Translate this page into:
Natural occurrence, infection dynamics, and molecular characterization of nucleopolyhedrovirus (SpfrNPV) infecting fall armyworm, Spodoptera frugiperda (J. E. Smith) from maize ecosystems in Gujarat, India
⁎Corresponding author. raghumic2@gmail.com (Boodanur Lingaiah Raghunandan),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
This comprehensive study investigates the stage-wise infection dynamics of Nucleopolyhedrovirus (NPV) in fall armyworm, Spodoptera frugiperda (J. E. Smith) larvae across distinct stages of maize crop in Panchmahal and Mahisagar districts of Gujarat, India during the kharif season in 2021. The examination of month-wise mean data reveals a pronounced pattern of NPV prevalence, with the highest infection observed during the cob formation stage (28.74 %), followed by flowering and tasseling (20.23 %), and the vegetative stage (13.93 %). Block specific analyses highlight variations in infection rates, with notable differences between districts and stages. Correlation coefficient analysis indicates a significant and positive relationship between specific crop stages and NPV infection. Regression equations are established, demonstrating a positive increase in natural NPV infection during different crop stages. Weather parameter analysis reveals significant correlations between temperature, humidity, sunshine hours, and NPV infection. The phylogenetic analysis of NPV polyhedrin sequences indicates species-specific clustering within S. frugiperda and a broader genus-specific association. Motif pattern analysis identifies conserved motifs in NPV polyhedrins. Protein structure predictions and Molecular docking studies predicted interactions between polyhedrin and chitinase, suggesting a potential role in the insect's defense against NPV. The bioassay results indicated increased susceptibility of early larvae to NPV, emphasizing the potential for optimizing biocontrol strategies. Overall, this study contributes valuable insights into the ecological and molecular aspects of NPV infection dynamics, offering a significance understanding for developing effective pest management strategies and also underscores its significance in promoting sustainable agriculture.
Keywords
Survey
Natural occurrence
NPV
Fall armyworm
Molecular assay
Docking
- FAW
-
Fall armyworm
- NPV
-
Nucleopolyhedrovirus
- OB
-
Occlusion bodies
- AAU
-
Anand Agricultural University
- AICRP
-
All India Coordinated Research Project
- SDS
-
Sodium dodecyl sulfate
- CTAB
-
cetyltrimethylammonium bromide
- SDW
-
sterile distilled water
- polh
-
polyhedrin gene
- ORFs
-
open reading frames
- LC50
-
lethal concentrations
- CDS
-
coding region
- GO
-
Gene Ontology
Abbreviations
1 Introduction
Maize (Zea mays L.) have originated in the western hemisphere around 7000–10000 years ago. Renowned as the 'queen of cereals' due to its versatility, it is the main staple food for the tribal communities in the Panchmahal and Mahisagar districts of middle Gujarat. Traditionally, residents consume maize in the form of chapati as their primary food source. Key maize-growing districts in Gujarat include Panchmahal, Dahod, Mahisagar, Vadodara, Chhotaudepur, Arvalli, Sabarkantha, Banaskantha, and Anand. The introduction of recent pests, such as the fall armyworm (FAW) or Spodoptera frugiperda (J. E. Smith), has become a global challenge (Kenis et al., 2022). Originally native to the neotropics, FAW was first identified as an invasive pest in Africa in the 1960s, spreading to various parts of the continent and targeting a wide range of plant species, with a preference for those in the Poaceae family (Abbas et al., 2022). Maize production in India reached 28.7 million metric tonnes in 2017, but the fall armyworm's impact led to a 3.2 % reduction in production (Deshmukh et al., 2021). The management of S. frugiperda in maize predominantly relies on chemical insecticides, which pose significant drawbacks despite their widespread use. These chemical solutions lack specificity, exhibit high toxicity, and raise concerns regarding their long-term impact. The sustained use of these insecticides can result in adverse consequences, including developing resistant pest populations, declining beneficial insects, and detrimental environmental effects (Abbas et al., 2023). One noteworthy concern is the potential emergence of resistant populations of S. frugiperda due to the continuous use of chemical insecticides. Additionally, the indiscriminate application of these chemicals can reduce beneficial insects, disrupting the ecosystem's natural balance. In light of these issues, there is a growing recognition of the need for alternative and more sustainable approaches to pest control in maize cultivation. Exploring methods such as biological control, integrated pest management, and environmentally friendly pesticides can offer effective solutions while minimizing the negative consequences of the continuous use of chemical insecticides (Abbas et al., 2023). Biopesticides, with their minimal residual effects and lower threat of bioaccumulation, are gaining acceptance, particularly in organic farming. The nucleopolyhedrovirus (NPVs) from the Baculoviridae family are gaining attention as potential microbial pesticides. Several NPVs have successfully controlled insect pests in agriculture (Sivakumar et al., 2020). Considering the detrimental effects of fall armyworm on both fodder and food crops in India, where the use of insecticides is discouraged, evaluating the natural presence of nucleopolyhedrovirus on fall armyworm becomes paramount importance for maize cultivation. Fall armyworm infestations significantly jeopardize maize crops, resulting in considerable economic losses. This study aims to investigate the influence of weather parameters, including temperature, humidity, and sunlight exposure on the natural occurrence of Nucleopolyhedrovirus (NPV) on fall armyworm populations within maize ecosystems. Additionally, characterizing the genetic structure, phylogeny, and protein interactions of SpfrNPV strains can provide insights into their effectiveness as biocontrol agents. Bioassays help assess the efficacy of isolated NPV strains in controlling fall armyworm populations, contributing to sustainable pest management practices in agriculture.
