Translate this page into:
Exploring major bioactive phytocompounds of Ficus racemosa and its key pharmacological activities
⁎Corresponding authors at: Department of Biotechnology, Saveetha School of Engineering, Saveetha Institute of Medical and Technical Sciences (SIMATS), Thandalam, Chennai 602105, Tamil Nadu, India (A. Thirunavukarasou). bupeshgiri@gmail.com (Giridharan Bupesh), anand.t@baatral.com (Anand Thirunavukarasou), anicholasdaniel@gmail.com (Nicholas Daniel Amalorpavanaden) nicholasdaniel@mukuba.edu.zm (Nicholas Daniel Amalorpavanaden)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Ficus racemosa has long been used as a treatment for bacterial and fungal infections by people in India. The search for a safe alternative therapy from plant resources has gained interest in recent years due to herb’s more central place in contemporary medicine and their status as a vast global reservoir of potentially active pharmaceuticals. Using standard methods, we aim to evaluate the pharmacological and bioactive potential of F. racemosa compounds extracted from fruits. Our observation reveals that stigmasterol has the highest docking score across all targets, making it the most promising chemical overall. According to Lipinski's rule of thumb, the obtained compounds derived from F. racemosa were predicted to have high bioactivities, giving them a high drug-likeliness score. Target and compound docking scores provide insight into binding posture and critical amino acid residues in antibacterial and antifungal activity. This research confirms that F. racemosa fruit extract is effective against fungal infections. These findings will aid biochemists in their quest to produce effective phytochemicals by further testing multi-targeting drugs.
Keywords
Herbal medicines
Natural products
Drug design
Phytochemicals
Stigmasterol
1 Introduction
India is one of the top mega biodiversity nations, and its ethnic groups have vast information about how to treat various infections with plants (Heinrich, 2000; Nachiappan et al., 2022). The advantage over the numerous applications of plants as medicine is conferred by the traditional knowledge gained over centuries, which is of enormous relevance (Jose and Manchikanti, 2022; Shankar and Rawat, 2013). The use of herbal medicine is drastically increasing in wealthy nations (Dobos and Tao, 2011; Yuan et al., 2016). The World Health Organization (WHO) has listed 20,000 medicinal plants utilized worldwide (Tagboto et al., 2001). The Ficus racemosa Linn. (Moraceae) plant is well-known and can be found in Southeast Asian parts. It has many medicinal properties and is often used to treat wounds. Parts of the plant, like the fruit, leaves, and bark, have been tested for their pharmacological properties (Shi et al., 2018). These include antidiarrheal, anti-inflammatory, antipyretic, antifungal, antibacterial, hypolipidemic, antifilarial, and hepatoprotective (Velayutham et al., 2012; Veerapur et al., 2012). Surprisingly, people have used the plant's fruits to treat leprosy, menorrhagia, leucorrhoea, and blood disorders, as well as burns, intestinal worms, dry cough, and urinary tract infections (Mousa et al., 1994; Keshari et al., 2016).
F. racemosa is a moderate-to-large, spreading, lactiferous, deciduous tree that is 15–––18 m high and has no obvious aerial roots (Bhalerao et al., 2014). Ficus is a very large genus of tropical plants. It has more than 700 species that grow in warm parts of Asia, Africa, America, and Australia (Olaoluwa et al., 2022). In Ayurveda, all parts of this plant are considered to have medicinal value. It has been used extensively to treat biliary disorders, jaundice, dysentery, diabetes, diarrhoea, and conditions that cause inflammation (Deep et al., 2013). Numerous phytochemicals, physicochemical compounds, and minerals may be found in the fruit, bark, roots, and latex of F. racemosa (Ca, K, Mg, P, Fe) (Ahmed and Urooj, 2010). Trees provide enough nutrition in their fruit and bark (crude protein, total lipids, crude fat, sugars, and starch). Rich in reducing sugars, flavonoids, and minerals, F. racemosa leaves are a nutritional powerhouse (Pahari et al., 2022). The fruit's easily absorbed carbohydrates, high-calorie content, and phytotherapeutic components make it a valuable food source (Joseph and Raj, 2010). Fasting blood glucose levels in normal, type 1, and type 2 diabetic rat models were used to assess the hypoglycemic and antioxidant properties of F. racemosa fruit extract on various solvents, with the type 2 diabetic model showing the highest levels of activity (Mousa et al., 1994). In vitro research comparing the hypoglycemic effects of fruits and leaves has shown that the former had the upper hand (Amboupe and Mustaqim, 2021). In parallel, molecular modeling and molecular dynamics serve as a important tool to aid the understanding of the fundamental concepts of structure- activity relationships, and to elucidate the mechanism of action of phytochemicals against different targets.
Clinical pathogens are usually dangerous for every living thing on earth (Wienhues, 2022). Scientists have long recognized viruses, bacteria, and fungi as potential agents of various disorders (Qadri et al., 2022). The microorganisms that cause diseases like Acquired immunodeficiency syndrome (AIDS) and tuberculosis are becoming more resistant to antibiotics and other treatments as they learn to survive in a shifting environment (Kishore Kumar et al., 2022). Competing microbes, such as Escherichia coli and Candida albicans, are thought to promote disease transmission and infection rates (Rueda-Robles et al., 2022). Although synthetic chemicals are effective in resisting bacterial growth, they are impractical and may negatively affect people's health (APB et al., 2022). Considering the medicinal significance of the microorganisms, this research focused on characterizing the phytochemicals present in F. racemosa fruit that aid in developing new drugs derived from plants.
2 Materials and methods
2.1 Fruits collection and preparation
The physiologically active chemicals and microbe-inhibiting characteristics of F. racemosa fruits were collected, cleaned, and preserved. The process mentioned above was used to dry and powder fruit. The powder was kept at room temperature. The solubility, characterization, and identification of the phytochemicals in the fruit extract were investigated by extracting the produced sample in several solvents, including diethyl ether, ethyl acetate, dichloromethane, and chloroform.
2.2 Fourier transform infrared spectroscopy (FTIR) analysis
Functional groups in F. racemosa fruit powder extracted with methanol were identified and characterized using FTIR. Twenty milligrams of fruit extract were coated with one hundred milligrams of KBr pellet to create a clear disc for testing. The FTIR spectroscopy was run with a scan range of 400 to 4000 cm−1 and a resolution of 4 cm−1, and dried powder was added randomly. The functional groups were detected in the 4000–400 cm−1 range.
