cu-NPs were used to check their potential of decolorization of RB-5. 50 ppm concentration of RB-5 was used for this purpose. Various concentrations of NPs such as (0.05, 0.1, 0.2, 0.5, 1, 2, 3, 4, 5 and 10 mg ml−1) were added to the solution of RB-5 and placed in solar irradiation for 4 h after shaking for about 10 min. After each hour, 1 ml of sample was taken from each concentration in a 1.5 ml Eppendorf tube. The samples were centrifuged for about 10 min at 7000 rpm to remove the Cu-NPs. Spectrophotometer was used to check the ability of different concentrations of nanoparticles by measuring the decolorization of RB-5. The optimized concentration (0.5 mg ml−1) of Cu-NPs was used for further analysis.
2.6.2 Decolorization of azo dyes using optimized concentration of NPs
The optimized concentration (0.5 mg ml−1) of nanoparticles was used for the decolorization of various other dyes such as MB, RB-5, BD, CR, MG, and RR-2 dyes at 25 and 50 ppm concentrations. For this purpose, optimized concentration of biosynthesized Cu-NPs (50 mg) was added to the solution (100 ml) of above-mentioned azo dyes. The reaction mixtures were then placed in sun light for incubation for about 4 h. 1 ml of sample was taken from each dye in 1.5 mL Eppendorf tube after specific time interval. The Eppendorf tubes containing samples were centrifuged for 10 min at 7000 rpm to remove the Cu-NPs. Spectrophotometer was used to evaluate the degradation of dyes. The decolorization of dyes namely MB, CR, BD, RR-2, RB-5, and MG was evaluated at 561, 478, 592, 540, 597, and 624 nm, respectively. The %age of dyes degradation was calculated using the formula written below, Whereas,
-
C is control
-
S is sample
2.6.3 Treatment of wastewater
Bacterial synthesized Cu-NPs were also used for the treatment of actual wastewater. The sample of wastewater was collected from paharang drain (North-31.526904 and East-73.118483) and Sargodha Road, Faisalabad, Pakistan. It was firstly centrifuged at 10,000 rpm for 5 min to remove the particulate matter. The collected wastewater was spiked with congo red (50 ppm) to have considerable concentration of the dye and color intensity. 50 mg (0.5 mg ml−1) of biosynthesized Cu-NPs were then added to 100 ml of wastewater by keeping the control without nanoparticles to evaluate the treatment of wastewater. Sample was vortexed and incubated in the presence of sunlight for about 7 h. After incubation, the samples were centrifuged for 10 min at 7000 rpm to remove the Cu-NPs. Different parameters like pH, chemical oxygen demand, electrical conductivity, phosphates, color intensity, and sulphates were checked before and after the treatment following the procedure described by Rump (1999) and Maqbool et al. (2016).
2.7 Statistical analysis
The experimental data of treatment was entered in Microsoft Excel 2016, and percentage decolorization, means, and standard deviation was calculated. For analyzing data, completely randomized design was used. Analysis of variance was calculated by using statistix 8.1 software. Principle component analysis and heat map was also constructed using the R-studio (Version 4.2.2).
3 Results and discussion
3.1 Isolation, molecular characterization and phylogenetic analysis
The selected isolate was purified by repeated streaking method on freshly prepared nutrient agar plates until single colony was obtained. The growth pattern of selected isolate on nutrient agar plate media has been represented in the Figure S1. The phylogenetic analysis of 16 S rRNA gene and BLASTn of isolate determined that the selected isolate belongs to genus Bacillus. In Phylogenetic tree (Fig. 1), selected strain clustered with Bacillus spp. The 16S rRNA gene sequence of strain (FB-1) was also compared with known sequences in database using BLASTn program of NCBI and the sequence showed more than 99 % resemblance with the bacterial strains belonging to genus Bacillus.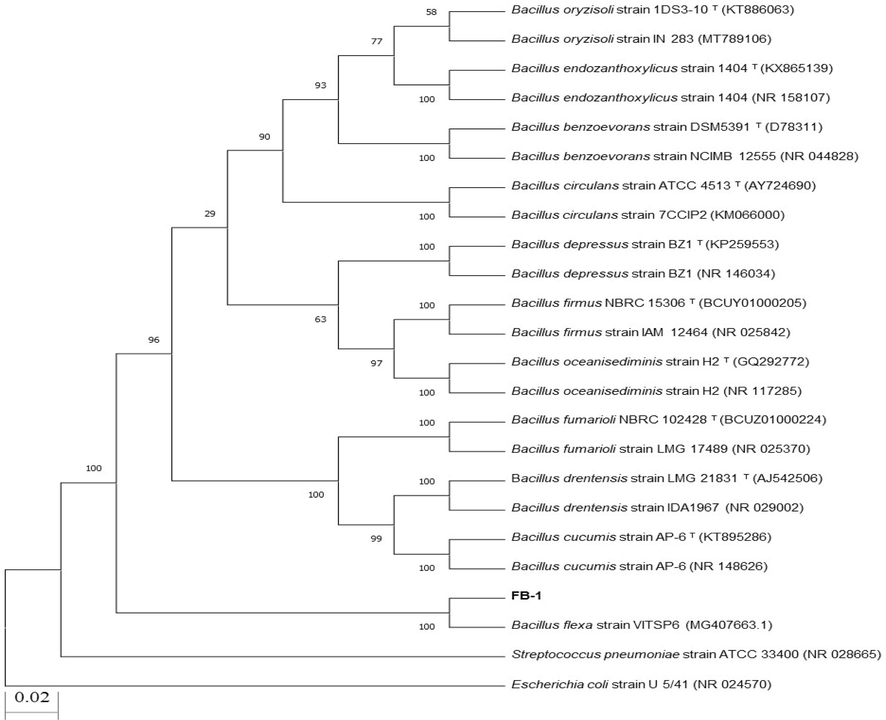
Neighbor joining phylogenetic tree of Bacillus spp. Strain FB-1.
