Translate this page into:
In silico view of MTA1 biochemical signatures in breast malignancy for improvement in immunosurveillance
⁎Corresponding authors. humaira.yasmin@comsats.edu.pk (Humaira Yasmin), faisalnouroz@gmail.com (Faisal Nouroz),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Metastasis Associated 1 (MTA1) chromatin modifier oncoprotein played a crucial role in both normal and genotoxic stress situations for genome maintenance. To investigates MTA1 regulatory pattern with drug resistance abilities in breast primary carcinoma to advanced invasive stages for identification of cancer adapting defense system. We design rationale in silico pipeline from data retrieval to analysis i.e. gene enrichment analysis performed by GeneCards Version 5.1: THE HUMAN GENE DATABASE (www.genecards.org), UALCAN database (www.ualcan.path.uab.edu) for analyzing MTA1 gene expression and promoter methylation in both breasts normal and cancerous tissue samples, cBioPortal for Cancer Genomics (www.cbioportal.org) database for MTA1 mutation analysis and finally analyzed MTA1 functional association with anticancer drugs in breast malignancy via online CCLE GDSC toolkit (www.public.tableau.com/CCLE_GDSC_Correlations). Our results revealed MTA1 overexpression aggressive behavior in stage II, stage III, TNBC-LAR, TNBC-M, TNBC-UNS, IDC, ILC, and post-menopause events of breast malignancy. MTA1 upregulation strongly promotes primary tumor transformation into invasive metastatic carcinoma by hijacking the host lymphatic system and cytokine signaling in both ductal and glandular breast cancers. MTA1 upregulation in the African-American population invites to design de novo model of cancer cell homeostasis under a reduced supply of vegetable nutrients, local-foreign stress, and replicative capacity for metastasis. MTA1 showed a hypomethylation profile that reflects regulatory strength under stress-mediated situations for higher events of transcription. In drug resistance analysis MTA1 has strong resistance towards 15 anti-cancer drugs that confirmed its previously reported behavior of genotoxic stress adaptation for metastasis. Our in silico evidence invites us to design a comprehensive strategy against MTA1 mediated stress managing proteome. In the future there is an urgent need to explore MTA1 shared stress coped protein networks for early diagnosis and better prognosis.
Keywords
MTA1
Breast invasive carcinoma
Genotoxic stress
Proliferation
Differentiation
Apoptotic signaling
1 Introduction
Advancements in biochemistry approaches greatly explored huge data regarding both macromolecules (nucleic acids, proteins, vitamins, lipids and carbohydrates) and micromolecles (ions and small molecules) in cellular microenvironment. Nowadays, biochemistry deeply focuses on mechanistic view of disease development from early onset to fatal behavior. Biochemistry findings are appilied in disease investigation, impact on cell–cell communication, influence on survival status, prediction of therapeutic stretetigies and development of prognostic kits. Biochemical omics technologies enables us to re-investigate molecular bases of tumor based diseases (Fiorentini et al., 2021; Barr, 2018; Hariharan and Sivakumar, 2017; Bhaumik and Patel, 2017; Coelho et al., 2013). Metastasis Associated 1 (MTA1) oncoprotein is a vital member of nucleosome remodeling/histone deacetylase complex (NuRD) that is involved in cell cycle regulation, genome stability and transcriptional mechanisms. In nucleolus MTA1-NuRD complex triggered transcription of pre-ribosomal RNA by association with nucleophosmin and nucleolin proteins (Liu et al., 2021). MTA1 act as chromatin modifier that supports cancer cell to stabilize the effects of genotoxic stress and hypoxia that leads to resistance towards anti-cancer drugs (Galluzzi et al., 2018). MTA1-NuRD complex overexpression stimulates breast invasive carcinoma to metastasis through transcriptional regulation of oncogenes by chromatin remodeling process (Sen et al., 2014). MTA1 regulates both co-activator and co-repressor activities for oncogenes regulation due to the specific association of the NuRD complex (Malisetty et al., 2017). MTA1 overexpression caused quantitative accumulation in the tumor microenvironment that reduces the effects of therapeutic regimes (Liu et al., 2020). MTA1 oncogenic activity managed via protein binding with OGT, OGA and components NuRD complex that affects a wide range of cellular processes including survival, proliferation and colony formation (Millard et al., 2016). In breast carcinoma, MTA1-OGT interaction produced S237, S241, and S246 serine residues post-translational modifications that lead to carcinoma cells adaptation towards genotoxic stress (Liu et al., 2020). During genotoxic stress conditions, MTA1 occupied downstream gene promoters for the protection of breast cancer cells that further translate into worse dispersion as metastasis (Jang et al., 2006). In carcinoma cells, MTA1 upregulation enhances oncogenesis effects by encouraging STAT3, WNT1, MYC and RAS signaling pathways that are active indicators of invasion, transformation, epithelial-mesenchymal transmission, inflammation and angiogenesis (Gao et al., 20182018). In breast cancer biology, the MTA1-NuRD complex drives fate of estrogen receptor (ER) mediated cancer progression. MTA1-HSF1-NuRD complex upregulation negatively regulates transcription of ER targeted genes that develops ER- phenotype which offered resistance towards tamoxifen (Khaleque et al., 2008). Similarly MTA1-NRIF3-CAK complex performs inhibitory actions towards ER-dependent genes that promote hormone independence and progression of triple-negative breast carcinoma (TNBC) (Talukder et al., 2003). The immune system capability to encounter mitogens or carcenogens for improvement in patient survival against tumor progression is known as immunosurveillance. In this phenomenon immune system activates anti-tumor cascades through both innate and adapative immune signals in response of oncogenes overexpression. The lack of comprehensive understanding regarding breast preinvasive to highly invasive triple-negative cancers interaction with immune system behavior offered limited success in drug development for better prognosis. Several studies reported increased level of leukocytes, neutrophils, microphages and other antibody proteins in higher stages of breast carcinoma than primary tumors. In breast malignancy combination of atezolizomab and nab-paclitaxel immunotherapeutic drugs are approved first time against triple negative disease (Gil Del Alcazar et al., 2020; Schreiber et al., 2011; Kwa and Adams, 2018; Adams et al., 2019). Biochemistry of metastasis played essential role in treatment failure by providing resistance towards anti-cancer drugs (Kachalaki et al., 2016). In metastatic cancer biochemical factors triggered several key events including tumor localization, migration, angiogenesis and aggressiveness (Icard and Lincet, 2012). The identification of biochemical markers in MTA1 mediated tumor biology offered active agents for blocking malignant capabilities in personalized medicine areana (Taddia et al., 2015). MTA1 protein is biochemical modifier that has excellent ability in either tumor expension or invasion on host immune system (Madureira et al., 2012).