2.3 Phytochemical profiling by gas chromatography–mass spectrometry (GC–MS) analysis
For the GC–MS analysis, we utilized a Shimadzu 2010 plus equipped with an AOC-20i autosampler and a gas chromatograph coupled to a mass spectrometer under the following settings: Injector temperature 270 °C; ion-source temperature 200 °C; column RTX 5Ms (Column diameter 0.32 mm; column length 30 m; column thickness 0.50 m); electron impact mode at 70 eV; Helium gas (99.999 %) used as carrier gas at a constant flow of 1.73 ml /min; injection volume 0.5 I (split ratio 10:1). To achieve the desired results, and the oven was set to gradually increase in temperature from 40 degrees Celsius (isothermal for 2 min) to 150 degrees Celsius, then at a rate of 8 degrees Celsius per minute to 250 degrees Celsius, and lastly for 20 min at a temperature of 280 degrees Celsius. The mass spectral data were gathered from collisions with fragments ranging in size from 40 to 450 Da, and the excitation energy was 70 eV, with a scanning interval of 0.5 s. The overall length of the GC is 51 min and 25 s. The percentage contributions of the individual components were determined using the ratio of the average peak area of each component to the sum of all areas. To manage mass spectra and chromatograms, Turbo Mass Version 5.2.0 was utilized (Krishnamoorthy et al., 2014). For GCMS analysis, scientists used the NIST database, which includes more than 62,000 patterns. The spectrum of the unknown component was compared to the spectra of known components housed in the NIST collection. The test material’s constituents' names, molecular weights, and structures were established.
2.4 Determination of antimicrobial activity
The antimicrobial activity was performed by the disc diffusion method followed by Awoyinka et al. (2007) (Awoyinka, 2007).
2.5 Preparation of Media
Nutrient Agar (NA-Himedia) Media for Bacteria. Media composition (Animal’s tissue 5.00 g, Sodium chloride 5.00 g, Beef extract 1.50 g, Yeast extract 1.50 g, Agar 15.0 g).
2.6 Preparation of medium
Suspend 28.0 g of nutrient agar in 1000 ml of distilled water. Heat to boiling and dissolve the medium completely. Sterilize by autoclaving at 15 Ibs pressure (121˚c) for 15 min. Mix well and pour into sterile Petri plates.
Potato Dextrose Agar (PDA-Himedia) Media for Fungi. Media composition (Potatoes infusion 200.00 g, Dextrose 20.00 g, Agar 15.00 g).
2.7 Preparation of medium
Put 39 g of PDA into a litre of distilled water and shake it up. Turn up the heat until the medium is fully dissolved. Autoclave at 15 Ibs pressure (121˚c) for 15 min to sterilize. When a pH of 3.5 is needed, acidify the medium with sterile 10 % tartaric acid and mix thoroughly before using. It takes around 1 ml of acid to treat 100 ml of cooled sterile medium. After adding acid, the medium should be kept from being heated.
3 Microorganisms
Escherichia coli (MTCC 732) and Candida albicans (MTCC 183) were used as bacterial and fungal microbial strains, respectively, in the respective biological experiments, sourced from the IMTECH (Institute of Microbial Technology) Microbial Type Culture Collection (MTCC) in Chandigarh, India.
3.1 Preparation of 24 h of pure culture
About 10 ml of physiological saline were used to suspend a loop full of each bacteria in a Roux bottle. All were streaked onto the proper culture slants, and after 24 h at 37 °C (except for the fungal culture, which needed 48 h at 25 °C), they were all incubated. Once the incubation time had ended and growth had been detected, the tubes were placed in 2–8C storage.
3.2 Preparation of samples solutions for the experiment
Standard solutions included chloramphenicol (25 mg/ml) and fluconazole (25 mg/ml) in distilled water, and each sample weighed 10 mg before dissolving it in 10 ml of solvent. They were stored in the fridge until needed for the experiment.
3.3 Preparation of dried filter paper discs
Four discs measuring 6 mm in diameter were cut from Whatman filter paper (No.1) and sterilized with hot air. The standard Chloramphenicol 30 ml and control 30 ml (ethanol) was used to compare the test solution. Discs were loaded with 50 ml, 100 ml, and 150 ml of each sample after sterilization. They were stored in the fridge until needed for the experiment.
3.4 ADME properties
Using qikprop, we double-checked the physiochemical characteristics of the GC–MS identified phytochemicals in the F. racemosa extract. Lipophilicity, solubility, bioavailability, drug-likeness, and human oral absorption of phytochemicals found in the PubChem and chemical database were assessed.
3.5 HOMO-LUMO energy prediction
To understand the energies of the border molecular orbital, salvation, the HOMO-LUMO gap, and the softness of a compound, the jaguar module was used to determine the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO). The energies of the phytocompounds were calculated using DFT/B3LYP, which considered the bond lengths, bond angles, and atom transfers. Predicted energy levels for a molecule were ranked according to the relative importance of its HOMO and LUMO energy gaps and their ability to form a solution. The negative eV values of tiny molecules showed compounds' stability and charge transfer.
3.6 Molecular docking
The extract's chemical compounds against the E. coli and C. albicans were investigated by molecular docking studies. The target’s crystal structure reveals the atomic basis for the sensitivity of DraE-CD55 binding to chloramphenicol, in contrast to other chloramphenicol-protein complexes (PDB:1USQ). Proteins of the eukaryotic CYP51 family (PDB:5V5Z) found in the endoplasmic reticulum are cytochrome P450 monooxygenases that are bitopic and mono-cross the membrane. An essential enzyme, lanosterol 14-demethylase (LDM), is involved in human and fungal cholesterol production. Finally, we considered the 3D structures of proteins from E. coli (PDB:1USQ) and C. albicans (PDB:5V5Z). Therefore, the Glide XP technique was chosen over full molecular docking of the protein–ligand (Friesner et al., 2004). To identify a molecule with high antimicrobial activity, we ranked and tabulated predictions of receptor-ligand binding affinity.