2 Materials and methods
During kharif,2021 survey was conducted in all seven blocks of Panchmahal (Godhra, Shehera, Halol, Kalol, Morwa, Jambughoda and Ghoghamba) and four blocks of Mahisagar district (Santrampur, Kadana, Khanpur and Lunawada) to study the relationship between weather parameters and natural NPV infection on larvae of fall armyworm, S. frugiperda infesting maize on farmer's fields during July, August, and September (Supplementary file 1; Table. S1; Fig. S1; Fig.S2). Among the different selected blocks, 5 villages were selected and in each village one maize field was randomly selected (Additional details in Supplementary file 3).
2.1 Isolation of NPV occlusion bodies and data analyses
The collected larvae were brought to the laboratory and stored at −20 °C and then evaluation was carried out to find percent infected larvae from collected larvae. The infected larvae were dissected and wet smears were prepared to examine under phase contrast microscope for the presence of occlusion bodies (OBs). The occlusion bodies (OBs) were isolated by following the standard methodology (Hussain et al., 2019) with few modifications (Additional details in Supplementary file 3).
2.1.1 Purification of polyhedron occlusion bodies (POBs) produced in the cadavers of Spodoptera frugiperda nucleopolyhedrovirus (SpfrNPV) infected larvae
Dead larvae infected with the virus were collected and homogenized in sterile water. Polyhedra inclusion bodies (PIBs) were purified through triple centrifugation andresuspended in sterile water with 0.002 % sodium azide, and homogenized in 0.5 % SDS at 37 °C for 2 h (Sivakumar et al., 2020) (Additional details in Supplementary file 3).
2.2 Virus DNA extraction
The total DNA was extracted from larvae exhibiting symptoms of NPV disease using the cetyltrimethylammonium bromide (CTAB) method, following the protocol (Sivakumar et al., 2020). Subsequently, all DNA extracts underwent a 1:100 dilution in sterile distilled water (SDW) before being utilized in RCA amplifications (Pavan et al., 2024). Quantification and adjustment of the isolated DNA concentration to 0.1 g/l were performed using a NanoDropTM 1000 Spectrophotometer from ThermoScientific, USA. Verification of the size and purity of the DNA was achieved by loading 1 µl of the sample DNA and 1 µl of control genomic DNA onto a 0.6 % agarose/EtBr gel, along with a 1 kb Plus DNA ladder. This gel electrophoresis confirmed the size and purity of the extracted DNA (Additional details in Supplementary file 3).
2.3 PCR amplification of polyhedrin gene of SpfrNPV strain AAUBC1 and AP2
In this study, the conserved polyhedrin gene (polh) of NPV was targeted using species-specific primers designed from the NCBI database. The PCR reaction, following Thermo Scientific's protocol, employed 35 cycles to amplify the marker (Sivakumar et al., 2020). The amplified products were visualized on a 1.2 % ethidium bromide-stained agarose gel, alongside Gene Ruler 100 bp DNA ladder, confirmed fragment sizes. Sequencing at Barcode Bioscience, Bengaluru, India, facilitated SpfrNPV identity confirmation. BLAST analysis against NCBI GenBank verified nucleotide sequence homology. The sequences were deposited in GenBank with accession numbers PP025827 and PP025828 for the isolated SpfrNPV strains AAUBC1 and AP2 (Additional details in Supplementary file 3).
2.4 Gene structure prediction
The gene structure, including exons and introns, was determined by comparing of open reading frames (ORFs) and the SpfrNPV polyhedrin genes (Guo et al. 2007). As described, the structural details were visualized using GSDS 2.0 (https://gsds.cbi.pku.edu.cn) Fig. 1.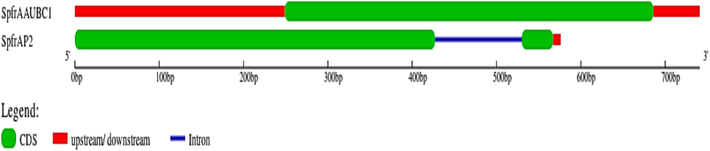
Gene structure schematic diagram for SpfrNPV AAUBC1 and AP2 polyhedrin genes. Exons were demonstrated by filled green boxes and untranslated region(UTR) was displayed red boxes, and introns represented by dark blue lines.
2.5 Phylogenetic analysis
A total of 102 polyhedrin sequences were utilized to construct the phylogenetic tree. The sequences were aligned using the Clustal W program, employing default gap penalty parameters with a gap opening of 10 and an extension of 0.2. Subsequently, a neighbor-joining tree was constructed using MEGA 6.0, adopting a p-distance model and implementing pairwise deletion of gaps (Tamura et al., 2013). To assess the robustness of the tree branches, bootstrap support was determined by resampling amino acid positions 1000 times. Additionally, conserved domains within the identified SpfrNPV strain AAUBC1 and AP2 were predicted using the SMART tool (https://smart.embl-heidelberg.de/), as outlined by Letunic et al. (2014) (Supplementary file 1; Fig.S3 & Fig. 2).
2.6 Motif analysis
Following the BLAST results, the protein identified as a key player in disease incidence, specifically the symptom-determinant protein, was subjected to motif discovery and pattern analysis. The MEME software, version 4.12.0, available on the online server (https://meme-suite.org/index.html), was employed for this purpose. In this analysis, the parameters used for motif discovery were set as follows: minimum width = 6, maximum width = 10, and the maximum number of motifs to find = 6. These parameters guided the MEME software in identifying and analyzing motifs within the protein sequence, providing valuable insights into potential functional patterns associated with the disease-related protein.