3.2 Biosynthesis and characterization of copper nanoparticles
Various microbial isolates have been reported describing the synthesis of Cu-NPs. In this study Bacillus spp. Strain FB-1was used for the extracellular synthesis of Cu-NPs at 10 mM concentration of copper sulphate salt. On addition of salt, the change in color (from bluish to blue greenish) firstly confirmed the biosynthesis process (Fig. 2A). Similar result was reported earlier by Tiwari et al. (2016) who synthesized Cu-NPs using Bacillus cereus strain at 10 mM concentration of copper sulfate salt. On the other hand, Noman et al. (2020) also reported the bacterial synthesis of Cu-NPs using Escherichia sp. strain SINT7 at 5 mM concentration of precursor salt. The Bacillus flexus strain was also used for the synthesis of silver nanoparticles (Priyadarshini et al., 2013). Furthermore, several other nanoparticles such as cadmium sulphide (Singh et al., 2011), iron oxide (Sundaram et al., 2012), titanium dioxide (Khan and Fulekar, 2016) and zinc oxide (Hamk et al., 2023) were synthesized from Bacillus spp. Hence, the bacterial strain Bacillus spp. FB-1 is a new addition as a bacterial member for the synthesis of nanoparticle.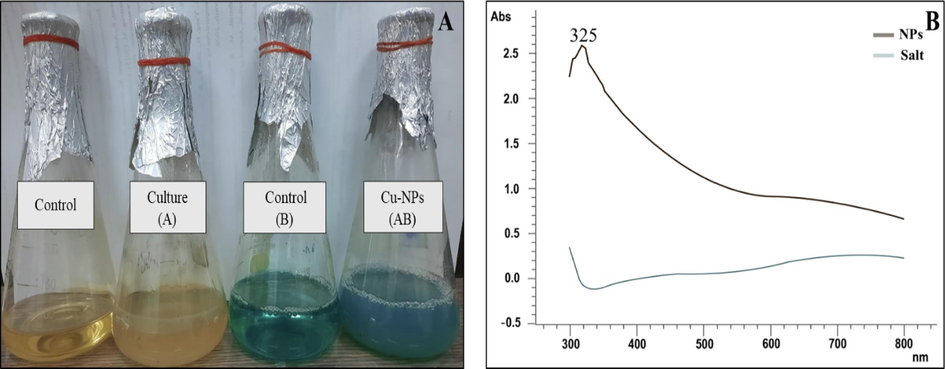
(A) Confirmation of biosynthesized copper nanoparticles by changing in color and (B) UV–vis spectroscopy. The brown line in UV–Vis spectrum confirmed the synthesis by showing an absorption peak at 325 nm while the blue line indicates the control. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The change in color was the first confirmation of biosynthesis of Cu-NPs. The cell free reaction mixture showed absorbance peak at 325 nm using UV–vis spectrophotometer which confirmed the synthesis of Cu-NPs as depicted in Fig. 2B. Similar result was reported by Noman et al. (2020) and concluded that peak in this region confirmed the reduction of precursor salt into nanoparticles. Moreover Singh et al. (2010) and Mamuru et al. (2016) also reported the confirmation peak of Cu-NPs at 310 nm. While Fathima et al. (2018) and Hassan et al. (2019) showed an absorption peak at 560 and 590 nm.
The SEM analysis revealed the morphology of biosynthesized Cu-NPs. The SEM image of the biosynthesized Cu-NPs (Fig. 3) indicated that the sample contained copper nanoclusters with irregular spherical shape and present in an agglomerated form. The analysis of SEM images revealed that the particles size was in range of 17–34 nm.Various reports described the different shapes of Cu-NPs such as cubical (Kuo et al., 2007), rhombic dodecahedral (Liang et al., 2009), 18-facet polyhedral (Lin et al., 2010), octahedral (Xu et al., 2011) and spherical (Noman et al., 2020), Bukhari et al., 2021).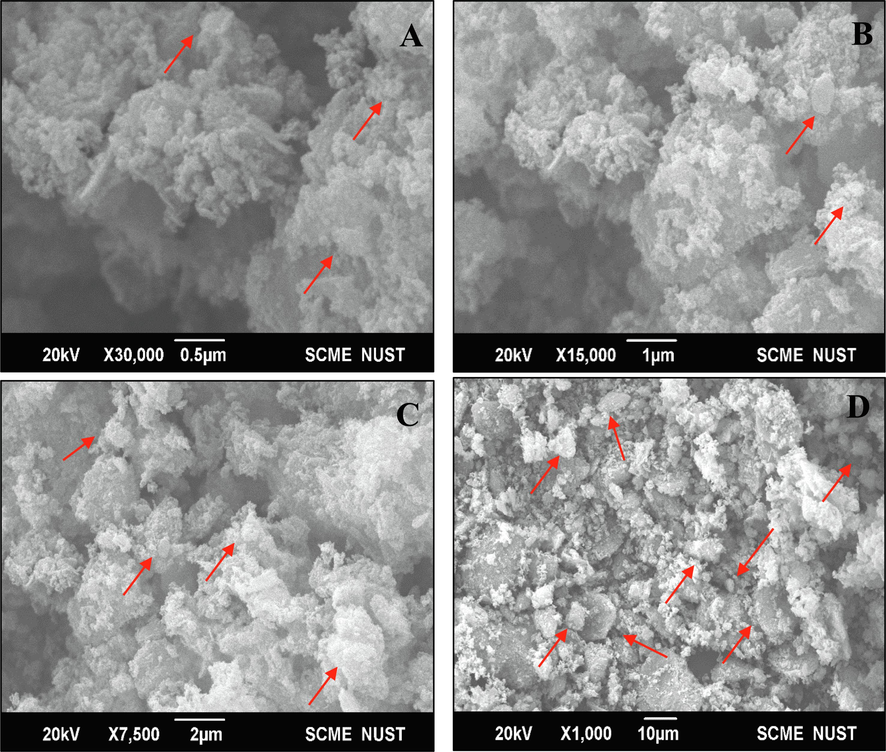
SEM images of biosynthesized copper nanoparticles under different magnifications such as 0.5 µm (A), 1 µm (B), 2 µm (C), and 10 µm (D).