3.7 Molecular dynamics simulation
Molecular dynamics simulation was applied to a complex file, including a protein and a phytocompound. The GROMACS 2019 package was started to capture each frame for molecular identification during trajectory formation (Pronk et al., 2013). To generate a topology, small molecules were subjected to PRODRG2 server run. The SPC water model and the GROMOS96 43a1 force field were used to arrange the protein topology (Jorgensen et al., 1983). Cubic boxes were used to contain the complex system and achieve solvation and energy reduction. The addition of NA and Cl ions deactivated the receptor-ligand complex system. To do this, we used the steepest descent integrator to minimize energy costs. Using a leap-frog integrator, we achieved thermal and pressure equilibrium in the complicated system at 300 Kelvin (K) and 1 bar, respectively. Molecular dynamics simulations were used to evaluate NVT and NPT. The frames were time-stamped to within 100 ns using Trajectory files.
3.8 Computation of binding free energy by MM-PBSA
Using the Molecular Mechanics/Poisson-Boltzmann Surface Area (MMPBSA) method, we calculated the free binding energy of protein–ligand complex structures from Molecular Dynamics trajectories (Wang et al., 2018). All of the three best-performing protein–ligand complexes had their MM-PBSA free energy predicted. The binding free energy was determined using the MM/PBSA method implemented in the g_mmpsba tool. The final 20 ns trajectory (20 frames from each nanosecond) of the 100 ns normal NPT MD simulation was used (Kumari et al., 2014).
4 Results and discussion
4.1 Plant sample solubility
The sample was dissolved in various solvents for 48 h, and the solubility was observed (Diethyl ether - Immiscible, Ethyl acetate- Immiscible, Dichloro methane- Partially Soluble, Chloroform-Immiscible).
4.2 FTIR spectroscopic analysis
For the FTIR analysis, the unique peaks between 400 and 4000 cm−1 and the related functional groups were identified using a Perkin Elmer Spectrophotometer system (Table S1 and Fig. 1). The highest FTIR values possible were recorded. To assure accuracy in the spectrum verification, every analysis was carried out twice (Arokiyaraj et al., 2012). Functional groups in the biosynthesized F. racemosa fruit extract were found using FTIR spectroscopy (in the 400–4000 cm−1 range). There were noticeable characteristic absorption peaks for the functional groups of secondary metabolites, including alcohol and phenols (O–H bonds, peak at 3404.72 cm−1), alkenes (C–H bonds, peak at 2977.22 cm−1), carboxylic acids (O–H bonds, peak at 2541.22 cm−1), alkynes (-C = C- stretch, peak at 2131.55 cm−1), and aromatic amines (C-N stretch, peak at 1254 cm−1). Alcohols, phenols, and alkenes were identified as the most frequently occurring functional groups in the extract based on the analysis of the FTIR test findings.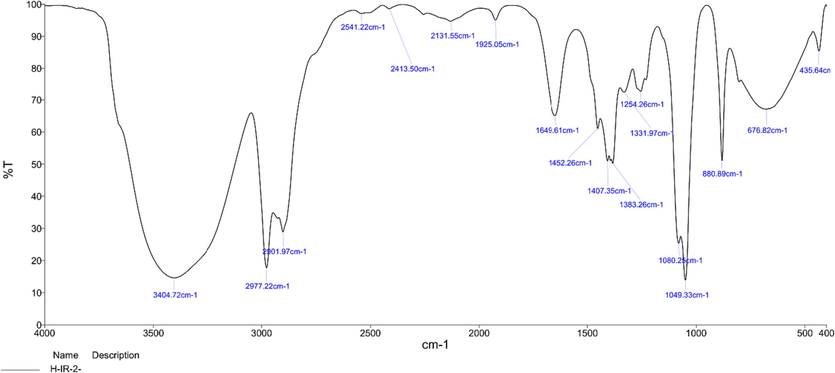
FTIR spectrum of Ficus racemosa fruit extract. Characteristic peaks indicated the presence of important phytochemicals.
4.3 Identification of bioactive compounds in your sample by GC MS analysis
Next, the chemical constituents of the F. Racemosa extract were identified by GC–MS, and the mass spectra of chemical constituents of active compounds were identified by searching in NIST and Wiley libraries. The GC–MS analysis of the sample extract revealed the presence of ten different chemicals. Table S2 lists the active ingredients together with their corresponding retention time (RT), molecular formula, molecular weight (MW), and concentration (%). Diethyl esters of 1,2-benzenedicarboxylic acid, hexadecanoic acid (10.031), pentadecanoic acid (14.256), and other predominate. Methyl ester of (Z)-9,12-octadecadienoic acid (Z,Z)-11,14-eicosatrienoic acid (Z,Z)-17 (Z)(16.072). A total of 9-Pentadecadien-1-ol (14.256), 4-Hexadecen-6-yne (16.168), and Nonacosane (16.067) were discovered in the sample (Fig. 2). The plant's chemical composition, which contains many bioactive compounds, supports how traditional healers have used it to cure various ailments (Vijayaram et al., 2016; Saravanan et al., 2022). Yet, a favorable outcome is guaranteed when certain phytochemical components are isolated, and their biological activity is tested.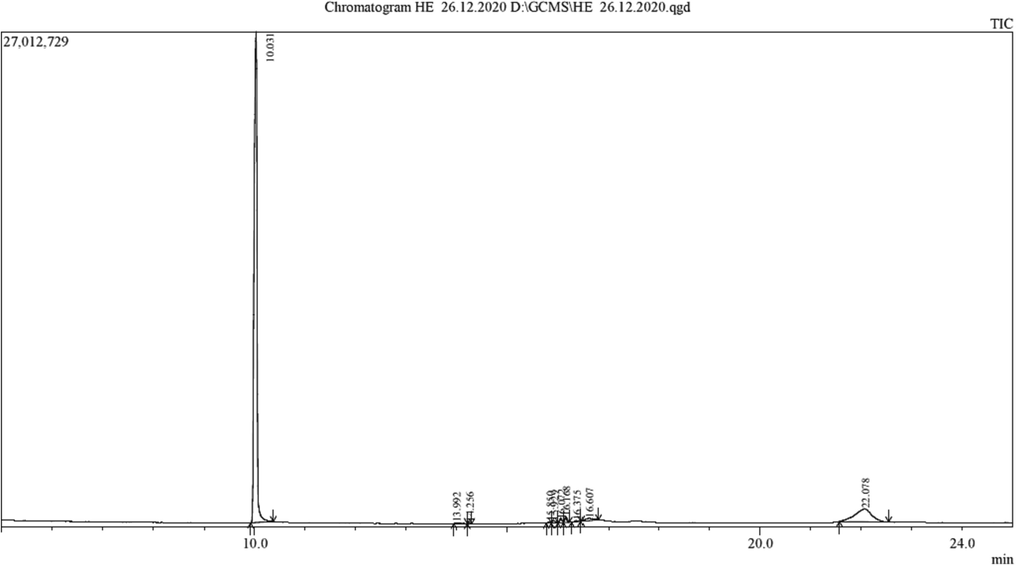
GC–MS spectrum of Ficus racemosa fruit extract.
