Translate this page into:
Despite the genetic variability: NS1 of different dengue serotypes has comparable affinity for various host protein in silico
⁎Corresponding authors at: Department of Microbiology, University of Swabi, Pakistan; Department of Health and Biological Sciences, Abasyn University Peshawar, Pakistan. hayatkhan@uoswabi.edu.pk (Hayat Khan), aminbiotech7@gmail.com (Amin Ullah)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background and Aim
Dengue infection is a global issue which is caused by insect transmitted, positive sense single-stranded RNA dengue virus (DENV). Around 50 million cases of dengue infection is being reported per year whereas population at risk of dengue of infection reaches about 2.5 billion in the tropical and subtropical regions of the world. The present study aimed to develop global consensus sequence of dengue NS1 protein and modelling interaction of NS1 with various host proteins of different cellular pathways.
Methods
Serotype specific consensus sequences of Dengue NS1 were generated using CLC-Work Bench software. The Consensus sequences of NS1 and Cellular proteins were modelled using Protein Model Portal (PMP). Comparative interaction of NS1 of various dengue serotypes were analyzed by means of Haddock server. The biomolecular complexes of dengue NS1 and various host proteins (PTBP1, NF-kB, TRAF6, PAIP1, SRFBP1, FGB, TCF7L2 and EIF4G1) thus formed were evaluated for interacting residues through Protein Interaction Calculator (PIC).
Results
The serotype-specific consensus sequences revealed high degree of conservation among the various NS1 domains of different isolates of same serotype but is more heterogeneous among different serotypes. The docking results demonstrated that most of the host proteins including (PTBP1, NF-kB, TRAF6, PAIP1, SRFBP1, FGB, TCF7L2 and EIF4G1) have comparable affinity for the NS1 of different serotypes despite considerable degree of variability among the serotypes. Analysis of interactive residues demonstrated involvement of different domains of NS1 for interaction with host partners even within the NS1 of different serotypes for a particular host protein.
Conclusion
The in silico study revealed that various NS1 domains of different isolates were relatively conserved among different serotypes but highly heterogeneous from serotype to serotypes. The Wing domain of NS1 protein plays a vital role in interaction with the host proteins.
Keywords
Dengue NS1
Host Proteins
Dengue Serotypes
Protein-protein interactions (PPI)
1 Introduction
Approximately 50 million peoples are being affected from Dengue infection per year while about 2.5 billion people are being at threat of the dengue infection in the subtropical and tropical region of the world (Guha-Sapir & Schimmer, 2005). The main transmission among the peoples of South-East Asia occurs by a species of Aedes mosquito family Flaviviridae (flavusis a Latin word which means yellow, because of yellow fever virus which induce jaundice) Aedes aegypti while the Aedes albopicts works as a secondary vector (Gratz, 2004; Muller and Young, 2013) So far there are no antiviral therapies available for the dengue infections. However, developments are underway for antiviral against dengue infection to overcome the attack of dengue infection and associated complications (Noble et al., 2010; Sampath & Padmanabhan, 2009). Dengue virus has four different serotypes named as Dengue Virus 1 (DENV1), Dengue Virus 2 (DENV2), Dengue Virus 3 (DENV3) and Dengue Virus 4 (DENV4).
Electron microscopy reveals a relatively smooth surface of lipid envelope around the dengue virion with an approximate diameter 500 A°. Genome of dengue virus is a single stranded RNA with positive-sense of 10.7 kb in size. The genome codes for three structural proteins that are arranged in the genome in the core (C, made up of 100 amino acids), membrane (M, consist of 75 amino acids), and envelope (E, consist of 495 amino acids) (Rastogi et al., 2016; Wiemann et al., 2001. Besides structural components of virus, the genomic RNA encodes information for the synthesis of seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) (Lindenbach et al., 2003; Gutsche et al., 2011). Expression of NS1 is on the surface of the infected cell initiates antibody dependent cell mediated cytotoxicity (ADCC) of infected cell (Jacobs et al., 2000). The immune evasion mechanism of dengue virus is also partly mediated by the dengue NS1 (secretory and membrane bound) through interaction with proteins of complement pathway and hence modulate or antagonizing their activities (Avirutnan et al., 2011; Fakioglu et al., 2008). various reports also reveals involvement of NS1 in the genome replication of dengue, interaction with NS4B and double-stranded RNA (Akey et al., 2014; Chatel-Chaix et al., 2015; Youn et al., 2013).
Viral infection is a complex interplay between the host and viral factors. Various protein factors of virus interact with cellular factors in order to establish a productive infection. Exploring protein–protein interaction (PPi) is very important in finding out drug targets in proteins (Pedamallu & Posfai, 2010).
The present study was designed to use the in silico method for the detection and comparative analysis of protein–protein interaction between the NS1 protein of different dengue serotypes and reported cellular proteins. For this purpose, the NS1 protein sequences of dengue serotypes reported from all the over world were retrieved and aligned in order to obtain the consensus sequence for each serotype. The consensus sequences of NS1 were modelled in to protein structure of their respective serotypes. The host proteins and NS1 proteins were then subjected to docking for detection of PPI. It is assumed that NS1 of different dengue serotypes will have different affinity towards host proteins during infection that needs to be explored. Moreover, the in silico protein–protein interaction will provide insight for the predictive discovery of common interacting surface of NS1 of various serotypes involved in the PPI with various cellular proteins. The common interacting surface thus determined could serve as a pan-serotypic drug target.
2 Materials and methods
2.1 Generation of serotype-specific and global consensus sequence of dengue NS1
For the retrieval of DENV NS1 protein sequences, NCBI database was searched. Protein sequences of DENV NS1 of all four serotypes were retrieved all over the world which includes a total of 66 protein sequences of DENV1 NS1 were selected from various region of world. For DENV2 NS1, 133 protein sequences were included in the proposed study. For DENV3 and DENV4, 118 and 64 were protein sequences of NS1 were retrieved respectively and included in the current study. The NS1 sequences of all four different serotypes of DENV retrieved from NCBI were fed to CLC main workbench software to generate consensus sequence of each serotype and then consensus sequences of different serotypes thus constructed were aligned in CLC main workbench software in order to generate global consensus sequence of NS1 of all serotypes.
2.2 Modeling of dengue NS1 and host proteins
Protein Modelling Portal (PMP) (https://www.proteinmodelportal.org/), an online server was used for the protein modelling of NS1 consensus sequences of each dengue serotype and various human host proteins. It has different partner sites (Swiss Model, I-TASSER, Phyre2, M4T, RaptorX etc.) which provided us the structure model of requested protein via email in the pdb format.
2.3 Docking of dengue NS1 and host proteins
The CPORT algorithm of Haddock was used for the prediction of protein–protein interface residues (Active and Passive residues) of docking proteins and then for the analysis of protein–protein interaction between the DENV NS1 of different serotype and various reported host proteins, Haddock (High Ambiguity Driven protein–protein DOCKing) webserver was used. The CPORT stands for Consensus Prediction Of interface Residues. As the name represents the CPORT was used to find out active and passive residues of protein-proteins interface before docking with Haddock. In this way, all biomolecular complexes of DENV NS1 of all four serotypes and reported host proteins were modelled.