2.7 Predicting protein structures for SpfrNPV AAUBC1 and AP2 polyhedrin from S. frugiperda
For the prediction of the structure of the identified SpfrNPV AAUBC1 and AP2 polyhedrin, we employed Phyre2 (Protein Homology/Analogy Recognition Engine V 2.0), following the methodology (Rudra Gouda et al., 2023). The prediction process was conducted using the online tool available at https://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index. The prediction involved the utilization of intensive mode and ab initio techniques to achieve comprehensive protein modeling and to explore potential functional roles. Batch processing in expert mode allowed for simultaneous sequence processing, enhancing efficiency. While alternatives such as the Swiss model and I-Taser were considered, Phyre2 was chosen for its superior modeling capabilities. The quality of the predicted models was assessed using various tools, including SAVES v5.0 (https://servicesn.mbi.ucla.edu/SAVES), ProSA, and Qmean. These assessments provided valuable insights into the reliability and accuracy of the predicted protein structures.
2.8 In-silico docking
2.8.1 Preparing protein of SpfrNPV AAUBC1 and AP2 polyhedrin from S. frugiperda
To unravel the molecular intricacies governing the interaction between NPV polyhedrin and the chitinase enzyme, computational docking was employed. The focus centered on chitinase, a pivotal enzyme in polyhedrin degradation within the insect gut during later instars, crucial for larval survival against NPV infection. The computational docking of S. frugiperda gut chitinase involved systematically constructing protein structures for SpfrNPV AAUBC1 and AP2 polyhedrin. The aim was to identify compounds with high-energy binding. Constructed protein structures adhered to a meticulously outlined methodology and underwent thorough structural validation, ensuring precision and integrity. Validation criteria included Ramachandran outliers, sidechain outliers, RSRZ outliers, clash score evaluation for steric clashes, and Rfree measurements for fit to experimental data. This comprehensive validation strategy enhances the study's credibility, utilizing a diverse set of tools and criteria for reliable findings (Supplementary file 1; Fig. S4). The protein structure from the Protein Data Bank (PDB) was downloaded in PDB format and prepared for docking using the Auto Dock Vina plugin for PyMOL, following the procedure outlined (Seeliger and de Groot, 2010) (Additional details in Supplementary file 3).
2.9 Bioassay
The determination of the median lethal concentration (LC50) of the isolated strain of SpfrNPV AAUBC1 on 2nd (3–5 days old), 3rd (6–7 days old), and 4th (6–7 days old) instar larvae was carried out through a leaf disc bioassay method (Magholi et al., 2014) with certain modifications (Additional details in Supplementary file 3). The mortality rates of the larvae were documented every 24 h for a duration of up to 9 days post-inoculation Median lethal concentrations (LC50) were determined by probit analysis using SPSS software. The mortality rate of targeted insects was corrected using Abbott’s formula (Abbott, 1925). SPSS 21.0 software was used for all the analyses.
3 Results
3.1 Stage-wise NPV infection of fall armyworm in maize of Panchmahal and Mahisagar district during kharif, 2021
An extensive study in the Panchmahal and Mahisagar districts assessed the prevalence of nucleopolyhedrovirus (NPV) infection in S. frugiperda larvae during maize crop development (Supplementary file 1; Table S2 & Table 2). NPV prevalence peaked during cob formation (28.74 %), followed by flowering and tasseling (20.23 %), and the vegetative stage (13.93 %) (Table 1). Block-specific analyses revealed variations, with Jambughoda having the highest vegetative stage infection (20.41 %) and Khanpur the lowest (9.09 %). Ghoghamba showed the highest NPV infection during flowering and tasseling (27.47 %), while Santrampur had the lowest (15.79 %). Godhra had the highest infection during cob formation (34.44 %), and Morwa had the lowest (24.66 %). Correlation analysis found significant positive relationships between NPV infection and different crop stages (Table 2). Regression equations quantified these relationships (Table 2). Month-wise mean data analysis elucidated the relationship between weather conditions and NPV infection, with the highest infection occurring in September (28.74 %), followed by August (20.23 %) and July (13.93 %) (Supplementary file 1; Table S3). SE: Standard Error; %-percentage; Data was subjected to analysis using SPSS software 21. Note: ** Significant at 1 % level; r- Correlation co-efficient; y- Dependent Variable; x- independent variable; R2- proportion of the variance in the dependent variable that is explained by the independent variables: Data was subjected to Correlation analysis using SPSS software 21.
Blocks
Vegetative Stage
Flowering and Tasseling Stage
Cob formation Stage
Mean No. of larvae collected (5 villages)
Mean No. of NPV infected larvae
observed (5 villages)Average NPV infection (%) (5 villages)
Mean No. of larvae collected (5 villages)
Mean No. of NPV infected larvae
observed (5 villagesAverage NPV infection (%) (5 villages)
Mean No. of larvae collected (5 villages)
Mean No. of NPV infected larvae
observed (5 villagesAverage NPV infection (%) (5 villages)
Godhra
8.4
1.2
14.55
15.6
3.6
23.08
18
6.2
34.44
Shehera
5.4
0.8
14.81
11.2
1.8
16.07
11.8
3.4
28.81
Santrampur
6.4
0.6
9.38
11.4
1.8
15.79
16.2
4.4
27.16
Lunawada
9
1.2
13.33
12
2.4
20
14
3.8
27.14
Halol
7.4
0.8
10.81
14
2.8
19.4
14
4
28.57
Kalol
6.2
0.6
9.68
13.8
2.4
17.39
17.8
4.8
26.97
Morwa
5.4
0.6
11.11
13.4
2.2
16.42
14.6
3.6
24.66
Kadana
7.6
1
13.16
12.4
2.2
16.13
12.6
3.4
26.98
Khanpur
4.4
0.4
9.09
13.6
2.8
20.59
17.2
4.6
26.74
Jambughoda
9.8
2
20.41
19.4
4.6
23.71
17.2
5.2
30.23
Ghoghamba
10.4
2
19.23
18.2
5
27.47
19.2
6.2
32.29
(Mean ± SE)
7.32 ± 0.558
1.02 ± 0.157
13.93 ± 1.101
14.09 ± 0.765
2.85 ± 0.310
20.23 ± 1.09
15.69 ± 0.695
4.51 ± 0.292
28.74 ± 0.807
Sl. No.