The XRD analysis of biosynthesized Cu-NPs showed various diffraction peaks at 44.5°, 51.5°, 74.75°as depicted in Fig. 4A. The described peaks at 44.5°, 51.5°, 74.75° were assigned to (111, 200 and 220) planes of copper. The peaks at above mentioned planes confirmed the crystalline nature of Cu-NPs. There are also some extra peaks that might be due to presence of some cupric oxide or copper oxide nanoparticles. The size of nanoparticles was less than 27.6 nm calculated by Debye-Scherrer’s formula. Tiwari et al. (2016) reported the similar results and showed diffraction peaks of Cu-NPs at 111, 200 and 220 synthesizing from Bacillus cereus spp. They also reported some extra peaks of copper oxide nanoparticles. Similarly, Noman et al. (2020) and Lv et al. (2018) also reported the similar results showing diffraction peaks at 111, 200, and 220 with a size of particles about 28.55 and 10–16 nm.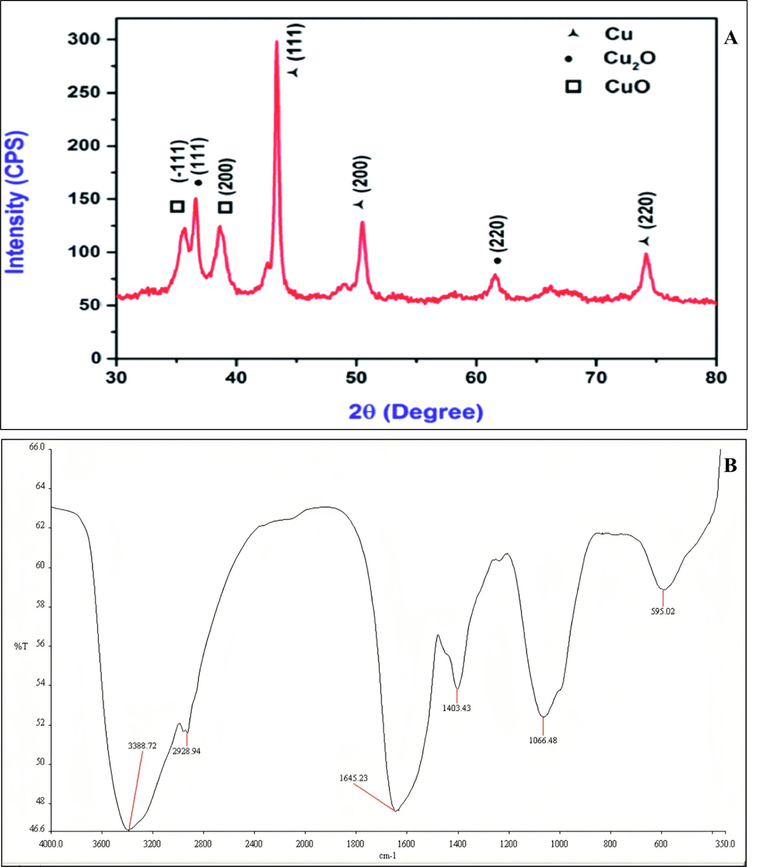
XRD (A) and FTIR (B) spectrum of biologically synthesized copper nanoparticles using Bacillus spp.
FTIR spectrum of biosynthesized Cu-NPs produced from Bacillus sp. FB-1 showed peaks of absorption at 3388.72, 2928.94, 1645.23, 1403.43, 1066.48, and 595.02 cm−1 as shown in Fig. 4B. The peaks at 3388.72 and 2928.94 cm−1 were due to presence of OH group of alcohol and C–H stretching. The peak at 1645.23 cm−1 was due to C⚌C stretching. Whereas peaks at 1403.43 and 1066. 48 cm−1 were due to bending of C–H and stretching of C-O group. The presence of bands at 595.02 cm−1 showed probabilities of existence of copper or copper oxide nanoparticles (CuO-NPs). The FTIR spectrum of biosynthesized Cu-NPs confirmed the presence of alcoholic group and proteins. Coating of protein around copper nanoparticles enhanced the long-term stability of particles (Valodkar et al., 2011). Whereas, Noman et al. (2020) reported the various absorption peaks at 3381.79, 2957.81, 1656.20, 1451.87, 1398.87 and 1111.12 cm−1. The peaks at 3381.79 and 2957.81 cm−1 were due to presence of OH group of alcohol and stretching of C–H bond. While Nieto-Maldonado et al., (2022) reported the plant based synthesis of copper nanoparticles and showed various absorption peaks at 3271, 2931, 1606, and 1029 cm−1. the peaks at 3271 and 2931 cm−1 were due to presence of O–H and C–H groups. On the other hand, the peaks at 1606 and 1029 cm−1 were due to C⚌O and C-O groups.