2.4 Analysis of interactive residues of biomolecular complexes
In order to examine the interacting residues of biomolecular complexes of dengue NS1 and various reported host proteins, Protein Interactions Calculator (PIC) an online webserve was used for this purpose. The PDB files of all biomolecular complexes modelled in the Haddock were uploaded and submitted in the PIC in order to find out the different types of interaction like Main Chain-Main Chain, Main Chain-Side chain and Side Chain-Side Chain interacting amino acid residues in the protein complexes.
3 Results
3.1 Serotype-specific and global consensus sequences of dengue NS1
Serotype-specific consensus sequences of protein NS1 of DENV was generated for each and every serotype with the help of CLC main workbench (Fig. 1). Global consensus sequence of protein NS1 was developed by aligning the consensus sequences of all four dengue serotypes (Fig. 2). The bars represent percent conservation of amino acid residues. The “X” shows highly variable residues whereas conserved amino acids residues are expressed by their symbols. Similarly, dengue NS1 proteins sequences of all serotypes reported from Pakistan were also included in the study. Variability and conservation were also analyzed among the various regions of global consensus sequence. The global and serotype-specific consensus sequences of dengue NS1 demonstrate that there is a high degree of variability among the different domains of NS1 of dengue serotypes but highly conserved among the isolates of the same serotype.
Serotype-specific consensus sequence of (A) DENV1 NS1, (B) DENV2 NS1, (C) DENV3 NS1 and (D) DENV4 NS1. Amino acids have been shown by their one letter code. The bars under the amino acids show percent amino acid conservation at their respective position.
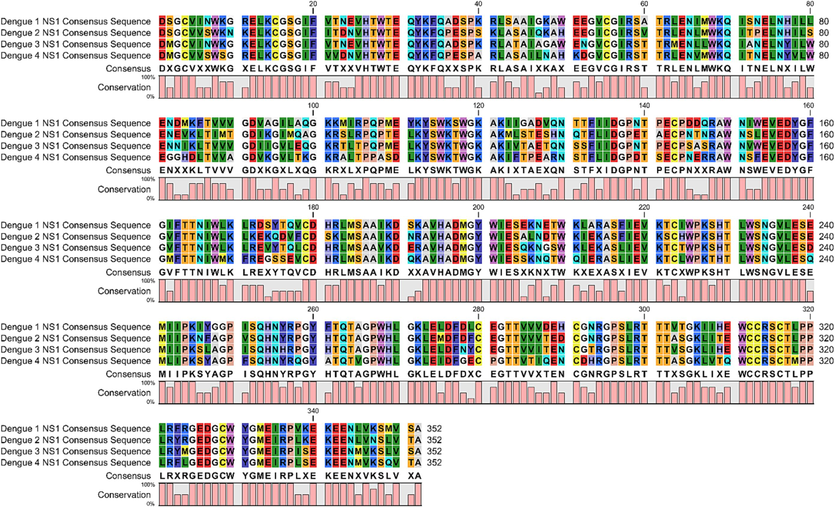
Global consensus sequence of DENV NS1. Serotype-specific consensus of dengue NS1 have been shown. The global consensus sequence has been shown by one letter amino acid codes at the bottom above the bars. The bars under the amino acids show percent amino acid conservation at their respective position.
3.2 Protein models of dengue NS1 and host proteins
The 3D structural models of NS1 of different DENV serotypes as well as the human host proteins selected for the docking were modelled using PMP online server. Protein models of NS1 of various dengue serotype have been shown in Fig. 3. Different domains of the protein have been represented by various colors; β-roll by black, wing by red and β-ladder by yellow.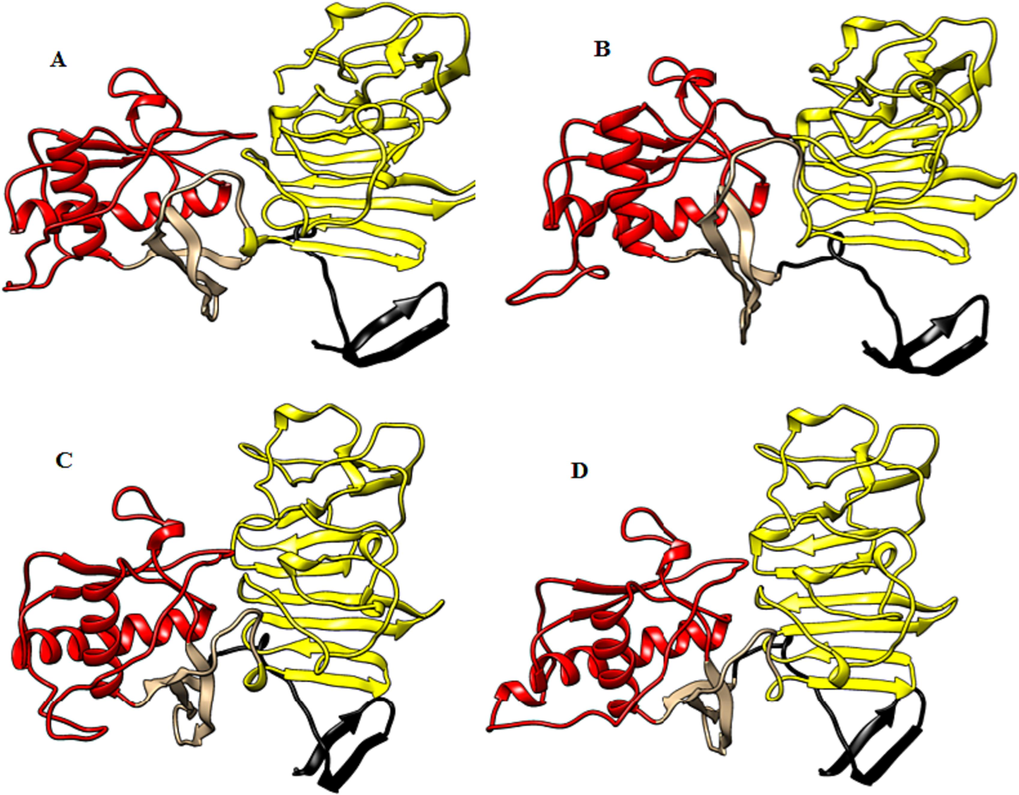
Protein structure of (A) DENV1 NS1, (B) DENV2 NS1, (C) DENV3 NS1 and (D) DENV4 NS1. The regions highlighted in black, red and yellow are β-roll, wing and β-ladder domains of NS1. Portions highlighted in gray represent connectors.