MTA1 positive performance promotes metastatic malignant trend in human carcinomas; despite these proteomic studies, the exact role of MTA1 regulation in tumor progression to metastasis in breast malignancy remains unclear. In this study, we investigated MTA1 expression, methylation, mutation and survival with resistance behavior towards breast cancer therapeutic strategies. Here, we provide MTA1 as a novel biomarker for the regulation of stress-mediated carcinogenesis.
2 Protocol
2.1 MTA1 gene enrichment analysis
We performed gene enrichment analysis from GeneCards Version 5.1: THE HUMAN GENE DATABASE (www.genecards.org) which is a user-friendly comprehensive integrative search engine for gene characterization. It incorporates the data from genomic, proteomic, transcriptomic, metabolomic and clinical resources. The GeneCards Version 5.1 contains 321,417 genes in which significantly 20,713 protein-coding genes and 19,195 disease genes are stored. Initially, we queried ‘MTA1’ in the search box that displayed plenty of valuable information including functions, domains, location, disorders, drugs, pathways and products.
2.2 MTA1 expression analysis in BRIC
We used an online, interactive and user-friendly UALCAN database (www.ualcan.path.uab.edu) for analyzing MTA1 expression in both breast normal and cancer tissue samples. It arranged the omics data from TCGA, CPTAC and MET500 omics platforms. It has a diverse variety of biomarkers for more than 31 human cancers through simple operating processes. Firstly we pressed ‘TCGA Analysis’ option that displayed a comprehensive and up-to-date list of human malignancies with the gene search box. We entered ‘MTA1’ as the gene in the gene search box, select ‘Breast Invasive Carcinoma (BRIC)’ as a disease of interest and finally pressed the ‘Explore’ option that presents vital information regarding differential expression in normal breast tissue and diverse carcinoma parameters including stages, race, gender, age, menopause state, TNBC subtypes and histological subtypes. All MTA1 gene expression profiles are displayed in box and whisker plots with multiple downloading options i.e. PNG, JPEG, PDF and SVG formats. The whisker box plots are generated on Transcript per Million (TPM) values that represent differential expression significance among different parameters. TPM value is used for comparisons across different RNA-seq gene expression samples. In cancer studies relative expression levels among normal and tumor samples are determined by TPM values calculation.
2.3 MTA1 promoter methylation analysis in BRIC
UALCAN database also allows us to the evaluation of MTA1 promoter methylation in both normal and tumor tissue samples. We select the ‘Methylation’ option that displayed whisker box plots based on beta-values that indicate 0 (fully unmethylated) and 1 (fully methylated). UALCAN database set cut-off value of 0.2–0.3 (hypomethylation) and 0.5–0.7 (hypermethylation). Here, we determined the impact of promoter methylation on gene expression across different group samples. The beta-values statistics is normally used in the estimation and quantification of DNA methylation status in microarray profiling studies.
2.4 MTA1 mutation analysis in BRIC
We used cBioPortal for Cancer Genomics (www.cbioportal.org) database for MTA1 mutation analysis in breast carcinoma. We queried ‘MTA1’ gene in the search box, selected all available 4 datasets including (1) Breast Invasive Carcinoma TCGA, Cell 2015 of 817 samples dataset (2) Breast Invasive Carcinoma TCGA, Firehouse Legacy of 1108 samples dataset (3) Breast Invasive Carcinoma TCGA, Nature 2012 of 825 samples dataset (4) Breast Invasive Carcinoma TCGA, PanCancer Atlas of 1084 samples dataset and pressed ‘submit’ option that displayed a wide range of results. We select ‘mutation’ tab that displayed graphical view and tubular information of MTA1 mutations, frequency and positions in breast cancer diverse samples. MTA1 mutation map consists of different mutation types including missense, truncating, splice site, insertions, and deletions with unique coloring patterns.
2.5 MTA1 drug sensitivity analysis in breast cancer
We analyzed MTA1 functional association with anticancer drugs in breast malignancy via online CCLE GDSC toolkit (www.public.tableau.com/CCLE_GDSC_Correlations). We entered ‘MTA1’ gene in the gene list and selected ‘EC50/IC50’ coefficient concentration from CCLE GDSC side options. The CCLE GDSC toolkit provides green color negative (non-resistant) and red color positive (resistant) characteristics. We obtained and arranged all available MTA1-drug sensitivity relationships in tabulated form for breast carcinoma.
3 Results
3.1 MTA1 gene enrichment analysis
Metastasis Associated 1 (MTA1) oncoprotein has 715 amino acids and 5 transcripts with 80786 Da molecular mass. MTA1 gene occupied 105,419,820–105,470,729 nucleotides of 50,910 bases at chromosome 14q32.33. It has 8 domains i.e. BAH, Znf/GATA, SANT/Myb, Homeobox-like, SANT, ELM2, MTA1_R1 and MIER-MTA domain. MTA1 regulates SNAI1, SNAI2 and CXCL1 while deregulates MTA3, EPHA2 and CDH1. The expression of PITX3 positively regulates MTA1 at the transcriptional level. MTA1 at the microenvironment level participates in plasma proteins and transcription factor protein classes. MTA1 resides in the cytoskeleton, cytosol and nucleus with low tissue specificity. MTA1 is a favorable prognostic marker in pancreatic cancer and an unfavorable prognostic marker in liver carcinoma. It is involved in SUMOylation, Acetylation, chromatin remodeling and RNA polymerase I mediated transcription, promoter clearance and epigenetic processes. MTA1 gene mutations are responsible for epileptic encephalopathy, tonsil malignancy and aggressive metastatic behavior towards cell lines of breast tissue adenocarcinoma (Fig. 1).