3.3 Photocatalytic degradation of azo dyes
3.3.1 Optimization of concentration of Cu-NPs for RB-5 decolorization
In the current work, different concentrations of biosynthesized Cu-NPs were used for the degradation of RB-5. After solar incubation of 4 h, decolorization of RB-5 with 0.05, 0.1, 0.2, 0.5, 1, 2, 3, 4, 5, and 10 mg ml−1 concentration of biosynthesized Cu-NPs was about 84.3 ± 0.3 %, 87.0 ± 0.5 %, 87.2 ± 0.5 %, 86.9 ± 1.4 %, 90.1 ± 1.8 %, 89.0 ± 1.6 %, 89.7 ± 2.6 %, 90.1 ± 1.2 %, 88.0 ± 3.1 %, and 88.5 ± 3.4 %, respectively (Fig. 5). From the rate of decolorization, it was clearly proved that each concentration shows maximum degradation of RB-5. However, it is noteworthy that, after 1 h incubation period, 24.9 ± 2.9 %, 45.0 ± 0.3 %, 60.4 ± 1.6 %, 80.5 ± 0.8 %, 80.8 ± 1.2 %, 80.8 ± 0.2 %, 81.9 ± 0.7 %, 80.9 ± 1.3 %, 81.8 ± 2.5 % and 75.5 ± 2.1 % decolorization of RB-5 was observed using 0.05, 0.1, 0.2, 0.5, 1, 2, 3, 4, 5, and 10 mg ml−1 concentration of Cu-NPs. So, 0.5 mg ml−1 of biosynthesized Cu-NPs was selected for the decolorization of other dyes as shown in Fig. 8. While Noman et al. (2020) used 1 mg ml−1 of Cu-NPs for the decolorization of various azo dyes. Furthermore Ahmad et al. (2018) decolorized the bromophenol blue (BPB) dye using 0.25 mg ml−1 of green synthesized Cu-NPs.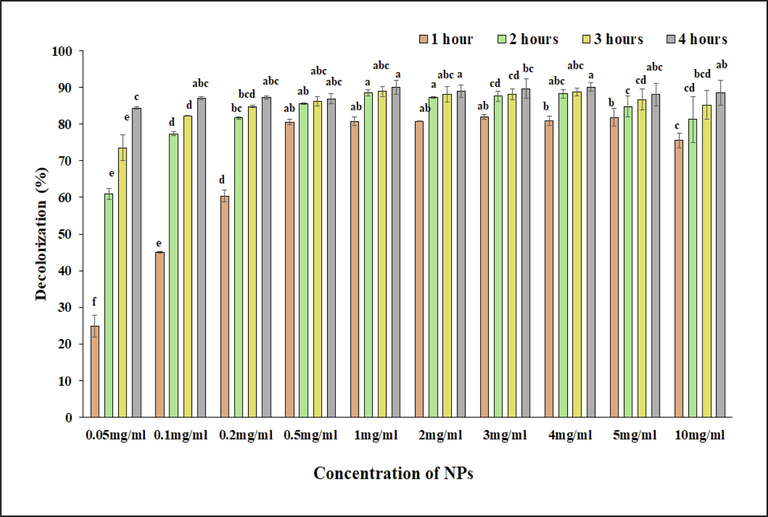
Photocatalytic degradation of RB-5 with different concentrations of biosynthesized copper nanoparticles (0.5, 0.1, 0.2, 0.5, 1, 2, 3, 4, 5, and 10 mg/ml) at 50 ppm of dye concentration, respectively.
Histogram correlation analysis was carried out for the selection of best concentration of nanoparticles for the decolorization of dyes. In a histogram (Fig. 6A) red color showed non-significant while pink, grey and blue colors showed significant results. After analyzing the histogram, it was observed that among all concentrations, 0.5 mg ml−1 showed exceptional results. When compared to 0.5 mg ml−1 concentration, lower concentrations (0.05–0.2 mg ml−1) showed significant decolorization but not effective as 0.5 mg ml−1 concentration. On the other hand, higher concentrations (1–10 mg ml−1) did not showed a proportional increase in decolorization potential, especially the minimum increase was observed after the 3 h. While, 0.5 mg ml−1 showed a higher initial decolorization rate and maintained throughout the time, thus providing an optimum balance between effectiveness and nanoparticles concentrations. So, the 0.5 mg ml−1 was selected that not only conserve the resources but also go with environmental and economic considerations, potentially reducing the ecological footprint and material costs associated with the process of decolorization of dyes.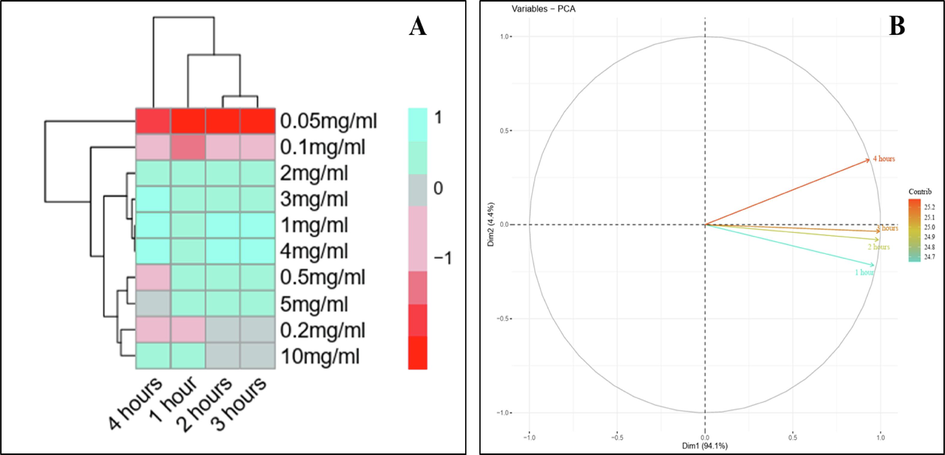
Heat map (A) and Principle component analysis (B) using different concentrations of nanoparticle for the decolorization of RB-5 dye under solar irradiation of 4 h.
The loading plots of PCA to check the effect of different concentrations of nanoparticles on rate of decolorization have been presented in Fig. 6B. In the whole database, Dim-1 and Dim-2 showed maximum contribution and occupy more than 98.5 % of all databases, among which Dim-1 showed 94.1 % and Dim-2 showed about 4.4 %. It was observed that concentration of nanoparticles showed a significant effect on rate of decolorization.