3.3 Biomolecular complexes of NS1 and host proteins
The biomolecular complexes between the NS1 of various dengue serotypes and host proteins (PTBP1, NF-kB, TRAF6, PAIP1, SRFBP1, FGB, TCF7L2 and EIF4G1) were generated through Haddock online webserver (Figs. 4–11). Best complexes were selected based on the Haddock their Haddock and Z-scores (Figs. 4–11). Docking results have been shown in Table 1.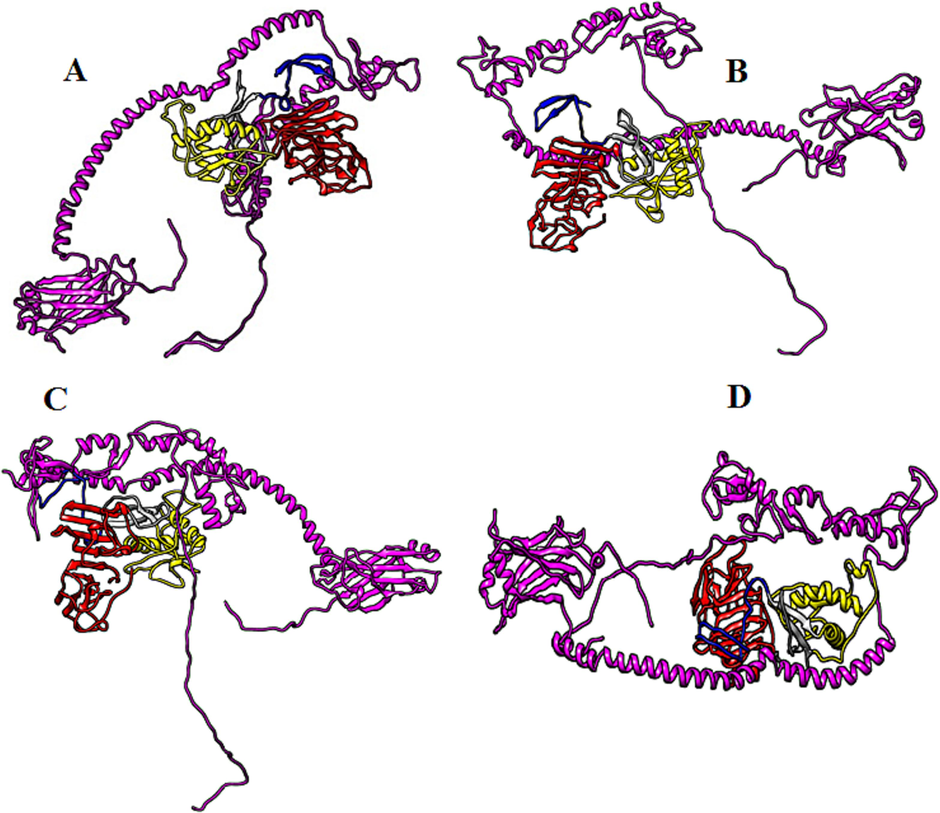
Biomolecular complexes of (A) DENV1 NS1-TRAF6, (B) DENV2 NS1-TRAF6, (C) DENV3 NS1-TRAF6 and (D) DENV4 NS1-TRAF6. Protein highlighted by multicolor represent DENV NS1 whereas TRAF6 protein has been shown by purple color in the complex.
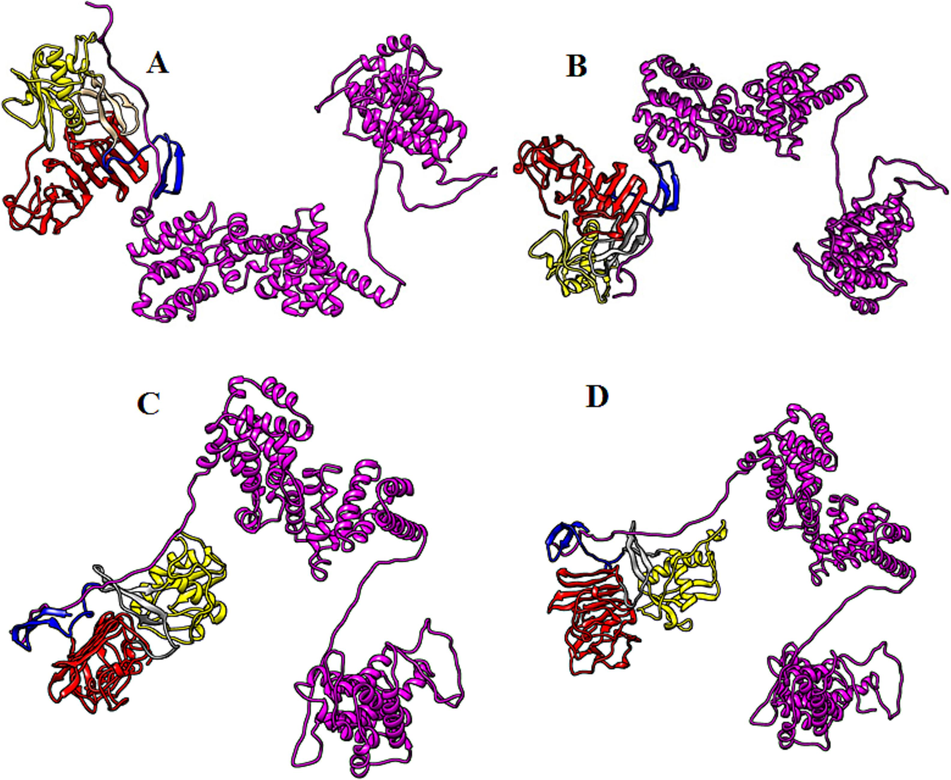
Biomolecular complexes of (A) DENV1 NS1-EIF4G1, (B)DENV2 NS1-EIF4G1, (C) DENV3 NS1-EIF4G1 and (D) DENV4 NS1-EIF4G1. Protein highlighted by multicolor represent DENV NS1 whereas EIF4G1 protein has been shown by purple color in the complex.
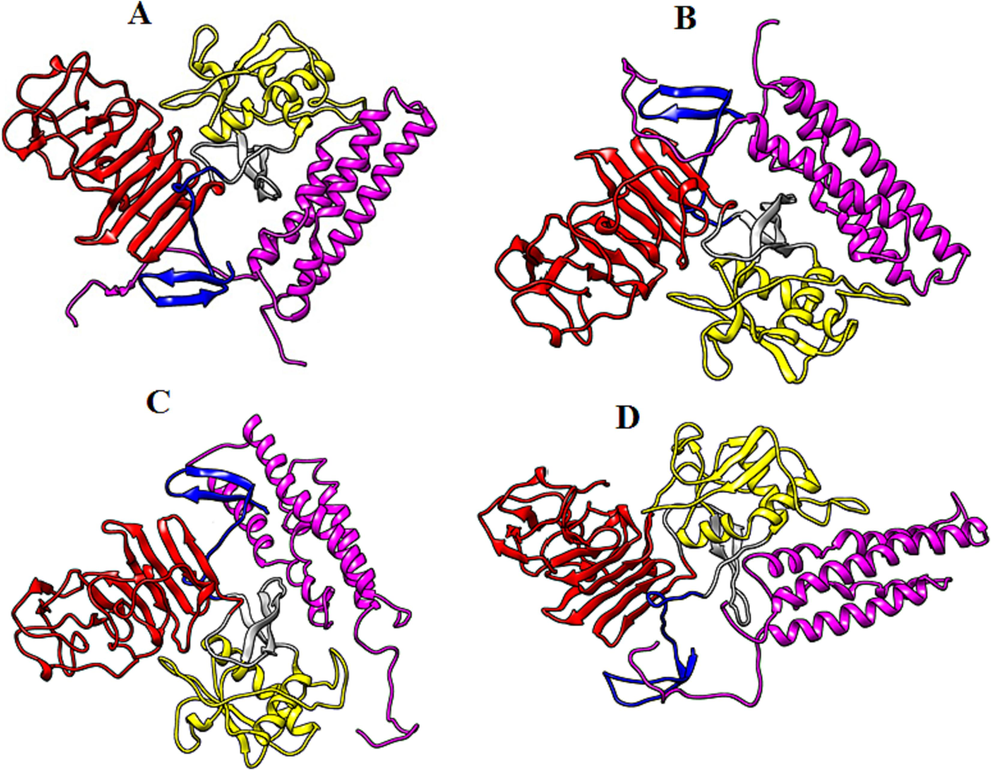
Biomolecular complexes of (A) DENV1 NS1-SRFBP1, (B)DENV2 NS1-SRFBP1, (C) DENV3 NS1-SRFBP1 and (D) DENV4 NS1-SRFBP1. Protein highlighted by multicolor represent DENV NS1 whereas SRFBP1 protein has been shown by purple color in the complex.