Red line at q32.33 indicates. Location of MTA1.
3.2 MTA1 expression analysis in BRIC
MTA1 differential expression analysis reveals lower expression (57.56% TPM) in breast normal tissue 114 samples and higher expression in breast cancer stage I 183 samples (78.02% TPM), stage II 615 samples (87.87% TPM), stage III 247 samples (83.71% TPM) and stage IV 20 samples (62.77% TPM). MTA1 has elevated expression in different populations including African-American 179 samples (90.51% TPM), Asian 61 samples (84.02% TPM) and Caucasian race 748 samples (83.71% TPM). MTA1 has higher expression in female gender 1075 samples (85.67% TPM) than male 12 samples (49.73% TPM). MTA1 also has overexpression in age-related factors i.e. 21–40 years 97 samples (83.71% TPM), 41–60 years 505 samples (85.96% TPM), 61–80 years 431 samples (84.54% TPM) and 81–100 years 54 samples (75.74% TPM). MTA1 has higher expression in breast malignancy-related parameters i.e. stages, race, age and gender cases than normal tissue expression. Here MTA1 has overexpression in stage II, African-American race, female gender and 41–60 years age group that reflects its link with the initiation of the aggressive mode of the tumor with specific ethnic groups (Fig. 2).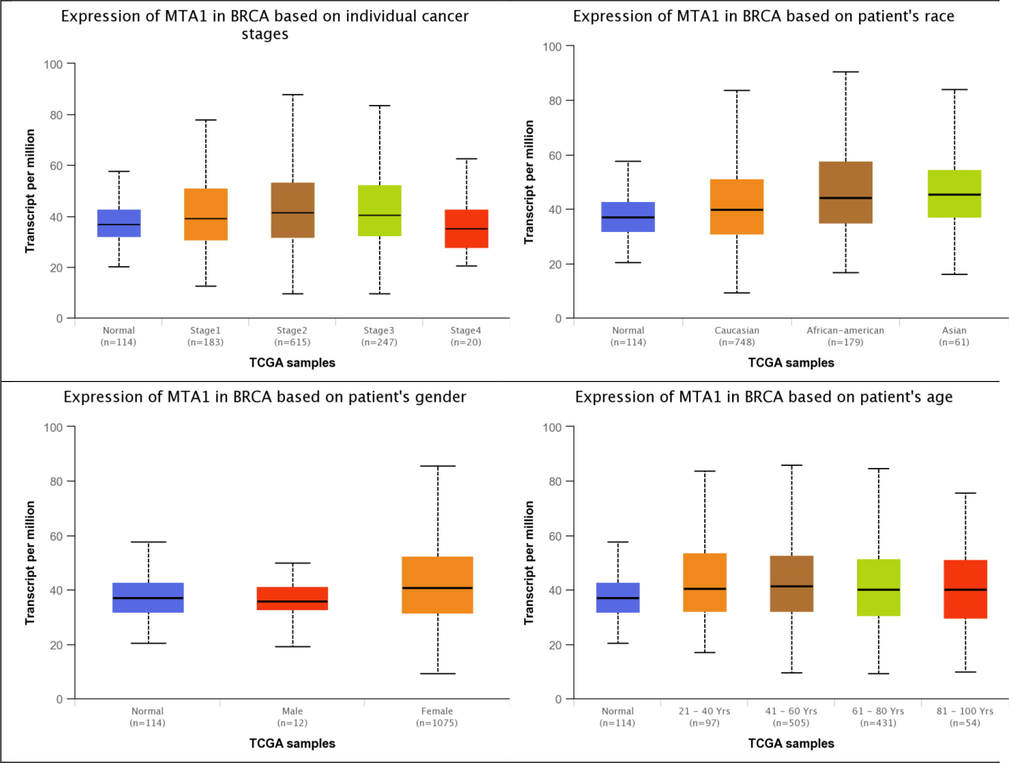
MTA1 differential expression analysis in normal, cancer stages, race, gender and age-related factors of breast tissue.