3.3.2 Decolorization of azo dyes using optimized concentration of NPs
After optimization, 0.5 mg ml−1 concentration of biosynthesized Cu-NPs was used for the decolorization of various other azo dyes. The Cu-NPs were observed to carry out an effective decolorization of different azo dyes (Fig. 7). The results indicated that, just after solar irradiation of 1/2 h, Cu-NPs decolorized about 84.5 ± 1.1 %, 64.8 ± 3.9 %, 38.7 ± 4.3 %, 46.9 ± 2.2 %, 20.4 ± 3.8 % and 30.7 ± 2.1 % of RB-5, CR, MG, MB, RR-2, and BD. However, 71.9 ± 1.9 %, 30.5 ± 1.4 %, 61.9 ± 3.9 %, 8.1 ± 0.5 %, 15.7 ± 2.2 % and 15.9 ± 1.6 % of RB-5, CR, MG, MB, RR-2, and BD were decolorized at 50 ppm concentration of dyes just after 1/2 h. However, after 1 h of solar incubation, decolorization of RB-5, CR, MG, MB, RR-2, and BD was increased to 86.3 ± 0.7 %, 90.3 ± 2.4 %, 54.2 ± 9.4 %, 58.9 ± 3.5 %, 36.7 ± 3.3 % and 45.8 ± 2.1 % at 25 ppm concentration. Similarly, at 50 ppm concentration, the decolorization of RB-5, CR, MG, MB, RR-2, and BD was also increased to 84.2 ± 0.6 %, 71.6 ± 2.5 %, 71.6 ± 3.9 %, 46.1 ± 2.8 %, 26.8 ± 1.6 % and 48.5 ± 5.5 %, after 1 of solar incubation. It is noteworthy that the decolorization of the above-mentioned dyes was more at 25 ppm (Fig. 7A) concentration of dyes than 50 ppm (Fig. 7B) concentration. Finally after 4 h of solar irradiation, the copper nanoparticles decolorized about 96.8 ± 1.0 % of RB-5, 92.2 ± 2.2 % of CR, 96.9 ± 1.1 % of MG, 72.5 ± 0.6 % of MB, 58.5 ± 1.2 % of RR-2 and 78.7 ± 2.7 % of BD at 25 ppm concentration and 88.2 ± 0.2 % of RB-5, 88.5 ± 1.9 % of CR, 90.4 ± 1.8 % of MG, 64.8 ± 1.0 % of MB, 46.8 ± 1.6 % of RR-2, and 67.3 ± 2.9 % of BD at 50 ppm concentration. The difference in catalytic potential of biosynthesized Cu-NPs for different dyes might be due to change in activity in aqueous media, structure of dye, and difference in intensity of solar radiation (Noman et al., 2020). The visual representation of decolorization of above-mentioned dyes has been shown in Figure S2. Dyes were decolorized after the addition of biosynthesized Cu-NPs and was proved from the decrease in absorption values at λmax of each dye. While, (Noman et al., 2020) reported the 97.07 %, 83.61 %, 88.42 %, and 90.55 % decolorization of 25 ppm concentration of congo red, reactive black-5, direct blue-1, and malachite green respectively. On the other hand, Ghaffar et al. (2021) reported the 73.5 % decolorization of Disperse yellow-125 using 0.01 % concentration of dye and 0.05 % of green synthesized Cu-NPs.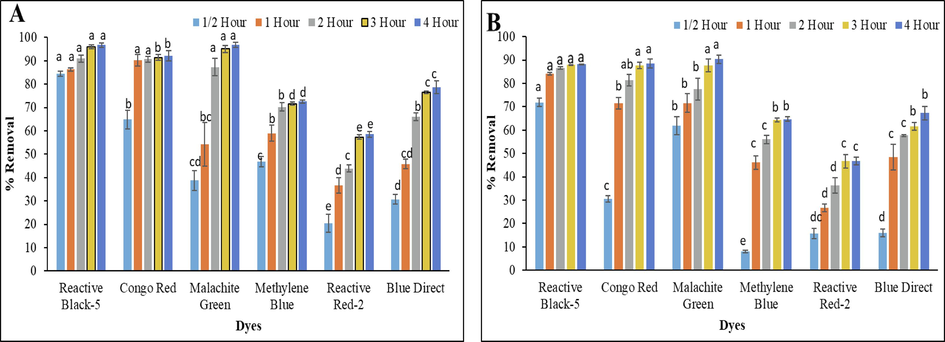
Decolorization of RB-5, CR, MG, MB, RR-2 and BD under solar irradiation of 4 h at 25 (A) and 50 (B) ppm concentration.
While, the photocatalytic activity of Cu-NPs for the degradation of various azo dyes such as, congo red, methyl red, methylene blue (Fathima et al., 2018), and methyl orange (Li et al., 2015) was also reported (Al-Hakkani, 2020). Moreover, the catalytic potential of biosynthesized nanoparticles can be used for the removal of pollutants present in actual wastewater.
The heat map analysis was also carried out to check the effect of different concentration of dyes over time. In a histogram (Fig. 8A) red color showed non-significant while pink, grey and blue colors showed significant results. After analysis it was observed that for both concentrations (25 and 50 ppm), the decolorization potential of biosynthesized Cu-NPs generally increased over time, with some variances among dyes. At 25 ppm concentration, RB-5, CR, and MG showed high decolorization potential, reaching over 90 % after 4 h. The decolorization of MB and BD also increased over time, while RR-II has the least decolorization potential. On the other hand at 50 ppm concentration, RB-5 showed a consistent increase in decolorization potential, but slightly lower than the 25 ppm concentration. While decolorization potential of CR, MG, and BD showed significant improvements over time, and MB and RR-II have moderate to low decolorization potential, thus indicating that higher concentrations do not lead to better decolorization for these dyes.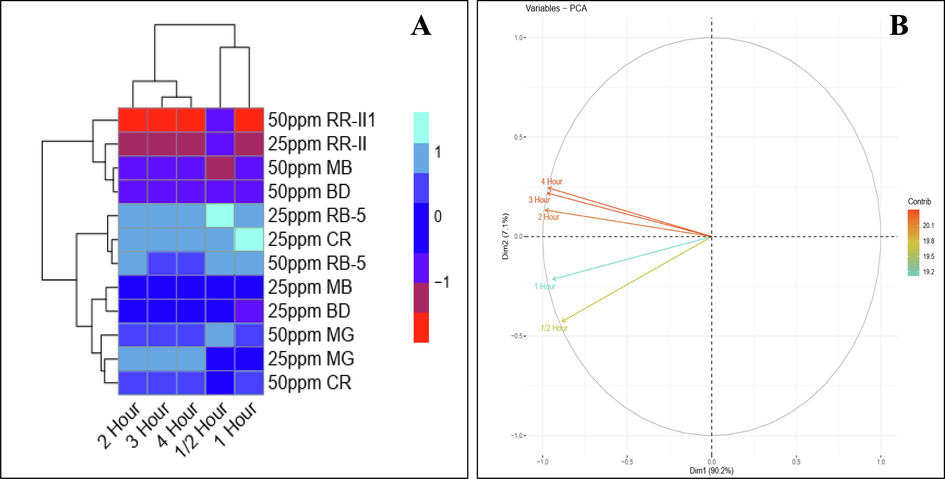
Heat map (A) and Principle component analysis (B) using different concentrations of dyes under solar irradiation of 4 h.