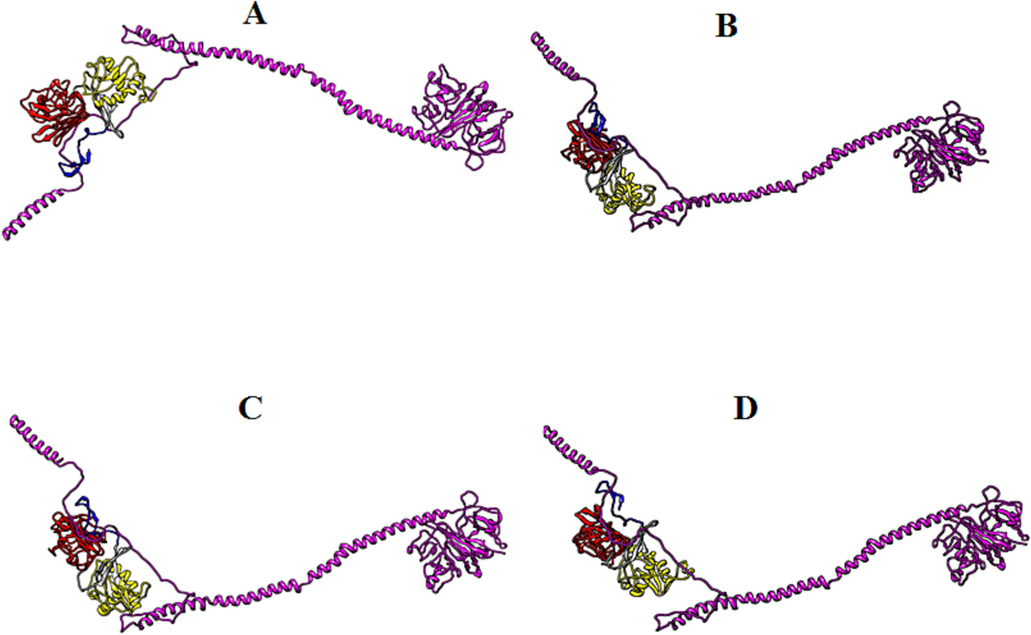
Biomolecular complexes of (A) DENV1 NS1-FGB, (B) DENV2 NS1-FGB, (C) DENV3 NS1-FGBand (D) DENV4 NS1-FGB. Protein highlighted by multicolor represent DENV NS1 whereas FGB protein has been shown by purple color in the complex.
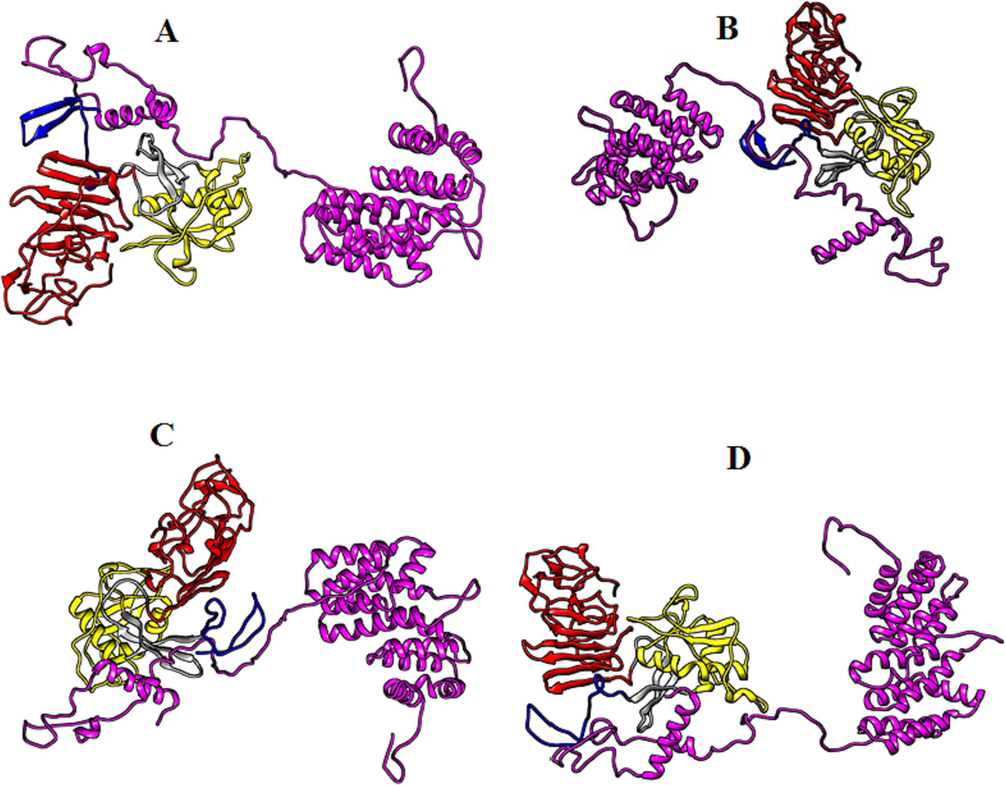
Biomolecular complexes of (A) DENV1 NS1-PAIP1, (B) DENV2 NS1-PAIP1, (C) DENV3 NS1-PAIP1 and (D) DENV4 NS1-PAIP1. Protein highlighted by multicolor represent DENV NS1 whereas PAIP1 protein has been shown by purple color in the complex.
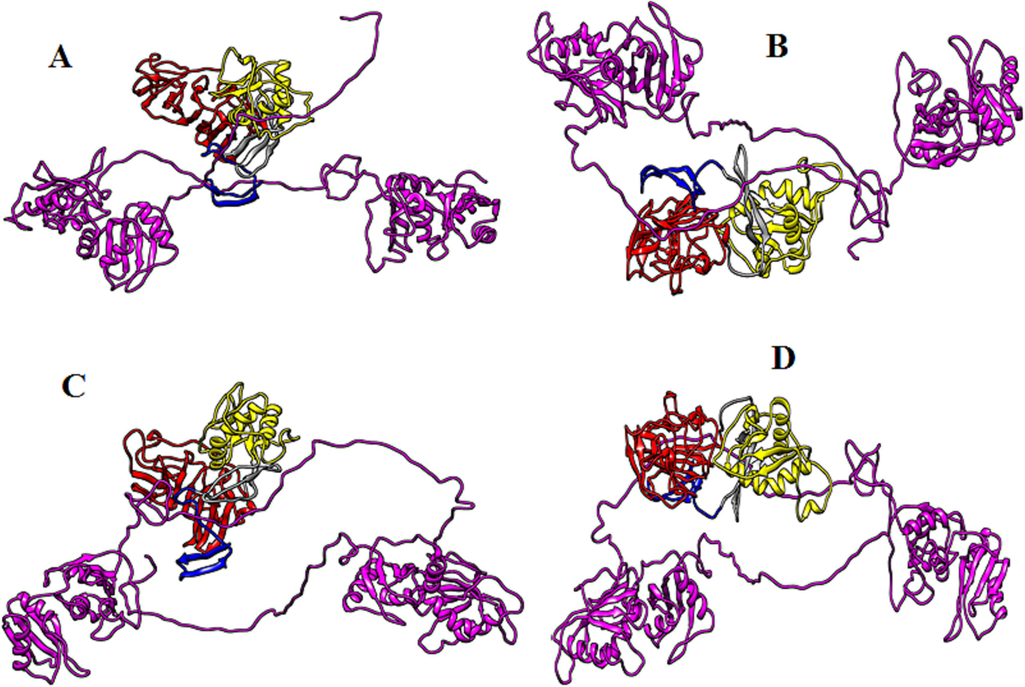
Biomolecular complexes of (A) DENV1 NS1-PTBP1, (B) DENV2 NS1-PTBP1, (C) DENV3 NS1-PTBP1and (D) DENV4 NS1-PTBP1. Protein highlighted by multicolor represent DENV NS1 whereas PTBP1 protein has been shown by purple color in the complex.