Our study reveals MTA1 higher expression in various breast carcinomas subtypes i.e. luminal 566 samples (81.78% TPM), HER2 + 37 samples (78.59% TPM) and TNBC 116 samples (95.87% TPM). MTA1 showed overexpression in TNBC subtypes including luminal 566 samples (81.78% TPM), HER2 + 37 samples (78.59% TPM), TNBC-BL1 13 samples (61.74% TPM), TNBC-BL2 11 samples (54.87% TPM), TNBC-IM 20 samples (76.92% TPM), TNBC-LAR 8 samples (144.93% TPM), TNBC-MSL 8 samples (77.66% TPM), TNBC-M 29 samples (109.26% TPM) and TNBC-UNS 27 samples (111.27% TPM). MTA1 showed upregulation in menopause conditions i.e. pre-menopause 230 samples (78.39% TPM), peri-menopause 37 samples (74.88% TPM) and post-menopause 700 samples (84.68% TPM). Breast carcinomas histological subtypes has overexpression of MTA1 including IDC 784 samples (81.21% TPM), ILC 203 samples (86.29% TPM), Mixed 29 samples (87.87% TPM), others 45 samples (73.53% TPM), Mucinous 17 samples (73.12% TPM) and Metaplastic 9 samples (109.26% TPM) while lower expression in INOS 1 sample (34.49% TPM) and medullary 6 samples (25.04% TPM). MTA1 has observable high expression in TNBC-BL1-IM-M-LAR-MSL-UNS related cancer scenarios with menopause conditions showed its contribution in worse prognosis malignancy. The IDC, ILC, mucinous and medullary types also showed overexpression that are strong signs of MTA1 expression link with tubular-glandular system-mediated invasiveness (Fig. 3).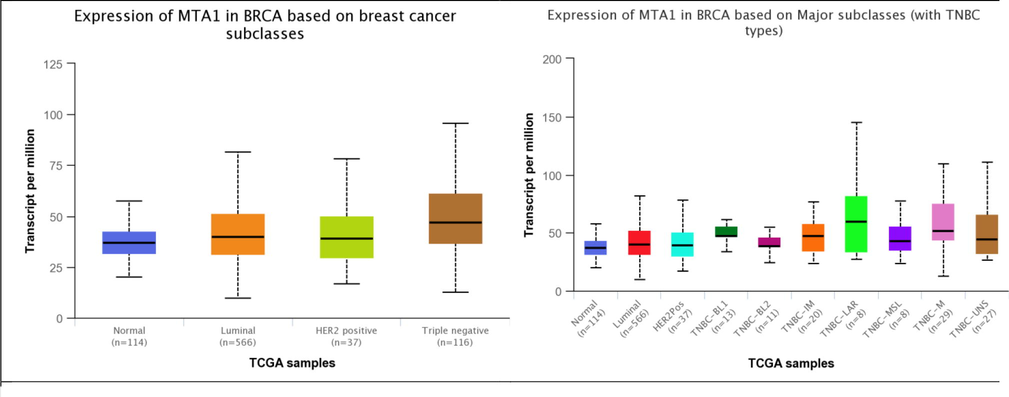
MTA1 differential expression analysis in normal, cancer subtypes, TNBC subtypes, menopause conditions and histological subtypes of breast tissue.
We performed MTA1 copy number variation analysis in all existing breast invasive carcinoma datasets in cBioPortal database. In overall in silico experiment 46 samples showed alterations in which 33 samples indicates amplification mediated overexpression while 13 samples present deep deletion mediated loss of function. MTA1 gene depicts functionally aggressive behavior towards the synthesis of metastasis-promoting factors (Fig. 4).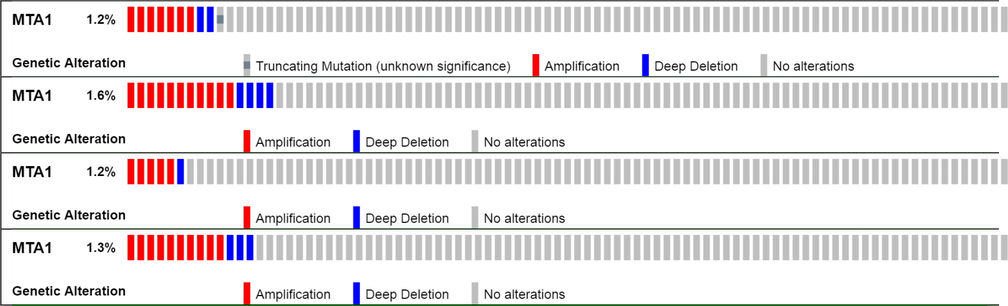
MTA1 gene copy number variation analysis in all available 4 breast cancer datasets.
MTA1 showed overexpression in several human malignancies including colon adenocarcinoma, pancreatic adenocarcinoma, lung squamous cell carcinoma, head and neck squamous cell carcinoma, esophageal carcinoma, liver hepatocellular carcinoma, rectum adenocarcinoma, pheochromocytoma and paraganglioma, cholangiocarcinoma and stomach adenocarcinoma. MTA1 aggressive behavior in multiple malignancies depicts its global role in cancer therapeutics. MTA1 also showed sensitive functional association with kinase coding genes, cell cycle genes, apoptosis-related genes, and metastasis-associated genes, oncogenes, DNA damage response genes and integrin coding genes. MTA1 inhibition is an ideal opportunity to reduce the risk of metastatic colony formation.