Stages of crop
Correlation co-efficient (r)
Regression equation
R2
1
Vegetative
r = 0.783**
Y = 1.95 + (1.540) x
0.613
2
Flowering and Tasselling
r = 0.856**
Y = 2.46 + (1.22) x
0.732
3
Cob formation
r = 0.484
Y = 19.71 + (0.56) x
0.234
3.2 Correlation between weather parameters and NPV infection in larvae of fall armyworm, S. frugiperda, infesting maize
The detailed analysis of weather parameters during the kharif season in 2021 sheds light on the correlation between environmental conditions and nucleopolyhedrovirus (NPV) infection in fall armyworm (S. frugiperda) larvae (Table 3). Maximum temperature displayed a non-significant but negative correlation (r = −0.959), suggesting that elevated temperatures are associated with decreased natural NPV infection. In contrast, minimum temperature exhibited a negative and significant correlation (r = −0.998*), indicating an inverse relationship with lower temperatures and enhanced NPV infection rates. Morning relative humidity showed a highly significant positive association (r = 0.999**), emphasizing the crucial role of high morning humidity in facilitating increased NPV infection rates. Evening relative humidity exhibited a positive association (r = 0.899), though non-significant, suggesting that higher evening humidity is linked to elevated NPV infection. Conversely, a negative but non-significant correlation between sunshine hours and NPV infection implies that increased sunlight exposure may be associated with decreased natural NPV infection. The analysis of rainfall indicated a non-significant negative correlation (r = −0.768), suggesting that higher rainfall is not strongly associated with enhanced NPV infection in fall armyworm larvae. Regression equations for temperature, humidity, and sunshine hours provide quantitative relationships (Table 3). For example, the equation Y = 153.26 + (−4.044) × (R2 = 0.920) for maximum temperature indicates a corresponding decrease in natural NPV infection for every unit increase in temperature. Similarly, equations for morning and evening relative humidity express an increase in NPV infection for every unit rise in humidity. The equation Y = 40.86 + (−6.77) × (R2 = 0.99) for sunshine hours conveys a decrease in natural NPV infection with every unit increase in sunlight exposure. Note: * = Significance at 5 %, ** = 1 % level; °C- Degree Celsius; %- Percentage; hr.- hours; mm- milli meter; r- Correlation co-efficient; y- Dependent Variable; x- independent variable; R2- proportion of the variance in the dependent variable that is explained by the independent variables: Data was subjected to Correlation analysis using SPSS software 21.
Sr. No.
Weather parameters
Correlation
co-efficient (r)Regression equation
R2
1
Temperature (°C)
Maximum
−0.959
Y = 153.26 + (−4.044) x
R2 = 0.920
Minimum
−0.998*
Y = 335.46 + (−12.07) x
R2 = 0.997
2
Relative humidity (%)
Morning
0.999**
Y = −210.64 + (2.60) x
R2 = 0.999
Evening
0.899
Y = −65.016 + (1.17) x
R2 = 0.808
3
Sunshine (hr.)
−0.996
Y = 40.86 + (−6.77) x
R2 = 0.99
4
Rainfall/week (mm)
−0.768
Y = 24.51 + (−0.06) x
R2 = 0.59
3.3 Prediction of domains and their function in SpfrNPV strain AAUBC1 and AP2 in S. frugiperda
Utilizing SMART bioinformatics and subsequently validating through the conserved domain search tool of NCBI, conserved domains in the polyhedrin of SpfrNPV strain AAUBC1 and AP2 from S. frugiperda were predicted. The analysis, conducted on the amino acid sequences of the coding region (CDS), revealed the presence of two functional domains in both polyhedrins. The predominant domains identified in SpfrNPV AAUBC1 and AP2 polyhedrins were associated with the polyhedrin pfam family. This suggests a conserved structural element within the polyhedrin protein. The inference drawn from this observation is that the polyhedrin family serves a Gene Ontology (GO) function, specifically contributing to structural molecule activity. This functional annotation was corroborated by SMART Bioinformatics (Letunic et al., 2014), supporting the structural role of the polyhedrin family. To visually represent these domains and their functions, the Quick GO EMBL tool was employed, and the annotated domains are illustrated (Supplementary file 1; Fig.S3). This figure comprehensively depicts the predicted domains and their attributed functions within the context of structural molecule activity. These findings contribute to understanding the functional aspects of polyhedrin in the context of S. frugiperda Nucleopolyhedrovirus.
3.4 Gene structure of SpfrNPV AAUBC1 and AP2 polyhedrin genes
The gene structure prediction for the SpfrNPV AAUBC1 and AP2 polyhedrin genes was accomplished using GSDS 2.0 software. The intron structure was deduced in this process, and exon boundaries containing functional domains (polyhedrin) were identified in the NPV polyhedrin gene. The genomic organization of polyhedrin genes was elucidated through this analysis. Remarkably, the majority of polyhedrin genes are covered by functional exons. A Single functional exon was found in SpfrNPV AP2 polyhedrin. However, SpfrNPV AAUBC1 polyhedrin has two exons separated by an intron, constituting a non-coding region. This structural feature, commonly referred to as interrupted genes, is highlighted (Fig. 1). Investigating the exon–intron arrangement within the polyhedrin gene of NPV provides a promising avenue for gaining new insights into the evolutionary trajectory of this gene family.