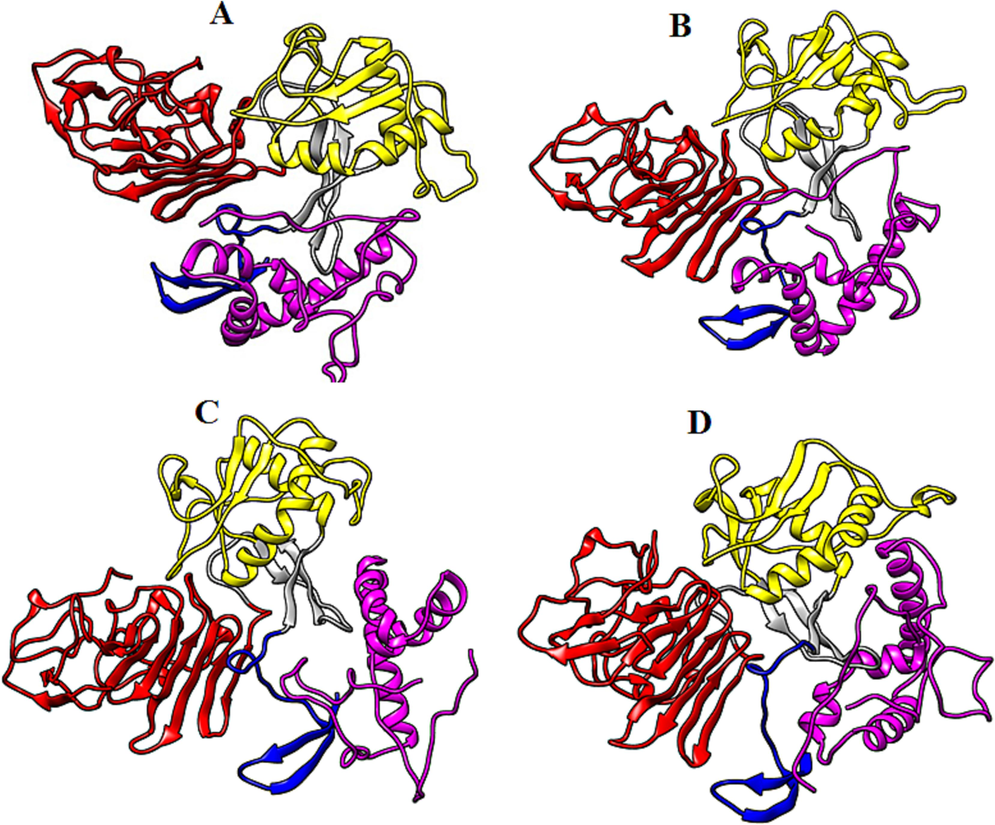
Biomolecular complexes of (A) DENV1 NS1-TCF7L2, (B) DENV2 NS1-TCF7L2, (C) DENV3 NS1-TCF7L21 and (D) DENV4 NS1-TCF7L2. Protein highlighted by multicolor represent DENV NS1 whereas TCF7L2 protein has been shown by purple color in the complex.
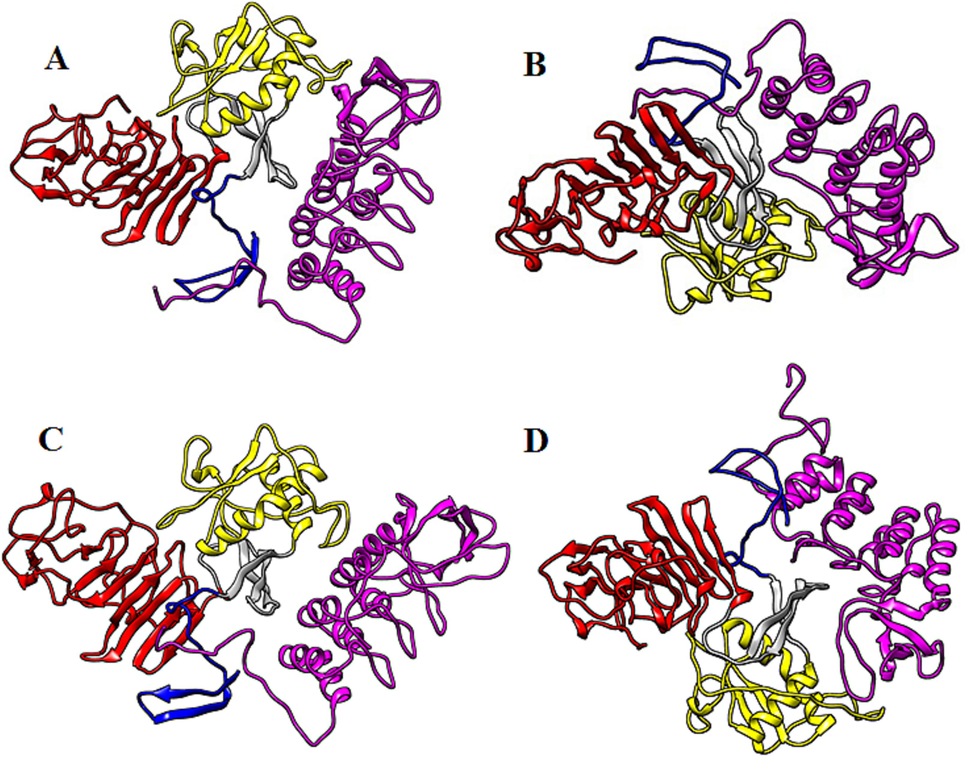
Biomolecular complexes of (A) DENV1 NS1-NF-kB, (B) DENV2 NS1-NF-kB, (C) DENV3 NS1-NF-kB and (D) DENV4 NS1-NF-kB. Protein highlighted by multicolor represent DENV NS1 whereas NF-kB protein has been shown by purple color in the complex.