4.4 Antimicrobial activity
The material was used to create an antibiogram using the disc diffusion technique. Thirty millilitres of NA/PDA medium were poured onto Petri dishes for the experiment. Using a micropipette, the test organism was inoculated onto a plate of solidified agar, where it was disseminated and left to dry for 10 min. Bacteria from a broth culture were used to inoculate the medium surfaces. Nutrient agar /PDA plates are inoculated by swabbing the whole surface with a sterile cotton swab dipped into a test suspension of a standardized microorganism. Various microorganisms were seeded onto Nutrient agar/PDA plates using inoculums. The infected agar plate was placed on a flat surface, and sterile filter sheets (6 mm in diameter) holding 30 l of sample and 30 l of Standard solution were placed on top using sterile forceps. At 37 degrees Celsius, the plates harboring the bacteria were left to grow for 24 h, while those harboring the yeast were left to do so for 48 h. All samples were analyzed three times.
4.5 Measurement of the zone of inhibition
The antibacterial potential of the test compounds was determined by calculating the average millimeter-sized zone of inhibition around the disc (Fig. 3). The samples' capacity to restrict the development of the microorganisms employed in the experiment was measured on a millimetre scale. Escherichia coli and Candida albicans were used as test organisms to determine the antibacterial activity of plant extracts from F. racemosa. For the examined bacteria, the extract had the highest level of antibacterial activity with a minimum inhibitory concentration in the range of 50 μl to 150 μl. According to these investigations, the higher concentrations of alcohol and phenol found in these extracts were responsible for the improved antibacterial activity shown. The antibacterial activity of the F. racemosa fruit extract was shown by the considerable slowing of E. coli growth (Bagyalakshmi et al., 2019). Nevertheless, further research is necessary to determine if any additional secondary metabolites contribute to the antibacterial action (Pingale et al., 2019). Second, the most extensively researched types of bacteria in antimicrobial research are those used to assess their antimicrobial activity. Therefore more studies into multi-drug resistant strains should be done (Chaware et al., 2020). Although this plant species' antibacterial activity has only been the subject of a small number of research, its dose-dependent antimicrobial activity against other microorganisms should be taken seriously.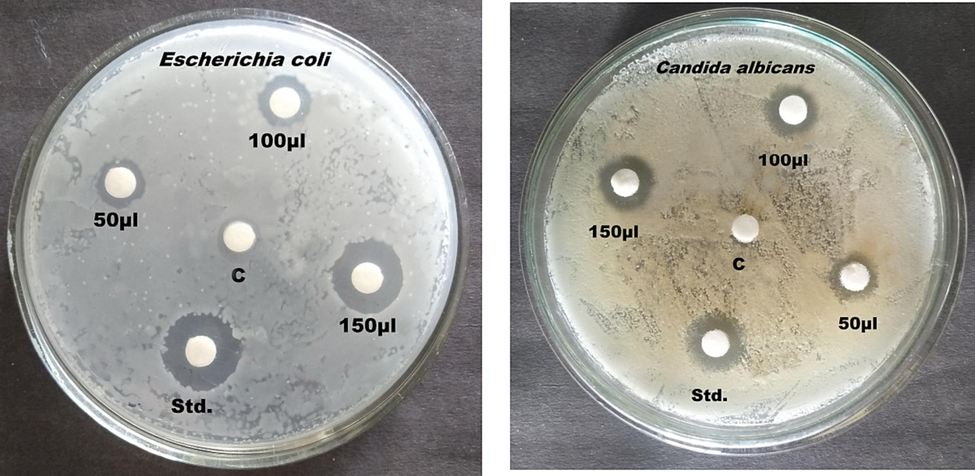
Anti-microbial activity of Ficus racemosa plant sample.
4.6 ADME properties
Additionally, the physicochemical characteristics and drug-likeness of phytochemicals revealed by GC–MS analysis were investigated. A compound's adherence to the Lipinski rule of five (RO5) is a potential prerequisite for its selection as a lead molecule. The phytochemicals were ranked according to RO5. Fig. 4 shows that after thorough inspection, it was determined that the compounds in question had the absolute qualities and the proportion of human oral absorption anticipated by the ADME properties. Phytochemicals 1, 2-benzenedicarboxylic acid diethyl ester and trans-stigmasta-5, 22-dien-3.beta show better qualities in in-vitro investigations than the gold standards (chloramphenicol and flucnazole).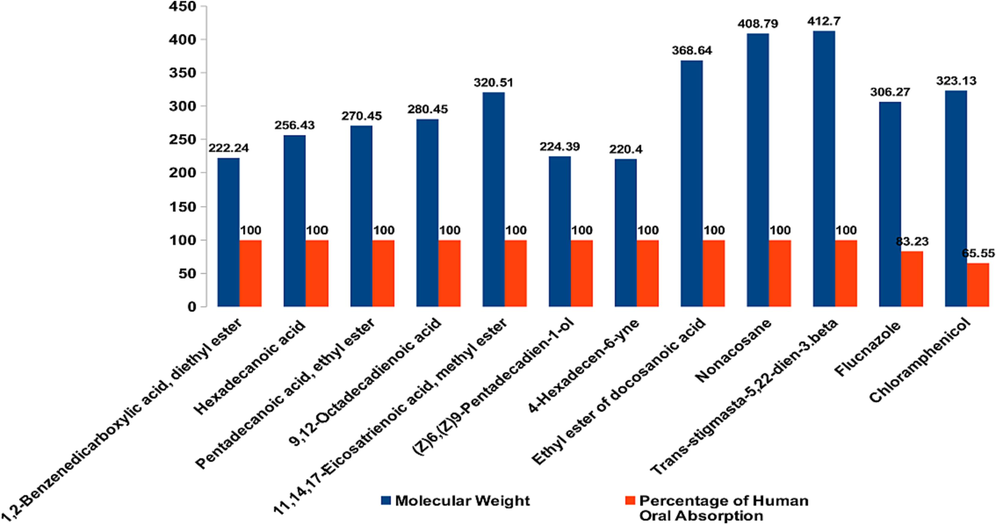
Molecular weighted percentage of human oral absorption is plotted to show the rule of five (RO5) for lead molecule identification.