Similarly PCA was also used to check the effect of different concentrations of dyes on rate of decolorization (Fig. 8B). In the whole database, Dim-1 and Dim-2 showed maximum contribution and occupy more than 97.3 % of all databases, among which Dim-1 showed 90.2 % and Dim-2 showed about 7.1 %. It was observed that concentrations of dyes showed significant effect on rate of decolorization over time.
3.3.3 Treatment of textile wastewater
The optimized concentration of Cu-NPs was also used for the treatment of textile wastewater added with CR dye (50 ppm). Before any treatment, the wastewater shows high values for various parameters such as sulphates, phosphates, pH, EC, TDS, COD, and color intensity. Whereas the treated wastewater showed significant reduction in all above-mentioned parameters (Table 1). Few studies already reported the treatment of wastewater via metal nanoparticles (Bibi et al., 2019, Dlamini et al., 2019, Al-Hakkani, 2020; Noman et al., 2020, Siddique et al., 2021). The results of present study showed that the pH of the wastewater was reduced from 8.6 to 6.9 after the treatment as compared to non-treated wastewater. The EC of treated wastewater was also reduced by 59.23 %. Whereas, other parameters such as TDS, COD, sulfates, phosphates, and color intensity were also reduced by 57.68 %, 39.66 %, 43.16 %, 49.49 %, and 95.97 % in comparison to non-treated wastewater as depicted in Table 1. Few previous studies have also reported the treatment of wastewaters by exploiting the potential of different types of nanoparticle (Bibi et al., 2019, Noman et al., 2020, Siddique et al., 2021; Rasool et al., 2023; Pham-Khanh et al., 2023), For example, the findings of Siddique et al., (2021) reported the reduction in various parameters such as pH, EC, TDS, COD, and color removal with the help of biologically and chemically synthesized zinc oxide nanoparticles. Furthermore, Fouda et al. (2022) also reported the treatment of tanning wastewater using zinc oxide nanoparticles. They reported the 96.5, 88.8, 88.5, and 91.5 % reduction in TSS, COD, BOD, and conductivity of wastewater treated with zinc oxide nanoparticle. Moreover, zinc oxide nanoparticles also lowered the Cr (VI) level from wastewater by 93.4 % (Fouda et al., 2022). On the other hand, Noman et al. (2020) also treated the textile wastewater with the help of Cu-NPs and discussed the decrease in different parameters like pH, turbidity, COD, TDS, TSS, hardness, chloride, and sulphates before and after the treatment. The reduction efficieny of Cu-NPs from wastewater can be attributed to functional groups, surface structure and chemical components that are attached to them (Dlamini et al., 2019; Al-Hakkani, 2020). So, the abovementioned results suggested that biosynthesized Cu-NPs might serve as an excellent photocatalyst for the treatment of textile wastewater loaded with dyes.
Wastewater treatment
pH
EC
(dS m−1)
TDS
(mgL−1)
COD
(mgL−1)
Sulphates
(mgL−1)
Phosphates
(mgL−1)
Color intensity
(nm)
Pre-Treatment
8.6
7.31
1940
1644
182.34
27.64
0.4
Post-Treatment
6.9
2.98
821
992
103.65
13.96
0.02
% Removal
−
−
57.68
39.66
43.16
49.49
95.97
4 Conclusion
Based on the findings of the current study, it might be concluded that biosynthesized Cu-NPs might serve as potential agent not only for photocatalytical degradation of different textile azo dyes but also for treatment of actual wastewaters by lowering sulphates, phosphates, color intensity, pH, EC, TDS, and COD. Therefore, the current study emphasizes on photocatalytic application of biosynthesized Cu-NPs in the decolorization of azo dyes on commercial level.
Ethics approval
This article does not contain studies with human participants or vertebrates performed by any of the authors.
Consent for publication
All authors give consent for publication of this original article.
Funding
The funds for the present research work were provided by Higher Education Commission (HEC) of Pakistan under the project No. 8206/Punjab/NRPU/R&D/HEC/2017 and the project number (RSP2024R469), King Saud University, Riyadh, Saudi Arabia.
CRediT authorship contribution statement
Fatima Batool: Writing – original draft, Formal analysis, Data curation. Muhammad Shahid: Writing – review & editing, Formal analysis, Conceptualization. Faisal Mahmood: Writing – review & editing, Resources, Methodology, Formal analysis. Tanvir Shahzad: Writing – review & editing, Methodology, Formal analysis. Farrukh Azeem: Writing – review & editing, Methodology, Investigation, Conceptualization. Sabir Hussain: Writing – review & editing, Supervision, Methodology, Funding acquisition, Formal analysis, Conceptualization. Tahani Saad Algarni: Writing – review & editing, Conceptualization. Mohamed S. Elshikh: Writing – review & editing, Funding acquisition, Data curation, Conceptualization. Wed A. Al.Onazi: Writing – review & editing, Conceptualization. Sadia Mustafa: Writing – review & editing, Resources, Data curation.
Acknowledgement
The authors extend their appreciation to the researchers supporting project number 8206/Punjab/NRPU/R&D/HEC/2017 funded by the Higher Education Commission of Pakistan and the project No. RSP2024R469 funded by King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Green synthesis of copper oxide nanoparticles for biomedical application and environmental remediation. Heliyon. 2020;6:e04508.
- [Google Scholar]
- Biogenic copper nanoparticles and their applications: a review. SN Appl. Sci.. 2020;2(3):505.
- [Google Scholar]
- Remediation of wastewater using various nano-materials. Arab. J. Chem.. 2019;12(8):4897-4919.
- [CrossRef] [Google Scholar]
- Effects of operational parameters on the removal of acid blue 25 dye from aqueous solutions by electrocoagulation. Appl. Chem. Eng.. 2019;30:742-748.