Biomolecular Complex
Haddock Score
Van der Waals energy
Electrostatic energy
Desolvation energy
Restraints violation energy
Z-Score
Dengue 1 NS1-NF-kB
−115.2 +/- 13.8
−120.2 +/- 5.4
−342.3 +/- 19.6
−37.6 +/- 5.3
1111.2 +/- 81.26
−2.3
Dengue 2 NS1-NF-kB
−84.0 +/- 2.1
−120.2 +/- 10.1
−258.0 +/- 39.8
−34.8 +/- 14.2
1225.2 +/- 109.08
−1.9
Dengue 3 NS1-NF-kB
−87.2 +/- 4.7
−95.1 +/- 3.9
–233.0 +/- 60.2
−38.7 +/- 9.9
932.3 +/- 79.99
−1.8
Dengue 4 NS1-NF-kB
−87.2 +/- 11.8
−112.5 +/- 9.5
−351.9 +/- 67.0
−38.1 +/- 14.9
1337.6 +/- 152.54
−1.6
Dengue 1 NS1- EIF4G1
−142.1 +/- 15.3
−107.2 +/- 5.2
−400.4 +/- 31.2
−70.8 +/- 23.4
1159.6 +/- 69.03
−2.3
Dengue 2 NS1- EIF4G1
−127.3 +/- 6.9
−109.8 +/- 10.3
−532.8 +/- 92.4
−40.5 +/- 11.8
1296.2 +/- 210.30
−1.7
Dengue 3 NS1- EIF4G1
−111.1 +/- 19.4
−104.3 +/- 6.3
−205.8 +/- 32.6
−89.0 +/- 16.6
1233.3 +/- 82.37
−1.2
Dengue 4 NS1- EIF4G1
−129.7 +/- 6.9
−110.0 +/- 6.7
−356.5 +/- 55.9
−64.9 +/- 9.9
1164.6 +/- 105.83
−1.2
Dengue 1 NS1-PAIP1
−147.8 +/- 15.1
−97.1 +/- 2.7
−483.7 +/- 31.9
−61.4 +/- 2.2
1075.1 +/- 97.42
−1.2
Dengue 2 NS1-PAIP1
−132.3 +/- 8.5
−95.3 +/- 6.6
−481.3 +/- 61.1
−32.1 +/- 16.5
912.9 +/- 153.03
−1.4
Dengue 3 NS1-PAIP1
−163.9 +/- 10.6
−101.2 +/- 4.7
−407.0 +/- 24.1
−48.5 +/- 20.0
672.0 +/- 122.10
−2.0
Dengue 4 NS1-PAIP1
−187.7 +/- 13.0
−128.8 +/- 6.4
−417.5 +/- 14.5
−73.0 +/- 11.5
975.0 +/- 74.00
−2.1
Dengue 1 NS1-PTBP1
69.3 +/- 9.3
−82.7 +/- 10.1
−166.6 +/- 26.4
−56.1 +/- 5.3
2415.0 +/- 139.74
−1.5
Dengue 2 NS1-PTBP1
62.7 +/- 9.3
−92.5 +/- 9.3
−110.8 +/- 14.5
−52.1 +/- 11.6
2294.0 +/- 100.03
−2.2
Dengue 3 NS1-PTBP1
82.9 +/- 29.1
−71.8 +/- 11.7
−67.2 +/- 29.2
−57.8 +/- 7.3
2259.4 +/- 224.47
−2.0
Dengue 4 NS1-PTBP1
52.3 +/- 34.0
−109.2 +/- 16.2
−160.0 +/- 19.6
−44.4 +/- 6.0
2379.7 +/- 171.42
−2.1
Dengue 1 NS1- SRFBP1
−85.6 +/- 18.4
−116.2 +/- 10.2
−402.9 +/- 91.5
−10.2 +/- 15.1
1214.4 +/- 108.10
−1.7
Dengue 2 NS1-SRFBP1
−128.7 +/- 18.6
−120.4 +/- 15.9
−486.4 +/- 26.3
−14.5 +/- 9.3
1034.6 +/- 74.60
−2.1
Dengue 3 NS1- SRFBP1
−133.7 +/- 28.5
−82.5 +/- 10.2
−641.9 +/- 96.6
−36.6 +/- 11.3
1137.1 +/- 98.82
−0.9
Dengue 4 NS1- SRFBP1
−111.2 +/- 5.9
−85.6 +/- 7.1
−479.1 +/- 34.7
−59.4 +/- 8.4
1296.8 +/- 132.49
−1.9
Dengue 1 NS1- TCF7L2
−156.2 +/- 15.5
−127.5 +/- 12.9
−299.6 +/- 28.4
−58.5 +/- 15.8
896.9 +/- 114.88
−1.9
Dengue 2 NS1- TCF7L2
−158.1 +/- 20.1
−120.1 +/- 8.0
−278.1 +/- 22.4
−55.9 +/- 14.8
735.3 +/- 61.47
−1.7
Dengue 3 NS1-TCF7L2
−168.5 +/- 4.0
−104.8 +/- 6.9
−387.9 +/- 26.3
−40.4 +/- 7.7
542.0 +/- 26.52
−2.5
Dengue 4 NS1-TCF7L2
−119.9 +/- 4.1
−113.4 +/- 10.4
−291.3 +/- 27.9
−34.8 +/- 9.4
865.0 +/- 88.87
−1.6
Dengue 1 NS1-FGB
66.2 +/- 4.1
−88.8 +/- 6.7
−202.1 +/- 38.5
−63.9 +/- 9.3
2593.3 +/- 194.33
−1.5
Dengue 2 NS1- FGB
73.9 +/- 12.0
−68.6 +/- 6.8
−232.3 +/- 64.2
−65.2 +/- 11.7
2541.3 +/- 204.74
−1.7
Dengue 3 NS1-FGB
55.2 +/- 23.3
−75.8 +/- 6.1
−264.3 +/- 80.8
−43.8 +/- 16.5
2276.9 +/- 105.94
−1.8
Dengue 4 NS1-FGB
43.4 +/- 23.4
−105.6 +/- 3.4
−221.2 +/- 23.6
−46.9 +/- 7.9
2402.3 +/- 159.12
−2.1
3.4 Interactive residues in biomolecular complexes
Protein-Protein interacting Calculator (PIC) was used to determine the interactive surfaces in the biomolecular complexes of NS1 and host proteins. The PIC determined different types of interactions (main chain-main chain interaction, main chain-side chain and side chain-side chain interaction) in the clusters (Supplementary Tables 1–8).
4 Discussion
The non-structural 1 (NS1) protein of all Flavivirus is a 48-kDa multifunctional protein and during translation translocate to the ER lumen. A small first domain is β-roll (1–30 residues) that results from the inter-wined of two beta hairpins. Second domain is Wing (37–152 residues) formed from an alpha–beta subdomain and a connector which lies against the first domain. Beta-ladder is the third domain (181–352 residues) that consists of nine anti-parallel beta-stands of each monomer forming an uninterrupted beta sheet along the entire dimer. The beta-roll and connector subdomain of wing form a projection which is believed to interact with transmembrane domains of other dengue proteins and also face towards ER membrane (Akey et al., 2014 and Youn et al., 2012).
The serotype specific consensus sequence developed from the 66 NS1 protein sequences of DENV1 isolates demonstrates that all regions (β-roll, Wing and Beta-ladder) of NS1 are highly conserved among the isolates of DENV1 included in the study. Similarly, different domains of NS1 are also highly conserved in the consensus sequence of 133 DENV2 protein sequences. Likewise, the serotype specific consensus sequences reveal conservation of various domains of NS1 among the 118 and 164 isolates of DENV3 and DENV4 respectively. In short, the NS1 is highly conserved among the isolates of same dengue serotype. In order to analyze the variability and conservation in the NS1 of different dengue serotypes, the serotype-specific consensus sequences were compared with each other by aligning them together to generate a representative global consensus sequence. The NS1 is relatively less conserved among the isolates of different serotypes as compared to the isolates of the same serotype. Among the different regions of NS1 of different serotypes, wing domain is relatively more heterogonous whereas relatively high proportion of beta-roll domain is conserved as can be find in the global consensus sequence.