4.7 HOMO-LUMO energy calculation
HOMO-LUMO energy computation plays a crucial role in the nucleophilic and electrophilic transfer and associations. Density functional theory was used to examine phytocompounds for their physicochemical properties and optimizing their border molecular orbitals and geometric structures (DFT). We used the conventional and widely-adopted DFT/B3LYP approach in Schrodinger to do our calculations. Phytochemicals with the most optimal shape were found, and they are now thought to be promising lead molecules for a wide range of biological functions.. The HOMO and LUMO energies predicted for the phytochemicals are shown in red and blue, respectively, in Fig. 5. 1, 2-Benzenedicarboxylic acid, diethyl ester (21.348 Kcal/Mol) and Trans-stigmasta-5, 22-dien-3.beta (46.406 Kcal/Mol) have greater solvation energies than benzene (C5H5C6H5O2). All phytochemicals have minimal HOMO-LUMO energy preference gaps for maximum action and stability.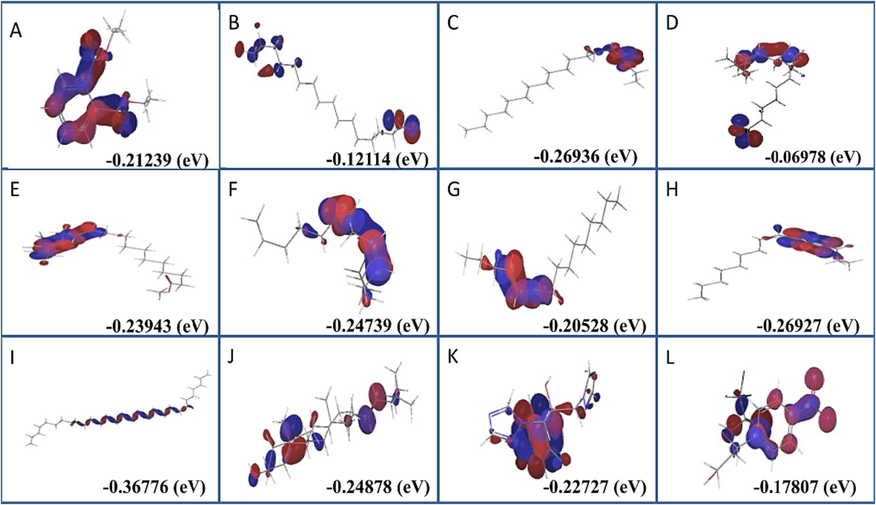
Homo-lumo energy preference of phytochemical identified in Ficus racemosa was in blue and red color. (A) 1,2-Benzenedicarboxylic acid, diethyl ester, (B) Hexadecanoic acid, (C) Pentadecanoic acid, ethyl ester, (D) 9,12-Octadecadienoic acid, (E) 11,14,17-Eicosatrienoic acid, methyl ester, (F) (Z)6,(Z)9-Pentadecadien-1-ol, (G) 4-Hexadecen-6-yne, (H) Ethyl ester of docosanoic acid, (I) Nonacosane, (J) Trans-stigmasta-5,22-dien-3.beta, (K) Flucnazole, (L) Chloramphenicol.
4.8 Molecular docking
The phytochemical docking score was compared to chloramphenicol-resistant bacteria and fluconazole-resistant fungus. As predicted, chemicals had a greater binding score for bacteria and fungus than controls, as presented in Tables S3 and Table S4. Different groups of researchers discuss the specific features and biological activities of each component isolated from the F. racemosa plant. Figs. 6 and 7 from the GC–MS report demonstrate that 1, 2-benzenedicarboxylic acid, diethyl ester and Trans-stigmasta-5, 22-dien-3, are abundant in the methanol extract and have high binding affinity compared to the standards chloramphenicol (-3.533 Kcal/Mol) and flucnazole (-5.391 Kcal/Mol). Despite this, substances that do not attach to the essential amino acids in the protein have been shown to have a greater binding affinity. Repeatedly, trans-stigmasta-5,22-dien-3 (stigmasterol) developed hydrogen bonding and hydrophobic contact with the aforementioned residues in C. albicans (-10.849 Kcal/Mol), respectively. From our docking results, we considered three compounds for further study based on glide score, GCMS results, and bioactivity predictions, such as Trans-stigmasta-5,22-dien-3, Ethyl ester of docosanoic acid, and 1,2-Benzenedicarboxylic acid, diethyl ester respectively. Discovery Studio Visualizer software was used to create Fig. 8, which depicts the interactions between the top three compounds in terms of hydrogen, hydrophobic, and other non-bonded terms. In the figure, the light green spheres represent van der Waals hydrophobic interactions, which are the dominant interaction between proteins and ligands. The figure's dark green spheres represent the typical hydrogen bonds contributed by amino acid residues. In the picture, the red orb represents unfavorable hydrogen bonding. Compared to the other two chemicals, the ethyl ester of docosanoic acid forms the most contact (24 interactions) with the protein through its amino acid residues, as seen in the figure. The chemicals Trans-stigmasta-5,22-dien-3 form 22 amino acid residue connections with protein whereas 1,2-Benzenedicarboxylic acid, diethyl ester form 15 amino acid residue interactions correspondingly.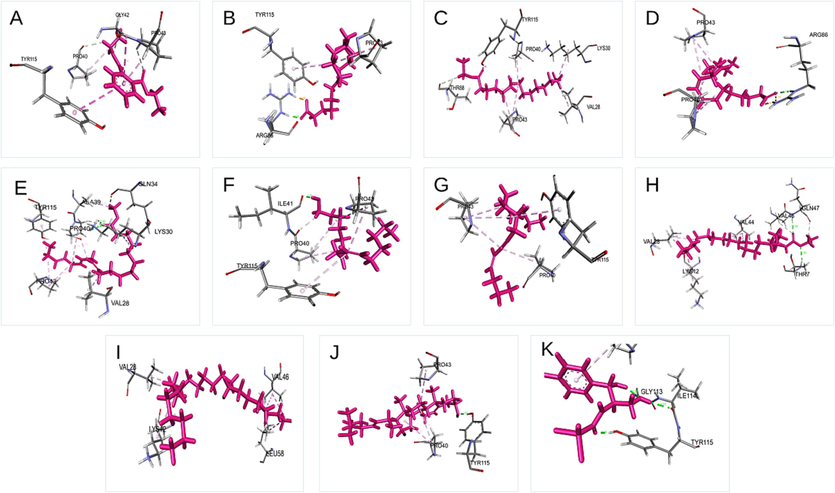
Schematic 3D interaction of phytochemical identified in Ficus racemosa with E.coli. (A) 1,2-Benzenedicarboxylic acid, diethyl ester, (B) Hexadecanoic acid, (C)Pentadecanoic acid, ethyl ester, (D)9,12-Octadecadienoic acid, (E) 11,14,17-Eicosatrienoic acid, methyl ester, (F) (Z)6,(Z)9-Pentadecadien-1-ol, (G) 4-Hexadecen-6-yne, (H) Ethyl ester of docosanoic acid, (I) Nonacosane, (J) Trans-stigmasta-5,22-dien-3.beta, (K) Chloramphenicol.