- [CrossRef] [Google Scholar]
- Ball, A.S., Patil, S., Soni, S., 2019. Introduction into nanotechnology and microbiology. In: Methods in microbiology, vol. 46, Academic Press, pp. 1-18.
- In vitro evaluation of antibacterial and antifungal activity of biogenic silver and copper nanoparticles: The first report of applying biogenic nanoparticles against Pilidium concavum and Pestalotia sp. fungi. Molecules. 2021;26(5402)
- [CrossRef] [Google Scholar]
- Introduction to industrial wastes containing organic and inorganic pollutants and bioremediation approaches for environmental management. Bioremediation Ind. Waste Environ. Saf. Vol. I Ind. Waste Its Manag.. 2020;1–18
- [Google Scholar]
- Green synthesis of iron oxide nanoparticles using pomegranate seeds extract and photocatalytic activity evaluation for the degradation of textile dye. J. Mater. Res. Technol.. 2019;8:6115-6124.
- [CrossRef] [Google Scholar]
- Analysis of aazoreductase gene harbored by Alcaligenes sp. YB4 capable of concurrent removal of sulphonated azo dye and hexavalent chromium. Asian J. Agri. Biol. 2023
- [Google Scholar]
- A facile synthesis of Cu2O and CuO nanoparticles via sonochemical assisted method. Curr. Nanosci.. 2019;15(2):209-213.
- [CrossRef] [Google Scholar]
- Biosynthesis of copper oxide nanoparticles using Streptomyces MHM38 and its biological applications. J. Nanomater.. 2021;2021:1-16.
- [CrossRef] [Google Scholar]
- Optimization and application of bioflocculant passivated copper nanoparticles in the wastewater treatment. Int. J. Environ. Res. Public Health. 2019;16:2185.
- [CrossRef] [Google Scholar]
- Reductive-degradation of carcinogenic azo dyes using Anacardium occidentale testa derived silver nanoparticles. J. Photochem. Photobiol. B Biol.. 2016;162:604-610.
- [CrossRef] [Google Scholar]
- Synthesis of eco-friendly copper nanoparticles for augmentation of catalytic degradation of organic dyes. J. Mol. Liq.. 2018;260:1-8.
- [CrossRef] [Google Scholar]
- Phyco-synthesized zinc oxide nanoparticles using marine macroalgae, Ulva fasciata Delile, characterization, antibacterial activity, photocatalysis, and tanning wastewater treatment. Catalysts. 2022;12(7):756.
- [CrossRef] [Google Scholar]
- Citrus paradisi fruit peel extract mediated green synthesis of copper nanoparticles for remediation of Disperse Yellow 125 dye. Desalin. Water Treat.. 2021;212:368-375.
- [CrossRef] [Google Scholar]
- Extracellular synthesis of copper nanoparticles using culture supernatants of Salmonella typhimurium. Orient. J. Chem. 2015;31(1):527-529.
- [Google Scholar]
- Green synthesis of zinc oxide nanoparticles using Bacillus subtilis ZBP4 and their antibacterial potential against foodborne pathogens. Prep. Biochem. Biotech.. 2023;53(3):255-264.
- [CrossRef] [Google Scholar]
- Endophytic actinomycetes Streptomyces spp mediated biosynthesis of copper oxide nanoparticles as a promising tool for biotechnological applications. JBIC J. Biol. Inorg. Chem.. 2019;24:377-393.
- [CrossRef] [Google Scholar]
- Zero-valent copper nanoparticles for effective dechlorination of dichloromethane using sodium borohydride as a reductant. Chem. Eng. J.. 2012;203:95-100.
- [CrossRef] [Google Scholar]
- Biodecolorization of reactive black-5 by a metal and salt tolerant bacterial strain Pseudomonas sp. RA20 isolated from Paharang drain effluents in Pakistan. Ecotoxicol. Environ. Saf.. 2013;98:331-338.
- [CrossRef] [Google Scholar]
- Microbial biotechnology for decolorization of textile wastewaters. Rev. Environ. Sci. Bio/technology. 2015;14:73-92.
- [CrossRef] [Google Scholar]
- Biosynthesis of titanium dioxide nanoparticles using Bacillus amyloliquefaciens culture and enhancement of its photocatalytic activity for the degradation of a sulfonated textile dye Reactive Red 31. J. Colloid Interface Sci.. 2016;475:184-191.
- [CrossRef] [Google Scholar]
- Biosynthesis and characterization of copper nanoparticles using Shewanella oneidensis: application for click chemistry. Small. 2018;14:1703145.
- [CrossRef] [Google Scholar]
- Synthesis of nickel nanoparticles using Citrullus colocynthis stem extract for remediation of Reactive Yellow 160 dye. Environ. Sci. Pollut. Res.. 2020;27:32998-33007.
- [CrossRef] [Google Scholar]
- Seed-mediated synthesis of monodispersed Cu2O nanocubes with five different size ranges from 40 to 420 nm. Adv. Funct. Mater.. 2007;17:3773-3780.
- [CrossRef] [Google Scholar]
- Enhanced decolorization of methyl orange using zero-valent copper nanoparticles under assistance of hydrodynamic cavitation. Ultrason. Sonochem.. 2015;22:132-138.
- [CrossRef] [Google Scholar]
- Facile synthesis and shape evolution of single-crystal cuprous oxide. Adv. Mater.. 2009;21:2068-2071.
- [CrossRef] [Google Scholar]
- Preparation and photocatalytic activity of Cu2O nanoparticles. Mater. Sci.. 2010;28:503-511.
- [Google Scholar]
- Biosynthesis of copper nanoparticles using Shewanella loihica PV-4 with antibacterial activity: novel approach and mechanisms investigation. J. Hazard. Mater.. 2018;347:141-149.
- [CrossRef] [Google Scholar]
- spectrophotometric evaluation of biosynthesized copper nanoparticles using Allium cepa, Azadirachta indica and Moringa oleifera plant extracts. Adamawa State University J. Sci. Res.. 2016;04(2):2251.
- [Google Scholar]
- Application of environmental bacteria as potential methods of azo dye degradation systems. Glob. J. Environ. Sci. Manag.. 2021;7:131-154.