3.3 MTA1 promoter methylation analysis in BRIC
MTA1 presents moderate methylation rates at gene promoter in both normal breast tissue 97 samples (0.38% β-value) and breast cancer specific stages i.e. stage I 127 samples (0.39% β-value), stage II 442 samples (0.42% β-value), stage III 200 samples (0.41% β-value) and stage IV 11 samples (0.40% β-value). MTA1 showed moderate methylation in diverse populations i.e. Caucasian 578 samples (0.42% β-value), African-american 160 samples (0.39% β-value) and Asian 38 samples (0.40% β-value). MTA1 has moderate methylation in both male 9 samples (0.40% β-value) and female 783 samples (0.42% β-value). MTA1 showed moderate methylation in all age groups i.e. 21–40 years 73 samples (0.40% β-value), 41–60 years 374 samples (0.42% β-value), 61–80 years 306 samples (0.42% β-value) and 81–100 years 37 samples (0.38% β-value). Here MTA1 promoter moderate methylation showed its influence towards methylation pathways that leads to aberrant expression in diverse conditions including cancer (Fig. 5).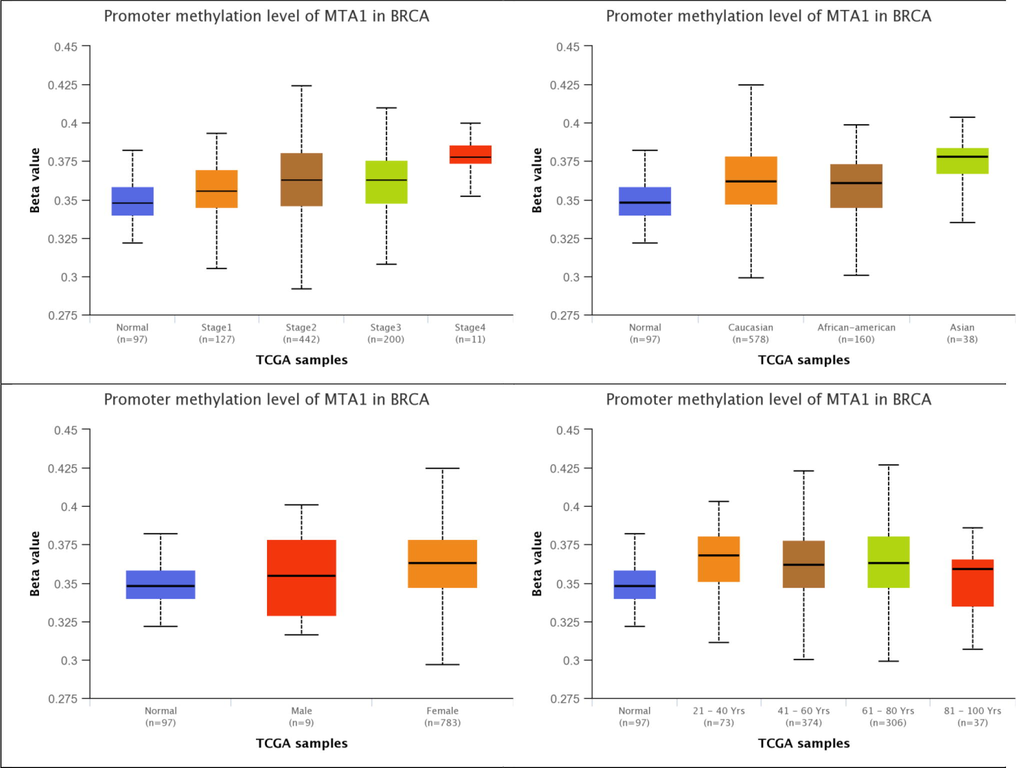
MTA1 promoter methylation in normal, cancer stages, race, gender and age-related factors of breast tissue.
MTA1 showed moderate methylation in various breast cancer histological forms including IDC 516 samples (0.42% β-value), ILC 182 samples (0.40% β-value), mixed 26 samples (0.41% β-value), others 35 samples (0.41% β-value), mucinous 15 samples (0.39% β-value), metaplastic 10 samples (0.37% β-value), INOS 1 sample (0.38% β-value) and medullary 6 samples (0.38% β-value). MTA1 presents moderate methylation in pre-menopause 171 samples (0.42% β-value), peri-menopause 23 samples (0.38% β-value) and post-menopause 506 samples (0.42% β-value). MTA1 moderate methylation in all histological types and menopause diverse states presents its interaction with methylation generating system that reduces the ratio of regulation in either normal or oncogenic conditions (Fig. 6).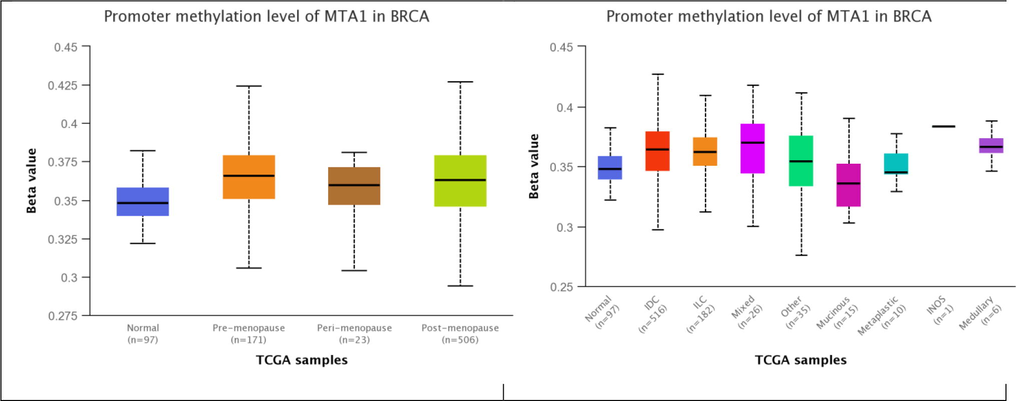
MTA1 promoter methylation in normal, menopause states and histological subtypes of breast tissue.