For the comparative interaction of NS1 of different dengue serotype with a panel of host proteins, serotype specific consensus sequences and host protein sequences were modeled in order to achieve their three dimensional structures. Interactive analysis of dengue NS1 of different serotypes and NF-kB was performed. Nuclear factor-kappa B (NF-kB) is a highly inducible transcription factor that plays an important role in the hepatic acute-phase response, innate/adaptive immunity, and cellular survival through the induction of genetic networks. The Haddock score shows that NS1 of different dengue serotypes has comparable interaction with the NF-kB demonstrating that NS1 of all four dengue serotypes interact NF-kB with equal affinity. This shows that the NS1 of all dengue serotypes is involved in the inhibition of interferon synthesis which is initiated from the interaction of viral component and toll-like receptors (TLRs). Thus activated NF-kB induce a set of genes which mediate both innate and adaptive immune responses (Lin et al., 2013). Inhibition of NF-kB has also been explored in the in vitro infection of dengue, where association of dengue infection also has been shown with low level of IFN-β and inflammatory cytokines such as IL-10, IL-12 and TNF-α (Chang et al., 2012). Furthermore, tumor necrosis factor receptor-associated factor 6 (TRAF6) also interact with NS1 of dengue virus. TRAF6 is a member of signaling pathways that originate from the toll-like receptors (TLRs), Interleukin-1 receptors (IL-1R) and tumor necrosis factor receptor (TNFR) and transduces the signal to the NF-kB and activator protein-1 (AP-1) transcription factors (Wang et al., 2015). Haddock analysis of dengue NS1 and TRAF6 does not reveal a prominent difference among the Haddock scores of biomolecular complexes of dengue NS1 of different serotypes and TRAF6. Interaction of dengue NS1 with TRAF6 could increase viral replication and could inhibit induction of interferon by interfering transduction of downstream signaling. Previous study highlights a fact that in vitro dengue infection suppresses stimulation of interfering by upregulating miR-146a which in turn downregulate TRAF6 expression as the infection progresses (Wu et al., 2013). This also supports our in silico data that it might be possible that dengue virus target TRAF6 both directly through NS1 and indirectly by means of miR-146a. Furthermore, comparable interaction of NS1 of different dengue serotypes with TRAF6 highlight the fact that targeting this host factor is critical for the viral replication of dengue as reported earlier (Wu et al., 2013).
Dengue viral RNA possess 7-methyl guanosine at 5′ end but does not contain poly A tail at 3′UTR. Eukaryotic translation initiation factor 4 gamma (EIF4G) is a large modular scaffolding protein and a member of cap-binding complex eIF4F, other components are eIF4E and helicase complex (eIF4A plus cofactor eIF4B) (Edgil et al., 2006). Role of dengue NS1 in the translation of viral RNA has been implicated earlier (Cervantes-Salazar et al., 2015). Our In silico analysis demonstrates that dengue NS1 of various serotypes interact with the EIF4G with slight different affinity from serotype to serotype. The NS1 of DENV1 has a relatively high affinity for the EIF4G as compared to the NS1 of rest dengue serotypes. Interaction of DENV3 NS1 with EIF4G is relatively weak. Interaction of dengue NS1 and EIF4G may leads us to predict that NS1 is not only involved in the translation of viral RNA but it may inhibit the translation of cellular mRNA by targeting EIF4G rendering it unable to bind with cellular mRNAs for the translation.
The in silico analysis demonstrate interaction of dengue NS1 with another host protein serum response factor binding protein 1 (SRFBP1), alternatively also known as p49/STRAP (SRF-dependent transcription regulation associated protein). SRFBP1 has been reported by various studies as one of many cofactors of SRF (serum response factor) (Zhang et al., 2004). Serum response factor is a transcription factor which is activated downstream in response to multiple signaling pathways such as mitogen activated protein kinase (MAPK). The SRF is very crucial for the regulation of set of genes responsible for the cell growth and differentiation, tissue injury, hematopoiesis, neutrophil migration etc (Taylor and Halene, 2015). Interruption of SRF pathway leads to myelodysplasia and immune impairment (Taylor & Halene, 2015). The Haddock scores for the NS1 and SRFBP1 reveal low affinity of DENV1 for the SRFBP1. Rest of dengue serotypes show almost equal potency towards SRFBP1. Interaction of dengue NS1 may highlight the fact that dengue infection might leads to immune dysfunction by targeting SRF via SRFBP1. Another host protein called Fibrinogen beta chain (FGB) also interacts with the dengue NS1 in our study. During the injury the FGB is cleaved by thrombin protease and is polymerized with FGA (fibrinogen alpha) and FGG (fibrinogen gamma) to form insoluble fibrin complex that prevent bleeding. Haddock scores reveals a relatively high affinity of DENV4 NS1 for the FGB in contrast to that of NS1 of other dengue serotypes. As the dengue infection is associated with elevated pro-inflammatory cytokines, vasculitis, bleeding and endothelial permeability (Chen et al., 2006, Modhiran et al., 2015, Beatty et al., 2015), so the interaction NS1 with FGB may leads us to predict that this interaction is one the possible contributing factors in the hemorrhagic characteristic of dengue infection.
Productive viral infection is a complex process that requires interplay between viral and host factors. Mitogen-activated protein kinase kinase 7-interacting protein 2 (MAP3K7IP2) is an enzyme that might be involved in the development of dengue infection. This protein lies downstream in the signaling pathway induced by the IL-1 which ultimately activates NF-kB and Mitogen-activated protein kinase 8 also known as c-Jun N-terminal kinase 1 (MAPK8/JNK1) (Takaesu et al., 2000). In the present study, association of MAP3K7IP2 with dengue NS1 have been explored. If the interaction of MAP3K7IP2 with the NS1 of different serotypes is compared via Haddock scores, then this protein shows relatively greater affinity towards the NS1 of dengue serotype 4. The NS1 of dengue serotypes 1 and 2 has equal binding affinity for this protein as revealed from their Haddock scores. In silico binding of dengue NS1 with MAP3K7IP2 may provide the evidence for the immune evasion mechanism of dengue virus because the MAP3K7IP2 is activated in response to IL-1 trigger and IL-1 has a key role in the regulation of inflammatory and immune responses (Dinarello, 2011). The present study also reveals interaction of dengue NS1 with another host protein called Polyadenylate-binding protein-interacting protein 1 (PAIP1). This protein binds with eIF4A (cap-binding complex) and poly(A)-binding protein (Wiemann et al., 2011 and Roy et al., 2002). Interaction of PAIP1 with polyadenylate-binding protein and eIF4A exhibit involvement of this protein in the translation and stability of mRNA. The interaction of NS1 with this protein may also highlight this fact for the dengue RNA. Haddock scores show that NS1 of dengue serotype 4 and 3 has comparatively strong interactive potential for the PAIP1 demonstrating that the NS1 of these two serotypes are more potent in translating their viral RNA. Polypyrimidine tract-binding protein (PTBP1), also known as hnRNP I (heterogeneous nuclear ribonucleoprotein) or PTB is a splice regulator and is also involved in the RNA stability, translocation and alternative splicing. The NS1 of dengue shows interaction with PTB may be for stability of its own viral RNA or to prevent its binding with cellular mRNAs and thus impairing the stability of cellular mRNAs. Our results demonstrate a relatively high interacting potential of DENV4 NS1 for PTB. Interaction of NS1 with PTB can be one of the contributing factors for the inhibition of protein synthesis in dengue infection.