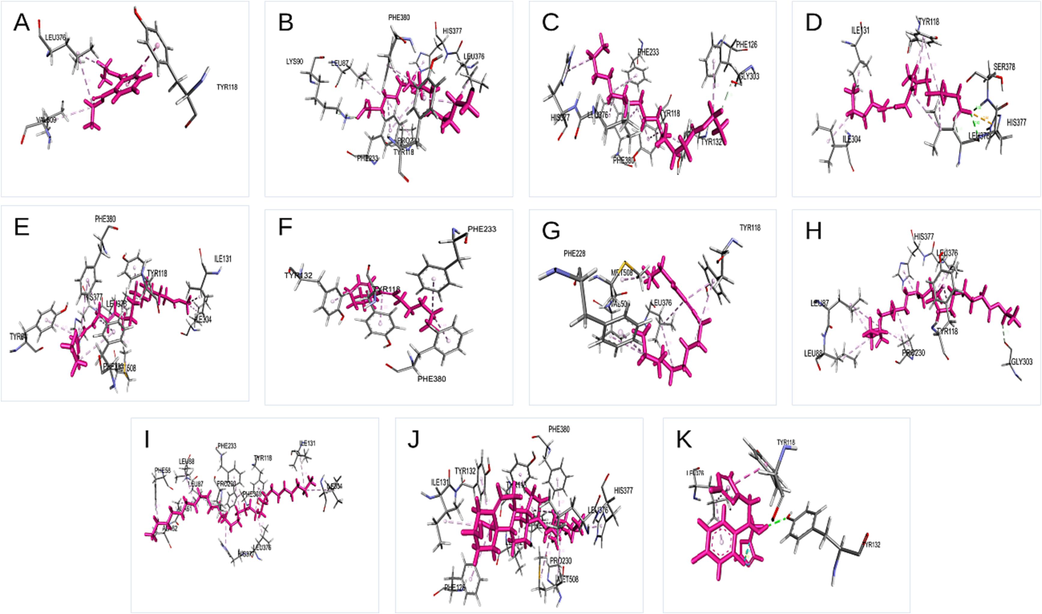
Schematic 3D interaction of phytochemical identified in Ficus racemosa with C. albicans structure. (A) 1,2-Benzenedicarboxylic acid, diethyl ester, (B) Hexadecanoic acid, (C) Pentadecanoic acid, ethyl ester, (D) 9,12-Octadecadienoic acid, (E) 11,14,17-Eicosatrienoic acid, methyl ester, (F) (Z)6,(Z)9-Pentadecadien-1-ol, (G) 4-Hexadecen-6-yne, (H) Ethyl ester of docosanoic acid, (I) Nonacosane, (J) Trans-stigmasta-5,22-dien-3.beta, (K) Fluconazole.
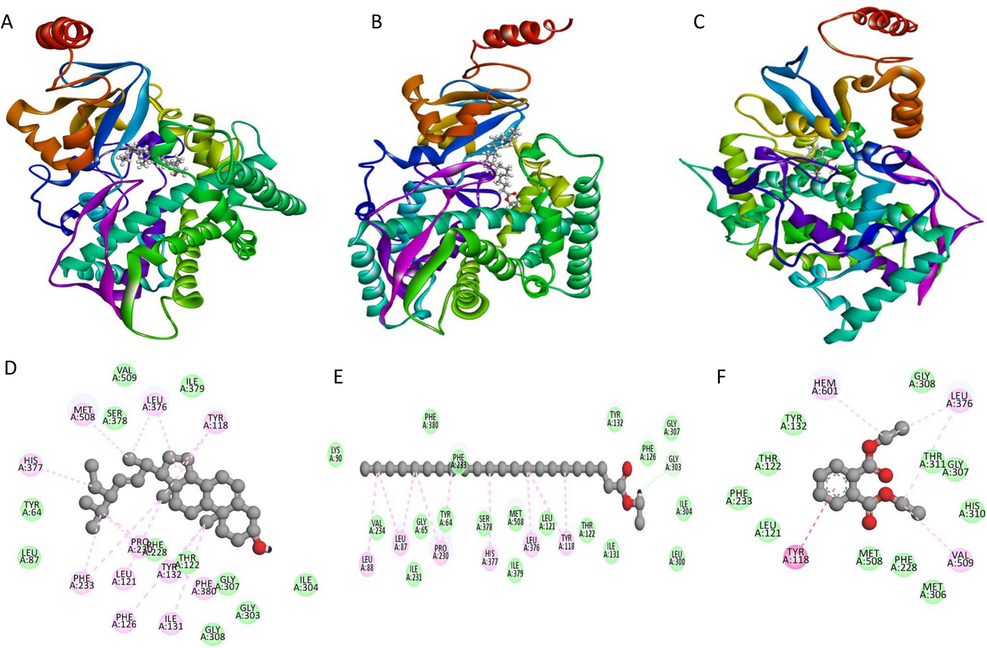
Schematic 3D (ABC) and 2D interaction diagram (DEF) of three compounds with CYP51 from the pathogen C. albicans. Dark green balls indicate van der Waals interactions whereas light green color indicates hydrogen bonds. Yellow and red color indicates cation-pi and unfavorable hydrogen bonds between protein and ligand [Trans-stigmasta-5,22-dien-3 (A), Ethyl ester of docosanoic acid (B), 1,2-Benzenedicarboxylic acid, diethyl ester (C), Trans-stigmasta-5,22-dien-3 (D), Ethyl ester of docosanoic acid (E), 1,2-Benzenedicarboxylic acid, diethyl ester.