3.4 MTA1 mutation analysis in BRIC
In all available breast cancer datasets, MTA1 gene has a mutation in only Breast Invasive Carcinoma TCGA, Cell 2015 of 817 samples dataset. In mutation analysis, the MTA1 gene has a single frameshift mutation R636Gfs13 in the GATA zinc finger (393-429AA) region. MTA1 has in which 3 important regions i.e. BHA domain from 4 to 164AA, ELM2domain from 167 to 220 AA and GATA zinc finger 393–429 AA in 3477 AA (Fig. 7A–D).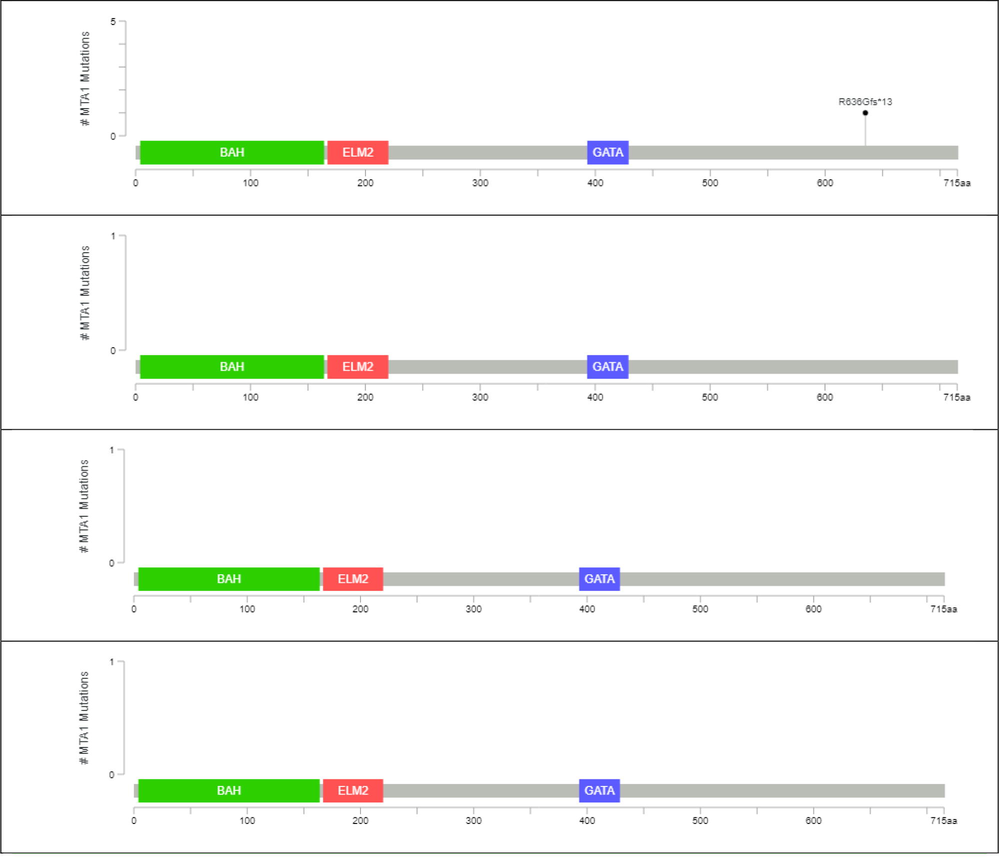
A–D: (A) MTA1 mutation analysis in Breast Invasive Carcinoma TCGA, Cell 2015 of 817 samples dataset. (B) MTA1 mutation analysis in Breast Invasive Carcinoma TCGA, Firehouse Legacy of 1108 samples dataset. (C) MTA1 mutation analysis in Breast Invasive Carcinoma TCGA, Nature 2012 of 825 samples dataset (D) MTA1 mutation analysis in Breast Invasive Carcinoma TCGA, PanCancer Atlas of 1084 samples dataset.
3.5 MTA1 drug sensitivity analysis in breast cancer
MTA1 showed significant crucial association with 17 anti-cancer drugs in which it has resistance towards 15 drugs efficiency and showed cooperative role to enhance the effectiveness of 2 drugs (Afatinib and AC-220). MTA1 has strong resistance towards influential drugs that are involved in damage repairing, growth inhibition, cell cycle arrest, activation of cell cycle checkpoints and metabolic pathways. MTA1 positive regulation provides ideal opportunities for breast cancer progression into metastasis by interrupting genotoxic effects (Table 1).
Drugs
Targets
Correlation
YK-4-279
RNAHA
Positive
Trametinib
MEK1, 2
Positive
SN-38
TOP1
Positive
Selumetinib
MEK1, 2
Positive
RDEA119
MEK1, 2
Positive
PD-0325901
MEK1, 2
Positive
Olaparib
PARP1, 2
Positive
Ispinesib Mesylate
KIF11
Positive
DMOG
Hydroxylase
Positive
Cyclopamine
SMO
Positive
CMK
RSK
Positive
Cetuximab
EGFR
Positive
Camptothecin
TOP1
Positive
Bleomycin
DNA damage
Positive
AZD-7762
CHECK1, 2
Positive
AC-220
FLT3
Negative
Afatinib
EGFR
Negative
4 Discussion
The term “Precision Medicine” is emerged due to day-to-day advancements in healthcare approaches for precise detection, diagnosis, treatment and prevention of diseases. Precision medicine biology modified the therapeutic regime regarding the old traditional view of a one-size-fits-all scheme. MTA1 multi-omics analysis also address parameter-specific attributes of breast malignancy that highlights exact therapeutic options under the context-dependent scenario. Globally above 3 million women are affected in the female population from breast carcinoma due to lack of awareness and late diagnosis (Waks and Winer, 2019). The exact mechanism of speedy forceful progression of the primary tumor to the invasive mode of carcinoma still remains a challenge in breast cancer therapy (Momenimovahed and Salehiniya, 2019 Momenimovahed and Salehiniya, 2019). Cancer cells' microenvironment proposed favorable adaptions towards genotoxic stress which is the basic philosophy of drug resistance (Ferrao et al., 2015). In carcinoma cellular homeostasis switched on the bulk of genes that are responsible for nucleotide damage repairing, detoxification and degradation of death signals (Hammerlindl and Schaider, 2018). MTA1 as a stress response protein primarily identified from breast malignant cells in higher quantity after induction of stress therapeutic agents (Lai and Wade, 2011). MTA1-NuRD complex modulates the estrogen-mediated genetic regulations that promote hormone independence based on drug resistance (Lovitt et al., 2018). MTA1 mediated stress response biology regulates cellular survival cascades for opposing the effects of apoptotic pathways during chemo-therapeutic approaches (Cong et al., 2011). In our findings, MTA1 has prominently overexpression in breast invasive carcinoma samples as compared to breast normal tissue samples. We found MTA1 mRNA higher levels are associated with most important stage II to stage III which are vital indicators of primary tumor transformation into invasive carcinoma. In stage II/stage III localized tumor cells dispersed into nearby lymph nodes that are key components of the lymphatic system which is the worse carrier of cancer clones. MTA1 aggressive behavior in stage II/stage III presents its oncogenic vitality that counteracts immune system response as well as hijacked lymphatic system transport for metastasis. MTA1 showed upregulation in TNBC-LAR, TNBC-M and TNBC-UNS that are involved in rapid proliferation by cytokine signaling pathway, differentiation, migration, epithelial-mesenchymal transmission and glycolysis. MTA1 upregulation in TNBC as compared to normal and hormone receptor-positive breast carcinoma proposed its contribution in a worse prognosis. MTA1 also showed higher expression in IDC, ILC and Metaplastic breast cancer that depicts its deep-rooted participation in both ductal and glandular tumorigenic dispersions. Here, MTA1elevated position in IDC/ILC displayed its transforming activity against homeostasis stress of local tissue immune system. MTA1 has higher expression in the African-American population that connects its upregulation with the low-level intake of fruits and vegetables which are strong risk factors for carcinogenesis. MTA1 upregulation invites to design de novo model of cancer cell homeostasis under a reduced supply of vegetable nutrients, local-foreign stress and replicative capacity for metastasis. We found MTA1 overexpression in 21–40, 41–60 and 61–80 years old age groups that highlight its regulatory power in any stage of life. MTA1 elevated expression in age groups addresses its influence at primary tumor