3.5 Phylogenetic analysis of polyhedrin of SpfrNPV strain AAUBC1 and AP2
A neighbour-joining tree was constructed to elucidate the evolutionary relationships among polyhedrin sequences, incorporating sequences from two SpfrNPV strains, AAUBC1 and AP2, in addition to 100 sequences from various insect species (Supplementary File S2). The resulting phylogenetic tree demonstrated a distinct clustering, where all polyhedrin sequences from S. frugiperda formed a separate group, indicating a species-specific divergence (Fig. 2). Intriguingly, both SpfrNPV strains, AAUBC1 and AP2, clustered together with polyhedrin sequences from the same S. frugiperda species. This suggests a close evolutionary relationship between the polyhedrins of these two strains, reinforcing their shared genetic lineage. Furthermore, the analysis revealed that polyhedrin sequences from S. frugiperda share a notable evolutionary affinity with polyhedrins from other Spodoptera genus species, such as exigua, litura, eridania, and cosomioides. This observation underscores the genus-specific nature of NPV groups, indicating a distinctive evolutionary history within each genus. Notably, the variation among species within the same genus is evident, highlighting contrasting evolutionary trajectories. In summary, the phylogenetic analysis provides insights into the evolutionary relationships of SpfrNPV polyhedrin, emphasizing both species-specific clustering within S. frugiperda and a broader genus-specific association with polyhedrins from other Spodoptera species. This information contributes to our understanding of the evolutionary dynamics of NPV polyhedrins across different insect species.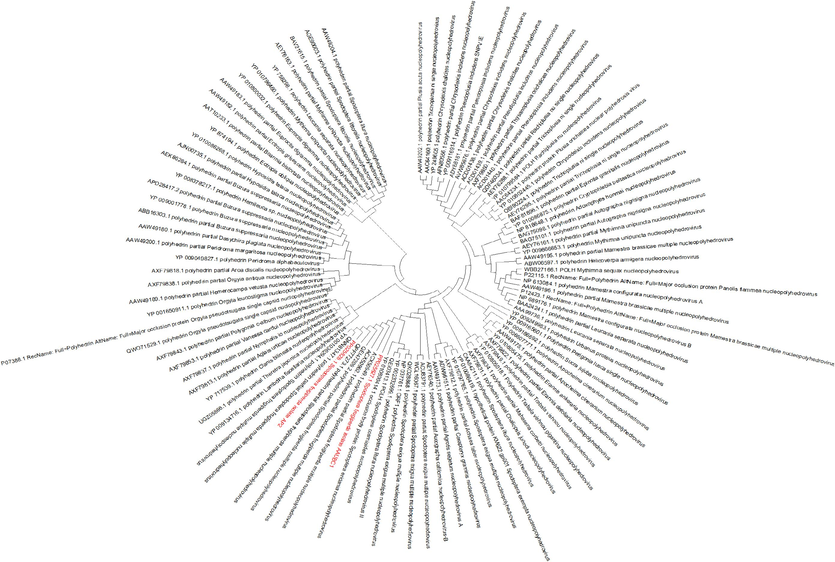
Phylogenetic analysis of the amino acid sequences of the NPV polyhedrin from different insect species (our accession is indicated in red color) in the context of various NPV Polyhedrin. The NPV polyhedrin was used to create a neighbor-joining tree, which was based on the nucleotide sequences of 102 different NPV isolates. Boots trap values were calculated with 1000 replications.
3.6 Motif pattern analysis of polyhedrin from SpfrNPV strain AAUBC1 and AP2
Motif analysis was conducted using MEME (version 4.12.0) with specific parameters to discern conserved motifs in the polyhedrin sequences of SpfrNPV strains AAUBC1 and AP2 from S. frugiperda. Employing a minimum width of six, a maximum width of 10, and limiting the identification to a maximum of six motifs ensured the stringent identification of biologically relevant motifs. The results revealed six distinct motifs in both NPV strain AAUBC1 and AP2 polyhedrins, denoted as AQHALRCDPD, MNLHSEYTHS, TWTRFMEDSF, SLAKKGGGCP, PIVNDQEIMD, and MRPTRPNRCF (Fig. 3A). Notably, all six motifs were conserved in both strains, indicating a shared motif pattern. Further analysis demonstrated an identical motif pattern in both strains, characterized by the presence of TWTRFMEDSF, PIVNDQEIMD, MRPTRPNRCF, AQHALRCDPD, SLAKKGGGCP, and MNLHSEYTHS. The statistical significance of this motif pattern was confirmed with p values of 4.61e-60 for NPV strain AAUBC1 and 1.11e-58 for AP2, underscoring the robust conservation of these motifs (Fig. 3B). This consistent motif pattern suggests a functional significance in the polyhedrin protein of SpfrNPV strains AAUBC1 and AP2, possibly contributing to their structural and functional roles in the viral life cycle. The stringent parameters applied in the motif analysis enhance the confidence in the biological relevance of the identified motifs.
Motif and protein structural details of polyhedrin of SpfrNPV strain AAUBC and AP2 from S. frugiperda. 3A: Motifs identified in polyhedrin of SpfrNPV strain AAUBC and AP2 from S. frugiperda. Motifs were discovered by using MEME tool. Heights of the symbols within a stack indicate the relative frequency of the amino acids at that position. Parameters used for motif discovery were: minimum width = 6, maximum width = 10, maximum number of motifs to find = 6. 3B: Motif pattern identified in polyhedrin of SpfrNPV strain AAUBC and AP2. Color boxes in figures are motifs and the pattern of that motif is indicated in color coded box at bottom. Both the polyhedrin motif pattern are less than p- value 0.0001 and are statistically significant. Length of the bar of each of polyhedrin indicates length of CDS region used in drawing motifs. The overall height of a stack indicates the degree of sequence conservation at that position. 3C: Ligand (Chitinase in red) interaction with the binding pocket in protein (Polyhedrin of S. frugiperda NPV in blue). (I) Surface structure (II) Stick structure.