- [Google Scholar]
- Use of RSM modeling for optimizing decolorization of simulated textile wastewater by Pseudomonas aeruginosa strain ZM130 capable of simultaneous removal of reactive dyes and hexavalent chromium. Environ. Sci. Pollut. Res.. 2016;23:11224-11239.
- [CrossRef] [Google Scholar]
- The biosynthesis of nickel oxide nanoparticles: an eco-friendly approach for azo dye decolorization and industrial wastewater treatment. Sustainability. 2023;15(20):14965.
- [CrossRef] [Google Scholar]
- Cu nanoparticles synthesis using biological molecule of P. granatum seeds extract as reducing and capping agent: growth mechanism and photo-catalytic activity. Int. J. Biol. Macromol.. 2018;106:1203-1210.
- [CrossRef] [Google Scholar]
- Green synthesis of copper nanoparticles using different plant extracts and their antibacterial activity. J. Environ. Chem. Eng.. 2022;10(2):107130
- [CrossRef] [Google Scholar]
- Use of biogenic copper nanoparticles synthesized from a native Escherichia sp. as photocatalysts for azo dye degradation and treatment of textile effluents. Environ. Pollut.. 2020;257:113514
- [CrossRef] [Google Scholar]
- Microbial-mediated copper nanoparticles synthesis, characterization, and applications. In: Agri-Waste and Microbes for Production of Sustainable Nanomaterials. Elsevier; 2022. p. :507-533.
- [CrossRef] [Google Scholar]
- Pham-Khanh, N.-H., Huynh, N.-Q., Le, H.-N.-B., Ha, T.-K.-Q., 2023. Green sunthesis of zinc oxide microparticles using the leaf extract of Dolichandrone spathacea in sustainable agriculture: a new approach for protecting the legume plant (Vigna radiata) against the Cr(VI) stress. Asian J. Agri. Biol. https://www.asianjab.com/wp-content/uploads/2024/02/AJAB-2023-245.pdf.
- Synthesis of anisotropic silver nanoparticles using novel strain, Bacillus flexus and its biomedical application. Colloids Surf. B Biointerfaces. 2013;102:232-237.
- [CrossRef] [Google Scholar]
- Anti-bacterial and anti-biofilm properties of green synthesized copper nanoparticles from Cardiospermum halicacabum leaf extract. Bioprocess Biosyst. Eng.. 2020;43:1649-1657.
- [CrossRef] [Google Scholar]
- Effect of incubation time, CuSO4 and glucose concentrations on biosynthesis of copper oxide (CuO) nanoparticles with rectangular shape and antibacterial activity: Taguchi method approach. Nano Biomed. Eng.. 2018;10:25-33.
- [CrossRef] [Google Scholar]
- Rasool, M., Rasool, M.H., Khursheed, M., Aslam, B., 2023. Biogenic synthesis and characterization of silver nanoparticles: exploring antioxidant and anti-inflammatory activities and assessing antimicrobial potential against multidrug-resistant bacteria. Asian J. Agri. Biol. https://www.asianjab.com/wp-content/uploads/2024/03/AJAB-2023-364.pdf.
- Laboratory Manual for the Examination of Water, Waste Water and Soil. Vol No. Ed. 3. Wiley-VCH Verlag GmbH; 1999.
- Synthesis of silver nanoparticles using bacterial exopolysaccharide and its application for degradation of azo-dyes. Biotechnol. Rep.. 2017;15:33-40.
- [CrossRef] [Google Scholar]
- Comparative efficacy of biogenic zinc oxide nanoparticles synthesized by Pseudochrobactrum sp. C5 and chemically synthesized zinc oxide nanoparticles for catalytic degradation of dyes and wastewater treatment. Environ. Sci. Pollut. Res.. 2021;28:28307-28318.
- [CrossRef] [Google Scholar]
- Synthesis of stable cadmium sulfide nanoparticles using surfactin produced by Bacillus amyloliquifaciens strain KSU-109. Colloids Surf. B Biointerfaces. 2011;85(2):207-213.
- [CrossRef] [Google Scholar]
- An overview of textile dyes and their removal techniques: Indian perspective. Pollut. Res.. 2017;36:790-797.
- [Google Scholar]
- Biological synthesis of copper oxide nano particles using Escherichia coli. Curr. Nanosci.. 2010;6–4:365-369.
- [CrossRef] [Google Scholar]
- Souza, A.G.V., Maria, T.C., Saran, L.M., Alves, L.M.C., 2022. Enzymatic Bioremediation of Dyes from Textile Industry Effluents. Doi: 10.5772/intechopen.103064.
- Photocatalytic membranes: a new perspective for persistent organic pollutants removal. Environ. Sci. Pollut. Res.. 2022;29:12506-12530.
- [CrossRef] [Google Scholar]
- Extracellular biosynthesis of iron oxide nanoparticles by Bacillus subtilis strains isolated from rhizosphere soil. Biotechnol. Bioprocess Eng.. 2012;17:835-840.
- [CrossRef] [Google Scholar]
- Microbial degradation of azo dyes by textile effluent adapted, Enterobacter hormaechei under microaerophilic condition. Microbiol. Res.. 2021;250:126805
- [CrossRef] [Google Scholar]
- Biosynthesis of copper nanoparticles using copper-resistant Bacillus cereus, a soil isolate. Process Biochem.. 2016;51:1348-1356.
- [CrossRef] [Google Scholar]
- Biocompatible synthesis of peptide capped copper nanoparticles and their biological effect on tumor cells. Mater. Chem. Phys.. 2011;128:83-89.
- [CrossRef] [Google Scholar]
- 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol.. 1991;173(2):697-703.
- [CrossRef] [Google Scholar]
- Cu2O nanocrystals: surfactant-free room-temperature morphology-modulated synthesis and shape-dependent heterogeneous organic catalytic activities. J. Phys. Chem. C. 2011;115:15288-15296.
- [CrossRef] [Google Scholar]
- Synthesis of copper nanoparticles in presence of microorganisms. Eur. J. Mol. Clin. Med.. 2020;7:1141-1147.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103309.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