initiation to invasive dissemination into distant parts of the body. MTA1 showed upregulation in postmenopause state that is a strong indicator of hormone-independent generation of cancer microenvironment with poor prognosis. MTA1 overexpression in diverse breast cancer parameters proposed it excellent biomarker for diagnosis and treatment in females. Here, MTA1 promoter methylation analysis displayed insignificant methylation variations between normal and cancerous tissue samples. MTA1 hypomethylation profile indicates its regulatory strength under stress-mediated situations for higher events of transcription. MTA1 mutation analysis showed single frameshift deletion in the 20th exon which has insignificant effects on protein folding to functioning that is an ideal factor for the precise and accurate higher amount of protein synthesis. In MTA1 drug resistance analysis, it has strong resistance towards 15 anti-cancer drugs that confirmed its previously reported behavior of genotoxic stress adaptation for metastasis. MTA1 strongly opposed the anti-metastatic effects of Trametinib drug that assists the lymphatic system for better performance against the invasive mode of dispersion (Robert et al., 2012). MTA1 resists the effectiveness of Selumetinib which is used in nervous, gastric and kidney carcinomas to reverse the mutations regarding deformations and inflammations (Patel et al., 2017). MTA1 also reduces the efficiency of Olaparib which maintenance and nucleotide repairing drug in BRCA mutated breast and ovarian carcinomas (Farago et al., 2019). Here, our in silico findings of MTA1 overexpression validate previous studies that addressed its lethal oncogenic role in the progression of the primary tumor to higher invasive stages by reducing stress effects (Liu et al., 2018). MTA1 overexpression in all breast cancer variables indicates its high esteem capacity to initiate and promotes the aggressive nature of breast metastasis. MTA1 positive regulation boosts up inflammation, angiogenesis, invasion, EMT, survival, damage response, chemo-resistance and transformation (Li, 2010). In cancer cells MTA1 oncogenic property functionally correlated with STAT3, WNT1 and RAS signaling pathways (Ohshiro et al., 2010). MTA1-nucleosome complex played an influential role in chromatin decondensation during the cell cycle progressive phase (Kumar et al., 2010). In metastasis hyper-stimulation of extracellular mitogens, mediated signaling cascades reshape MTA1 functionality as the master organizer of chromatin material (Pakala et al., 2013). Under carcinogenic stress conditions, MTA1 responsible genes strongly develop metastatic lesions by governing a wide spectrum of cancer progressing processes. MTA1 onco-proteome assembly is controlled by post-translational modifications of several upstream signaling units. MTA1 upregulation act as a ‘hub’ gene that integrates and functionalizes the whole invasive machinery of the metastatic cell. MTA1 also participates in global complexes of both Corepressor and Coactivator factors (Pakala et al., 2013).
Our presented MTA1 in silico investigation confirmed its role as a novel biomarker for the regulation of stress-mediated carcinogenesis. MTA1 overexpression speedup breast tumor metastatic abilities through the forceful promotion of cancer cell dispersion, invasive attitude and resistance towards anti-neoplastic agents. Collectively, our in silico approach suggesting MTA1 mediated oncogenic shift in cellular response by interacting with the tumor microenvironment.
5 Conclusion
Our in silico protocol demonstrates MTA1 overexpression that enhances metastatic chances in multiple breast invasive carcinomas attributes. MTA1 upregulation presents a strong defense system of cancer cells against apoptotic pathways and drugs-mediated genotoxic stress. MTA1 positive regulation adds a layer of metastasis promoting members to derive the fate of error-prone gene pools. Our in silico evidence invites us to design a comprehensive strategy against MTA1 mediated stress managing proteome. In the future there is an urgent need to explore MTA1 shared stress coped protein networks for early diagnosis and better prognosis. MTA1 overactivation triggered the synthesis of both autocrine and paracrine signaling factors that are viable agents of invasive dispersion. MTA1 gene-centered microenvironmental changes ultimately lead to de novo chemo-resistant metabolome organization. In the future MTA1 mediated oncology reopens several successful opportunities in the cancer medicine discipline to limit the transformation of neoplastic cells into metastatic cells.
6 Declarations
Ethics approval: Not Applicable.
Consent to participate: All authors consent to participate in this manuscript.
Consent for publication: All authors consent to publish this manuscript in the Journal of King Saud University-Science.
Availability of data and material: Data will be available on request to the corresponding or first author.
Data availability statement: The data presented in this study are available in this article.
Acknowledgements
The findings of this study are a part of the PhD studies of Zafar Abbas Shah. The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP 2021/301), King Saud University, Riyadh, Saudi Arabia.
Conflicts of interest/Competing interests
Authors declare that no conflict of interest exists
References
- Current landscape of immunotherapy in breast cancer: a review. JAMA oncology. 2019;5:1205-1214.
- [Google Scholar]
- Immune escape during breast tumor progression. Cancer immunology research.. 2020;8:422-427.