3.7 Deducing the protein structure of SpfrNPV polyhedrin identified in S. frugiperda
Our study successfully employed the Phyre2 tool to predict the theoretical protein structure of S. frugiperda Nucleopolyhedrovirus (SpfrNPV) polyhedrin (Table 4). The SpfrNPV genes deduced structures, AAUBC1 and AP2, exhibited significant similarities, with 89 % and 83 % resemblance, respectively, to the crystal structure of the infectious baculovirus polyhedral protein from Wiseana signata NPV. The identified polyhedrin molecules displayed distinctive structural features, including an extended N-terminal alpha helix projecting from the main part of the molecule. Moreover, a compact β-sandwich domain with three-layered β sheets and peripheral small alpha helices was evident. These characteristics align with the structural findings reported by Mori et al. in 2010. As of now, the NMR/crystal structures of only one polyhedral NPV have been resolved and are accessible in Entrez's 3-D structure database at NCBI. However, the observed similarities with W. signata NPV polyhedral protein suggest a conserved structural pattern among polyhedrins in different NPVs. The limited availability of crystal structures emphasizes the need for further research to unravel the detailed structural features of polyhedrins. A more extensive database of crystal proteins will contribute to a comprehensive understanding of the structural intricacies, facilitating advancements in the study of NPV polyhedrins. Column 2 indicates the predicted NMR (nuclear magnetic resonance structure) of polyhedrin of SpfrNPV strain AAUBC1 and AP. Column 6 indicates the match of our polyhedrin of SpfrNPV strain AAUBC1 and AP protein structure to the respective NPV proteins of other insects. Column 8 shows the organism from which our polyhedrin structure matches the polyhedrin of that organism.
Gene name
NMR structure
Confidence
Coverage%
Identity%
Descriptor
Functional Keywords
Biological source
Total formula weight
Total number of polymer chains
SpfrNPV AAUBC1 and SpfrNPV AP2
100 %
81 %
89 % (AAUBC1)
83 % (AP2)Crystal structure of infectious baculovirus polyhedra
jelly-roll, disulfide bond, domain swapping, viral protein
Wiseana signata NPV
28801.88
1
3.8 Molecular docking studies
The interaction between the polyhedrin of S. frugiperda and a chitinase enzyme was investigated by in silico docking analysis. The binding energy was used as a criterion to evaluate the affinity between the polyhedrin protein and the chitinase enzyme. We have chosen the chitinase enzyme because when insects are infected by NPV, their immune system is activated to defend against the virus. Chitinase production is one aspect of the insect's defense mechanism (Abulikemu et al., 2021). The chitinase enzyme may play a role in breaking down the chitinous components of the virus, such as the proteinaceous matrix or the viral occlusion bodies, which are structures that protect and transmit the virus. The breakdown of chitin by chitinase could potentially hinder the spread and replication of the NPV within the insect host. Additionally, chitinase may be involved in the degradation of infected cells or viral particles, contributing to the insect's defense against the virus (Abulikemu et al., 2021). Our objective was to optimise these interactions to achieve the most favourable binding with the lowest energy requirements. Our results reveal that ligand compound established robust bonds with one or more amino acids within the active pockets of the polyhedrin of S. frugiperda. Furthermore, chitinase enzyme exhibited very high binding energies of 9.4 kcal/mol (Supplementary file 1; Table.S4 & Fig. 3C).
3.9 Bioassay
The bioassay conducted on the isolated strain of SpfrNPV AAUBC1 against 2nd, 3rd and 4th instars larvae of S. frugiperda in laboratory conditions revealed a notable variation in its biological activity (Supplementary file 1; Table.S5 & Table 5). In two experiments, the LC50 values of SpfrNPV exhibited an inverse correlation with the age of the larvae, with the highest LC50 values observed for third instars. The strain, originally isolated from S. frugiperda, demonstrated high effectiveness, particularly against 2nd, 3rd and 4th instar larvae. The calculated LC50 values were 2.6 × 106, 3.9 × 107 and 3.1 × 108 POB’s/ml for 2nd, 3rd and 4th larval instar, respectively. N-number of insects per treatment; χ2-chi-square value; CL-Confidence limit; LC50 Lethal concentration; The larvae were exposed to NPV treated leaf disc for 24 h, following which they were separated in culture vials and mortality was recorded after 72 h. The corrected mortality data was subjected to probit analysis using Polo Plus software.