4.9 Molecular dynamics simulation
Gromacs 2019, which records trajectories for 100 ns, started to examine the conformational change of protein–ligand complexes. Stigmasterol-bound fungal protein was studied further to elucidate their inhibitory effects. A root-mean-square deviation (RMSD) plot was created in the trajectory files using xm_grace. Evidence from many time-lapse recordings of a protein–ligand complex shows that the protein changes in predictable ways in response to the ligands. To calculate the protein–ligand complexes dynamic stability, we tracked the relative mean squared deviation (RMSD) as a function of simulation duration (Fig. 9). Through 100 ns simulations, total conformational dynamics were used to calculate the structural variability of protein complexes. The protein-Trans-stigmasta-5,22-dien-3 combination is very stable, as shown by the average root-mean-square deviation (RMSD) (0.5 nm) across a 100-ns molecular dynamics simulation. The other ligands have a much more dynamic RMSD (between 0.5 and 1 nm) than the protein (Figure RMSD = 1 nm). The complex's RMSD relative to a suitable reference, on the other hand, suggests that the ligand itself undergoes a little conformational change upon binding. Protein backbone RMSD is within the acceptable range for the top three complexes, shedding light on the sequence of conformational changes. All three ligands form an average of 3 hydrogen bonds with the protein during the simulation (Fig. 10).![The root mean square deviation of protein backbone and ligand [Trans-stigmasta-5,22-dien-3 (A), Ethyl ester of docosanoic acid (B), 1,2-Benzenedicarboxylic acid, diethyl ester (C)] for 100 ns simulation time is presented.](/content/185/2024/36/1/img/10.1016_j.jksus.2023.102956-fig10.png)
The root mean square deviation of protein backbone and ligand [Trans-stigmasta-5,22-dien-3 (A), Ethyl ester of docosanoic acid (B), 1,2-Benzenedicarboxylic acid, diethyl ester (C)] for 100 ns simulation time is presented.
![The number of hydrogen bonds formed between protein-[Trans-stigmasta-5,22-dien-3 (A), Ethyl ester of docosanoic acid (B), 1,2-Benzenedicarboxylic acid, diethyl ester (C)] for 100 ns simulation time is presented.](/content/185/2024/36/1/img/10.1016_j.jksus.2023.102956-fig11.png)
The number of hydrogen bonds formed between protein-[Trans-stigmasta-5,22-dien-3 (A), Ethyl ester of docosanoic acid (B), 1,2-Benzenedicarboxylic acid, diethyl ester (C)] for 100 ns simulation time is presented.
4.10 Binding free energy of protein–ligand complexes
Molecular mechanics—Poisson-Boltzmann surface area continuum salvation (MM-PBSA) is the most popular approach for verifying molecular docking results, which involves calculating the binding free energy of protein and ligand complex. Using the MM PBSA method, the physical properties of protein–ligand complexes were determined. From the MD trajectories, we were able to estimate the binding energies. Figure S1 displays that the free binding energy of the protein with rans-stigmasta-5,22-dien-3 is greater than that of the other two protein–ligand complexes. The figure also displayed several stabilizing physical free energies, including van der Waals, electrostatic, and Solvent Accessible Surface Area (SASA). Higher van der Waals, electrostatic, and SASA free energy are seen in the docosanoic acid combination protein-Ethyl ester. As seen in figure, our data demonstrate that all three protein–ligand complexes have lower free energies than predicted.
5 Conclusion
In the essentials of drug design and personalized medicine, invitro and insilico combined analysis has been accepted widely to save and serve human welfare. In the current study, the fruit extract of F. racemosa has been analyzed for phytochemical profiling and antimicrobial inhibition properties. FTIR studies have been exercised to mark the functional groups through spectroscopy analysis. Invitro analysis reported that the fruit extract administered on E. coli and C. albicans has better inhibition properties than the standard, chloramphenicol and flucnazole, respectively. GC–MS analysis revealed the phytocompounds in the fruit extract treated with methanol/ethanol. ADME properties, density functional theory, molecular docking, and dynamics have proved the compound's efficacy as a potential molecule to combat clinical pathogens such as E. coli and C. albicans. Phytochemical profiling with invitro and insilico analysis has recognized the Trans-stigmasta-5, 22-dien-3.beta (Stigmasterol) as the lead compound in the F. racemosa fruit. Further biophysical and biochemical experiments will aid to understand the activity of the compound against several pathogens.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research and Innovation, “Ministry of Education” in Saudi Arabia for funding this research (IFKSUOR3-185-2). The authors thank Saveetha School of Engineering, Saveetha Institute of Medical and Technical Sciences (SIMATS) and Mukuba University, Off Chingola Road, Itimpi, P.O.Box. 20382, Kitwe, Zambia for the support.
Authors’ contributions
All the authors contributed to this work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Traditional uses, medicinal properties, and phytopharmacology of Ficus racemosa: A review. Pharm. Biol.. 2010;48:672-681.
- [CrossRef] [Google Scholar]
- Amboupe, D.S., Mustaqim, W.A. 2021. Ficus racemosa L. Moraceae BT - Ethnobotany of the Mountain Regions of Southeast Asia. In: Franco FM, editor., Cham: Springer International Publishing, p. 465–70. https://doi.org/10.1007/978-3-030-38389-3_181.
- APB, B., Bhuvaneswari, S., Raj, L., Bupesh, G., Meenakshisundaram, K., KM, S. 2022. A Review on the Potential Species of the Zingiberaceae Family with Anti-viral Efficacy Towards Enveloped Viruses. J Pure Appl Microbiol 16, 796–813. https://doi.org/10.22207/JPAM.16.2.35.