It is also discovered in the present study that dengue infection interferes with the Wnt signaling pathway. The NS1 of dengue form biomolecular complex with transcription factor 7-like 2 (TCF7L2 or TCF4) which is a member of Wnt signaling pathway. Role of TCF7L2 has been described in type 2 diabetes as well as gestational diabetes (Jin & Liu, 2008; Zhang et al., 2013). The NS1 of all serotypes show almost equal potency towards TCF4 except NS1 of serotype 4 which has a relatively low affinity for the TCF4. It has been shown that glucose consumption is increased in the dengue infection because the replication of dengue requires glucose metabolism (Fontaine et al., 2015). Inhibition of TCF4 induces gluconeogenesis and glycogenolysis (glucose synthesis from glycogen and non-carbohydrate sources) via the upregulation pre-glucagon. Targeting of TCF4 via NS1 of dengue might be the possible mechanism for the elevation of blood glucose level in dengue infection because the TCF4 is involved in the repression of pre-glucagon synthesis.
In order to find out the interacting residues in the biomolecular complexes of dengue NS1 and host protein, the biomolecular complexes were subjected to PIC. The PIC analysis highlight interactive residues of various domains of dengue NS1 of different serotypes in a biomolecular complex. The PIC analysis shows that in majority of cases, different sets of interactive residues are involved in biomolecular complexes of NS1 of different serotype and a particular host protein. This shows that interaction with a particular host protein and NS1 of different serotypes is governed by different residues because of genetic heterogeneity among the NS1 of different serotypes.
5 Conclusion
The present study explored that there is a high degree of conservation among the various NS1 domains of different isolates of same serotype. However, the NS1 is relatively more heterogeneous among the isolates of different serotypes than that of the same serotype. The Wing domain of NS1 protein plays a vital role in interaction with the panel of host proteins.
CRediT authorship contribution statement
Jadoon Khan: Conceptualization, Methodology, Formal analysis, Writing – original draft, Visualization. Khalid Amin: Formal analysis. Hayat Khan: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. Sadia Butt: Software, Validation. Junaid Ahmad: Resources. Zafar Abbass Shah: Software, Investigation. Shubana Hayat: Investigation. Ajaz Ahmad: Writing – review & editing, Project administration, Funding acquisition. Neelma Hassan: Software, Data curation. Amin Ullah: Conceptualization, Resources, Data curation, Writing – review & editing, Supervision, Project administration.
Acknowledgement
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2024R350), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343(6173):881-885.
- [CrossRef] [Google Scholar]
- Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J. Immunol.. 2011;187(1):424-433.
- [CrossRef] [Google Scholar]
- Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med.. 2015;7(304):304ra141
- [CrossRef] [Google Scholar]
- Dengue virus NS1 protein interacts with the ribosomal protein RPL18: this interaction is required for viral translation and replication in Huh-7 cells. Virology. 2015;484:113-126.
- [CrossRef] [Google Scholar]
- Dengue virus serotype 2 blocks extracellular signal-regulated kinase and nuclear factor-κB activation to downregulate cytokine production. PLoS One. 2012;7(8):e41635
- [CrossRef] [Google Scholar]
- A combined genetic-proteomic approach identifies residues within Dengue virus NS4B critical for interaction with NS3 and viral replication. J. Virol.. 2015;89(14):7170-7186.
- [CrossRef] [Google Scholar]
- Interference with nuclear factor kappa B and c-Jun NH2-terminal kinase signaling by TRAF6C small interfering RNA inhibits myeloma cell proliferation and enhances apoptosis. Oncogene. 2006;25(49):6520-6527.
- [CrossRef] [Google Scholar]
- Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117(14):3720-3732.
- [CrossRef] [Google Scholar]
- Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol.. 2006;80(6):2976-2986.
- [CrossRef] [Google Scholar]
- Herpes simplex virus downregulates secretory leukocyte protease inhibitor: a novel immune evasion mechanism. J. Virol.. 2008;82(19):9337-9344.
- [CrossRef] [Google Scholar]
- Dengue virus induces and requires glycolysis for optimal replication. J. Virol.. 2015;89(4):2358-2366.
- [CrossRef] [Google Scholar]
- Critical review of the vector status of Aedes albopictus. Med. Vet. Entomol.. 2004;18(3):215-227.
- [CrossRef] [Google Scholar]
- Dengue fever: new paradigms for a changing epidemiology. Emerg Themes Epidemiol.. 2005;2(1):1.
- [CrossRef] [Google Scholar]
- Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci.. 2011;108(19):8003-8008.
- [CrossRef] [Google Scholar]
- Minireview: The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. J. Mol. Endocrinol.. 2008;22(11):2383-2392.
- [CrossRef] [Google Scholar]
- Tanshinone IIA inhibits breast cancer stem cells growth in vitro and in vivo through attenuation of IL-6/STAT3/NF-kB signaling pathways. J. Cell. Biochem.. 2013;114(9):2061-2070.
- [CrossRef] [Google Scholar]
- Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med.. 2015;7(304):304ra142
- [CrossRef] [Google Scholar]
- The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res.. 2013;98(2):192-208.
- [CrossRef] [Google Scholar]
- Strategies for development of dengue virus inhibitors. Antivir. Res. 2010;85(3):450-462.
- [CrossRef] [Google Scholar]
- Open source tool for prediction of genome wide protein-protein interaction network based on ortholog information. Source Code Biol. Med.. 2010;5(1):8.
- [CrossRef] [Google Scholar]
- Flavivirus NS1: a multifaceted enigmatic viral protein. Virol. J.. 2016;13(1):131.
- [CrossRef] [Google Scholar]
- Paip1 interacts with poly (A) binding protein through two independent binding motifs. Mol. Cell. Biol.. 2002;22(11):3769-3782.
- [CrossRef] [Google Scholar]
- Molecular targets for flavivirus drug discovery. Antivir. Res.. 2009;81(1):6-15.
- [CrossRef] [Google Scholar]
- TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell. 2000;5(4):649-658.
- [CrossRef] [Google Scholar]
- The regulatory role of serum response factor pathway in neutrophil inflammatory response. Curr. Opin. Hematol.. 2015;22(1):67.
- [CrossRef] [Google Scholar]
- Activation of the NF-κB pathway as a mechanism of alcohol enhanced progression and metastasis of human hepatocellular carcinoma. Mol. Cancer. 2015;14(1):10.
- [CrossRef] [Google Scholar]
- Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res.. 2001;11(3):422-435.
- [CrossRef] [Google Scholar]
- miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J. Infect.. 2013;67(4):329-341.
- [CrossRef] [Google Scholar]
- Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis. Virol. J.. 2013;10(1):339.
- [CrossRef] [Google Scholar]
- Identification of a novel serum response factor cofactor in cardiac gene regulation. J. Biol. Chem.. 2004;279(53):55626-55632.
- [CrossRef] [Google Scholar]
- Association of TCF7L2 gene polymorphisms with type 2 diabetes mellitus in Han Chinese population: a meta-analysis. Gene. 2013;512(1):76-81.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103108.
Appendix A
Supplementary material
The following are the Supplementary data to this article:
Supplementary data 1
Supplementary data 1
Interactive residues among the complexes of NS1 of various dengue serotypes and TRAF6