Instar
LC50
(POB’S/ml)Fiducial limit 95 %
Slope
Intercept
χ2
Df
Lower
Upper
Second
2.3 × 106
2.8 × 105
2.6 × 107
0.210714286
3.656428571
0.452
5
Third
1.6 × 107
1.4 × 106
3.6 × 108
0.009592461
0.069834169
0.176
5
Fourth
3.1 × 108
2.3 × 107
1.3 × 109
0.017128428
0.072558136
0.326
5
4 Discussion
The detailed investigation on the stage-wise dynamics of Nucleopolyhedrovirus infection in fall armyworm (S. frugiperda) larvae in maize fields of Panchmahal and Mahisagar districts during the kharif season in 2021 offers valuable insights into ecological and molecular aspects of NPV prevalence. The findings are discussed about key ecological implications and potential applications for pest management, supported by relevant references. The pronounced pattern of NPV infection, peaking during the cob formation stage, aligns with the fall armyworm's life cycle and NPV transmission dynamics. This confirms previous research emphasizing the significance of crop stages in influencing vulnerability to NPV infection (Cory and Myers, 2003; Raghunandan et al., 2019). Regression equations established provide quantitative tools for predicting NPV infection during different crop stages, supporting integrated pest management strategies (Fuxa, 2004). The observed block-specific variations in NPV infection rates suggest localized factors influencing virus prevalence, such as environmental conditions, agricultural practices, and the natural resilience of fall armyworm populations (Gao et al., 2021). Positive correlations identified between specific crop stages and NPV infection highlight the potential influence of these stages on larval vulnerability. Despite the highest infection percentage, the non-significant correlation during the cob formation stage indicates a complex interplay of factors influencing NPV transmission. The likely origin of infected larvae from preceding stages prompts further research into the temporal dynamics of NPV spread and its interaction with fall armyworm biology (Gao et al., 2021). The ecological implications of these findings are crucial for understanding NPV prevalence and informing pest management strategies. The positive increase in NPV infection rates aligns with the potential use of NPV as a biopesticide for controlling fall armyworm infestations. Warmer conditions may negatively impact the viability and effectiveness of NPV, making it less capable of infecting and spreading within the S. frugiperda larval population (Gross et al., 1994). Additionally, higher temperatures might accelerate the breakdown of NPV particles or reduce their ability to enter host cells, thus limiting their infection potential (Mori et al., 2010). Elevated humidity provides the necessary moisture for NPV particles to remain stable and infectious facilitating their persistence and ability to infect S. frugiperda larvae effectively (Narramore et al., 2019). Overall, the conducive environment created by elevated humidity levels supports NPV proliferation and infection, leading to higher infection rates in fall armyworm populations The negative correlation between sunshine hours and NPV infection rates suggests that higher sunlight exposure is linked to decreased natural NPV infection. One reason for this is the detrimental effect of ultraviolet (UV) radiation on NPV particles. UV radiation, particularly UV-B and UV-C, can damage the genetic material (DNA or RNA) of viruses, including NPV, thereby reducing their infectivity and ability to replicate within host cells. The month-wise analysis of weather parameters reveals a distinct correlation between environmental conditions and NPV infection, with the highest rates in September, characterized by high humidity and reduced sunlight. This aligns with studies indicating the significance of these factors in NPV efficacy (Lacey et al., 2008). Understanding seasonal variations in NPV prevalence is essential for informing targeted pest management strategies, particularly during periods when weather conditions favor the natural control of fall armyworm populations through NPV infection (Fuxa, 2004). The study delves into molecular aspects, including the prediction of conserved domains, gene structures, phylogenetic analysis, and motif pattern analysis of SpfrNPV polyhedrin. The shared motif pattern in different strains suggests a conserved functional significance in the polyhedrin protein, contributing to its structural and functional roles in the viral life cycle (Herniou et al., 2003). Phylogenetic analysis reveals a species-specific clustering within S. frugiperda and a broader genus-specific association with polyhedrins from other Spodoptera species, enhancing our understanding of the evolutionary dynamics of NPV polyhedrins (Lanzi et al., 2006). The deduced protein structures of SpfrNPV polyhedrin exhibit significant similarities, emphasizing conserved features among polyhedrins in different NPVs. Molecular docking studies with chitinase highlight potential interactions between polyhedrin and a key insect defense enzyme, shedding light on virus-host interaction dynamics (Fath-goodin et al., 2006). Further research into the structural and functional aspects of polyhedrin can provide valuable insights for developing targeted pest management strategies. Bioassay results demonstrate the efficacy of SpfrNPV against early instar larvae, emphasizing the importance of considering larval developmental stages and cuticular defenses in host-pathogen interactions (Vargas et al., 2012). The observed variation in biological activity against different larval instars aligns with studies indicating age-dependent susceptibility of larvae to NPV infection (Gothama et al., 1995). The investigation reveals a distinctive pattern of chitinase activity in response to Diter NPV infection across different larval instars of S. frugiperda. Higher instar larvae exhibit significantly elevated chitinase activity compared to initial instar counterparts, suggesting a potential adaptive response aimed at countering NPV infection. Conversely, initial instar larvae demonstrate comparatively lower chitinase activity, contributing to their increased vulnerability to NPV-induced pathogenicity (Abulikemu et al., 2021). This aligns with heightened susceptibility of early instar larvae to pathogenic infections, emphasizing the significance of larval developmental stages in determining vulnerability. The increased cuticular melanism in eldest larvae acts as a protective barrier limiting pathogen entry, consistent with studies by Wilson et al. (2001). This protective mechanism, combined with reduced chitinase response, renders early instar larvae more vulnerable to NPV-induced pathogenicity (Ayyub et al., 2019). The interplay between larval age, cuticular defenses, and susceptibility to pathogenic infections underscores the multifaceted nature of host-pathogen interactions. These insights contribute to a comprehensive understanding of factors influencing larval susceptibility to infections, providing valuable information for developing targeted pest management strategies. The identification of an optimal intervention window and the inverse correlation between initial viral concentrations and required doses offer practical considerations for optimizing biocontrol strategies (Vargas et al., 2012).
5 Conclusion
In conclusion, the findings presented in this study significantly contribute to our understanding of the stage-wise dynamics, ecological factors, and molecular characteristics of SpfrNPV infection in fall armyworm populations. In essence, this study advances our knowledge of NPV's role in pest management and underscores its significance in promoting sustainable agriculture. By exploiting the natural enemies of fall armyworms through informed and eco-friendly practices, we take a step towards resilient and sustainable agricultural systems, ensuring a harmonious balance between productivity and environmental well-being. Integrating ecological and molecular perspectives provides a holistic view of NPV prevalence, offering valuable insights for developing sustainable pest management practices in agricultural ecosystems.
Ethics approval
Not applicable.
Consent to participate
All authors consent to participate in the manuscript publication.
Consent for publication
All authors approved the manuscript to be published.
Availability of data and material
The data supporting the conclusions of this article are included within the article. Any queries regarding these data may be directed to the corresponding author. Additional data have been furnished in the Supplementary Files 1–3.
Funding
No external funding. This research received no external funding.