- Phytochemical screening, antibacterial and free radical scavenging effects of Artemisia nilagirica, Mimosa pudica and Clerodendrum siphonanthus – An in–vitro study. Asian Pac. J. Trop. Biomed.. 2012;2:S601-S604.
- [Google Scholar]
- Phytochemical screening and in vitro bioactivity of Cnidoscolus aconitifolius (Euphorbiaceae) J Med Plants Res. 2007;1:63-65.
- [Google Scholar]
- Studies on phytochemical analysis, antioxidant and antibacterial activity of Ficus racemosa L. leaf and fruit extracts against wound pathogens. Vegetos. 2019;32:58-63.
- [CrossRef] [Google Scholar]
- Bioactive Compounds, Pharmacological Activity and Food Application of Ficus racemosa: A Critical Review. Int J Fruit Sci. 2020;20:S969-S986.
- [CrossRef] [Google Scholar]
- Pharmacological Potentials of Ficus racemosa - A Review. Int J Pharm Sci Rev Res. 2013;22:29-34.
- [Google Scholar]
- The model of western integrative medicine: The role of Chinese medicine. Chin. J. Integr. Med.. 2011;17:11-20.
- [CrossRef] [Google Scholar]
- Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004
- [CrossRef] [Google Scholar]
- Comparison of simple potential functions for simulating liquid water. J. Chem. Phys.. 1983;79:926-935.
- [CrossRef] [Google Scholar]
- Jose, A., Manchikanti, P. 2022. Protection of Geographical Indication: The Interface with Traditional Knowledge BT - Geographical Indication Protection in India: The Evolving Paradigm. In: Bhattacharya NS, editor., Singapore: Springer Nature Singapore; p. 141–66. https://doi.org/10.1007/978-981-19-4296-9_6.
- Phytopharmacological and phytochemical properties of three ficus species-An overview. Int J Pharma Bio Sci 2010:1.
- [Google Scholar]
- Isolated flavonoids from Ficus racemosa stem bark possess antidiabetic, hypolipidemic and protective effects in albino Wistar rats. J. Ethnopharmacol.. 2016;181:252-262.
- [Google Scholar]
- Kishore Kumar, M.S., Kumar, V.A., Alphonsa, T., Rajendran, S., Rajamanickam, K., A, A. et al. 2022. COVID-19 and Tuberculosis: Two Knives in a Sheath. Coronaviruses 3, 1. https://doi.org/10.2174/2666796703666220705144250.
- Phytochemical profiling and GCMS study of Adhatoda vasica leaves. Int J Pharma Bio Sci. 2014;5:B714-B720.
- [Google Scholar]
- G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model.. 2014;54:1951-1962.
- [CrossRef] [Google Scholar]
- Nachiappan, K., Nallakaruppan, N., Alphonse, M., Sekaran, M., Veluchamy, C., Ramamoorthy, S, et al. 2022. Status, Conservation, and Sustainability on Medicinal Plant Resources of India BT - Plant Genetic Resources, Inventory, Collection and Conservation. In: Ramamoorthy S, Buot IJ, Chandrasekaran R, editors., Singapore: Springer Nature Singapore; p. 351–87. https://doi.org/10.1007/978-981-16-7699-4_17.
- Olaoluwa, O., Taiwo, O., Nahar, D.L., Sarker, S. 2022. Ethnopharmacology, phytochemistry and biological activities of selected African species of the genus Ficus 2022–68. https://doi.org/10.30495/tpr.2022.1939285.1219.
- Exploring the pharmacognostic properties and pharmacological activities of phytocompounds present in Ficus racemosa linn.: A concise review. Pharmacol Res - Mod Chinese Med. 2022;4:100137
- [Google Scholar]
- Antibacterial and Antifungal Approaches of Ficus racemosa. Pharmacogn J. 2019;11:355-357.
- [CrossRef] [Google Scholar]
- GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845-854.
- [CrossRef] [Google Scholar]
- Natural products and their semi-synthetic derivatives against antimicrobial-resistant human pathogenic bacteria and fungi. Saudi J Biol Sci. 2022;29:103376
- [Google Scholar]
- Effect of Probiotics on Host-Microbiota in Bacterial Infections. Pathogens 2022:11.
- [CrossRef] [Google Scholar]
- Structural basis for the inhibition of SARS-CoV2 main protease by Indian medicinal plant-derived antiviral compounds. J. Biomol. Struct. Dyn.. 2022;40:1970-1978.
- [CrossRef] [Google Scholar]
- Conservation and cultivation of threatened and high valued medicinal plants in North East India. Int J Biodivers Conserv. 2013;5:584-591.
- [Google Scholar]
- The genus Ficus (Moraceae) used in diet: Its plant diversity, distribution, traditional uses and ethnopharmacological importance. J. Ethnopharmacol.. 2018;226:185-196.
- [Google Scholar]
- Tagboto, S., Townson, SBT-A in P. Antiparasitic properties of medicinal plants and other naturally occurring products. vol. 50, Academic Press; 2001, p. 199–295. https://doi.org/https://doi.org/10.1016/S0065-308X(01)50032-9.
- Antidiabetic effect of Ficus racemosa Linn. stem bark in high-fat diet and low-dose streptozotocin-induced type 2 diabetic rats: A mechanistic study. Food Chem. 2012;132:186-193.
- [CrossRef] [Google Scholar]
- Protective effect of tannins from Ficus racemosa in hypercholesterolemia and diabetes induced vascular tissue damage in rats. Asian Pac. J. Trop. Med.. 2012;5:367-373.
- [Google Scholar]
- Preliminary phytochemical screening, Antibacterial potential and GCMS analysis of two medicinal plant extracts. Pak. J. Pharm. Sci.. 2016;29:819-822.
- [Google Scholar]
- Recent developments and applications of the MMPBSA method. Front. Mol. Biosci.. 2018;4:87.
- [CrossRef] [Google Scholar]
- Looking through the microscope: Microbes as a challenge for theorising biocentrism within environmental ethics. Endeavour. 2022;46:100819
- [Google Scholar]
- The Traditional Medicine and Modern Medicine from Natural Products. Molecules 2016:21.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102956.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