- [Google Scholar]
- Student’ s perspective for importance of biochemistry in medical sciences. Org. Med. Chem. Int. J.. 2017;1:13-15.
- [Google Scholar]
- Biochemistry of adipose tissue: an endocrine organ. Arch. Med. Sci.: AMS. 2013;2:191.
- [Google Scholar]
- SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function. J. Biol. Chem.. 2011;286(51):43793-43808.
- [Google Scholar]
- Combination olaparib and temozolomide in relapsed small-cell lung cancer. Cancer Discovery. 2019;9(10):1372-1387.
- [Google Scholar]
- Magnesium: biochemistry, nutrition, detection, and social impact of diseases linked to its deficiency. Nutrients. 2021;13(4):1136.
- [Google Scholar]
- Linking cellular stress responses to systemic homeostasis. Nat. Rev. Mol. Cell Biol.. 2018;19(11):731-745.
- [Google Scholar]
- Proteomic analysis of the OGT interactome: novel links to epithelial–mesenchymal transition and metastasis of cervical cancer. Carcinogenesis.. 2018;39:1222-1234.
- [Google Scholar]
- Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J. Cell Commun. Signaling. 2018;12(1):133-141.
- [Google Scholar]
- Importance of biochemistry to medical industries. Res. J. Pharm. Technol.. 2017;10(12):4457.
- [Google Scholar]
- A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochimica et Biophysica Acta (BBA)-Reviews on. Cancer. 2012;2:423-433.
- [Google Scholar]
- MTA1 overexpression correlates significantly with tumor grade and angiogenesis in human breast cancers. Cancer Science.. 2006;97:374-379.
- [Google Scholar]
- Cancer chemoresistance; biochemical and molecular aspects: a brief overview. European journal of pharmaceutical sciences. 2016;89:20-30.
- [Google Scholar]
- Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene.. 2008;27:1886-1893.
- [Google Scholar]
- Metastasis-associated protein 1 and its short form variant stimulates Wnt1 transcription through promoting its derepression from Six3 corepressor. Cancer Res.. 2010;70(16):6649-6658.
- [Google Scholar]
- Checkpoint inhibitors in triple-negative breast cancer (TNBC): Where to go from here. Cancer. 2018;124:2086-2103.
- [Google Scholar]
- Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat. Rev. Cancer. 2011;11(8):588-596.
- [Google Scholar]
- Revelation of p53-independent Function of MTA1 in DNA Damage Response via Modulation of the p21WAF1-Proliferating Cell Nuclear Antigen Pathway. J. Biol. Chem.. 2010;285:10044-10052.
- [Google Scholar]
- O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. Cell Death Dis.. 2018;9(5)
- [CrossRef] [Google Scholar]
- Proteomic profiling and genome-wide mapping of O-GlcNAc chromatin-associated proteins reveal an O-GlcNAc-regulated genotoxic. Nat. Commun.. 2020;11(1)
- [CrossRef] [Google Scholar]
- Chromatin modifier MTA1 regulates mitotic transition and tumorigenesis by orchestrating mitotic mRNA processing. Nat. Commun.. 2020;11(1)
- [CrossRef] [Google Scholar]
- Cancer metastasis-associated protein 1 localizes to the nucleolus and regulates pre-rRNA synthesis in cancer cells. J. Cell. Biochem.. 2021;122(2):180-188.
- [Google Scholar]
- Doxorubicin resistance in breast cancer cells is mediated by extracellular matrix proteins. BMC Cancer. 2018;18(1)
- [CrossRef] [Google Scholar]
- The biochemistry and regulation of S100A10: a multifunctional plasminogen receptor involved in oncogenesis. J. Biomed. Biotechnol.. 2012;2012:1-21.
- [Google Scholar]
- MTA1 expression in human cancers–Clinical and pharmacological significance. Biomed. Pharmacother.. 2017;95:956-964.
- [Google Scholar]
- The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Elife. 2016;5
- [CrossRef] [Google Scholar]
- Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer: Targets Therapy. 2019;11:151.
- [Google Scholar]
- Acetylation-dependent oncogenic activity of metastasis-associated protein 1 co-regulator. EMBO Rep.. 2010;11(9):691-697.
- [Google Scholar]
- MTA1 promotes STAT3 transcription and pulmonary metastasis in breast cancer. Cancer Res.. 2013;73(12):3761-3770.
- [Google Scholar]
- Population pharmacokinetics of selumetinib and its metabolite N-desmethyl-selumetinib in adult patients with advanced solid tumors and children with low-grade gliomas. CPT: Pharmacometr. Syst. Pharmacol.. 2017;6(5):305-314.
- [Google Scholar]
- Robert, C., Flaherty, K.T., Hersey, P., Nathan, P.D., Garbe, C., Milhem, M.M., Demidov, L.V., Hassel, J.C., Rutkowski, P., Mohr, P., Dummer, R., 2012. METRIC phase III study: Efficacy of trametinib (T), a potent and selective MEK inhibitor (MEKi), in progression-free survival (PFS) and overall survival (OS), compared with chemotherapy (C) in patients (pts) with BRAFV600E/K mutant advanced or metastatic melanoma (MM). LBA8509-LBA8509.
- Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565-1570.
- [Google Scholar]
- Role of MTA1 in cancer progression and metastasis. Cancer Metastasis Rev.. 2014;33(4):879-889.
- [Google Scholar]
- Inside the biochemical pathways of thymidylate synthase perturbed by anticancer drugs: Novel strategies to overcome cancer chemoresistance. Drug Resistance Updates. 2015;23:20-54.
- [Google Scholar]
- MTA1 interacts with MAT1, a cyclin-dependent kinase-activating kinase complex ring finger factor, and regulates estrogen receptor transactivation functions. Journal of Biological Chemistry.. 2003;278:11676-11685.
- [Google Scholar]
- Breast cancer treatment: a review. Jama.. 2019;321:288-300.