Declaration of Generative AI and AI-assisted technologies in the writing process
During the preparation of this work the author(s) used Chat GPT Open AI in order to improve language editing. After using this tool/service, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
CRediT authorship contribution statement
Joldalu Sajan Pavan: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Methodology, Investigation. Nainesh Balubhai Patel: Validation, Supervision, Methodology. Boodanur Lingaiah Raghunandan: Visualization, Validation, Supervision, Methodology, Investigation, Conceptualization. Madigiri Nagarajgouda Rudra Gouda: Data curation, Conceptualization. Ashraf M. Ahmed: Supervision, Investigation, Formal analysis. Saleh Alansi: Formal analysis, Data curation, Conceptualization.
Acknowledgement
We thank to Researchers Supporting Project number (RSPD2024R695), King Saud University, Riyadh, Saudi Arabia
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Biological control of fall armyworm, Spodoptera Frugiperda. Agron. J.. 2022;12(11):2704.
- [Google Scholar]
- Sublethal effects of spinetoram and emamectin benzoate on key demographic parameters of fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae) under laboratory conditions. Environ. Sci. Pollut. Res.. 2023;20:1-4.
- [Google Scholar]
- A method of computing the effectiveness of an insecticide. J. Econ. Entomol.. 1925;18(2):265-267.
- [Google Scholar]
- Comparison of the potential activities of viral and bacterial chitinases. Egypt J. Biol. Pest Control.. 2021;31(1):91.
- [Google Scholar]
- Individual and combined impact of nuclear polyhedrosis virus and spinosad to control the tropical armyworm, Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae), in cotton in Pakistan. Egypt J. Biol. Pest Control.. 2019;29(1):1-6.
- [Google Scholar]
- The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst.. 2003;34(1):239-272.
- [Google Scholar]
- Economic analysis of pesticide expenditure for managing the invasive fall armyworm, Spodoptera frugiperda (JE Smith) by maize farmers in Karnataka, India. Curr. Sci.. 2021;10:1487-1492.
- [Google Scholar]
- Polydnavirus genes that enhance the baculovirus expression vector system. Adv. Virus Res.. 2006;68:75-90.
- [Google Scholar]
- Ecology of insect nucleopolyhedroviruses. Agric. Ecosyst. Environ.. 2004;103(1):27-43.
- [Google Scholar]
- Sublethal effects of the microbial-derived insecticide spinetoram on the growth and fecundity of the fall armyworm (Lepidoptera: Noctuidae) J. Econ. Entomol.. 2021;114(4):1582-1587.
- [Google Scholar]
- Interactive effects of Steinernema carpocapsae and Spodoptera exigua nuclear polyhedrosis virus on Spodoptera exigua larvae. J. Invertebr. Pathol.. 1995;66(3):270-276.
- [Google Scholar]
- Design and application of a hive-mounted device that uses honey bees (Hymenoptera: Apidae) to disseminate Heliothis nuclear polyhedrosis virus. Environ. Entomol.. 1994;23(2):492-501.
- [Google Scholar]
- The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol.. 2003;48(1):211-234.
- [Google Scholar]
- First record of a nucleopolyhedrovirus infecting brown-tail moth larvae, Euproctis chrysorrhoea in India. Egypt J. Biol. Pest Control.. 2019;29(1):1-5.
- [Google Scholar]
- Invasiveness, biology, ecology, and management of the fall armyworm, Spodoptera Frugiperda. Entomol. Gen.. 2022;10:1127.
- [Google Scholar]
- Effect of temperature on long-term storage of codling moth granulovirus formulations. J. Econ. Entomol.. 2008;101(2):288-294.
- [Google Scholar]
- Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.) J. Virol.. 2006;80(10):4998-5009.
- [Google Scholar]
- SMART: recent updates, new developments and status in 2015. Nucleic Acids Res.. 2014;43:257-260.
- [Google Scholar]
- Effects of Helicoverpa armigera nucleopolyhedrosis virus (HaNPV) on the larvae of the diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae) Plant Prot. Sci.. 2014;50(4):184-189.
- [Google Scholar]
- Mori, H., Ijiri, H., Kotani, E., 2010. Application of insect virus polyhedra to protein nanocontainers. In: Proc. 2010 NSTI Nanotechnol. Conf. Expo (Vol. 3, pp. 254-257).
- New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase. Bioorg. Med. Chem.. 2019;27(16):3546-3550.
- [Google Scholar]
- Comparative efficacy of nucleopolyhedrovirus (NPV) alone and in conjunction with chemical insecticides against fall armyworm, Spodoptera frugiperda (JE Smith)(Noctuidae: Lepidoptera) under laboratory conditions. Int. J. Trop. Insect Sci. 2024:1-12.
- [Google Scholar]
- Natural occurrence of nucleopolyhedrovirus infecting fall armyworm, Spodoptera frugiperda (JE Smith)(Lepidoptera: Noctuidae) in Gujarat, India. J. Entomol. Zool. Stud.. 2019;7(2):1040-1043.
- [Google Scholar]
- Elucidating Whitefly Bemisia tabaci Asia II 1 Transmission of MYMIV and the Possible Role of ORF AC4 in YMD of Soyabean. Biol. Forum.. 2023;15(8):148-158.
- [Google Scholar]
- Conformational transitions upon ligand binding: Holo structure prediction from apo conformations. Biophys. J.. 2010;98(3):428a.
- [Google Scholar]
- Characterization and field evaluation of tetrahedral and triangular nucleopolyhedrovirus of Spilosoma obliqua (SpobNPV) strain NBAIR1 against jute hairy caterpillar. Egypt. J. Biol. Pest Control.. 2020;30:1-7.
- [Google Scholar]
- MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol.. 2013;30:2725-2729.
- [Google Scholar]
- Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol.. 2012;158(3):1342-1358.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary material to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103274.
Appendix A
Supplementary material
The following are the Supplementary material to this article:







