Translate this page into:
Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles
⁎Corresponding author at: Department of Medical Bacteriology, Faculty of Medical Sciences, Tarbiat Modares University, P.O. Box 14115-111, Tehran, Iran. Tel.: +98 (21) 82883862; fax: +98 (21) 82884555. mmmobarez@modares.ac.ir (Ashraf Mohabati Mobarez)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
This study aimed to investigate possible associations between virulence profiles and biofilm formation in Clinical UTI isolates. Isolates were collected from five university hospitals and identified and characterized for the presence of virulence factors by PCR. Biofilm assays were conducted in 96 well microtiter plates by reading the OD570 after crystal violet staining. 75% of isolates had esp gene, 38.77% had asa1, 84.18% had ace, 81.63% had efaA, 93.36% had ebpR, 34.18% had cylA, 81.63% had gelE and 17.35% had hyl. Biofilm experiences were done and isolates having asa1 or efaA genes produced more biofilms than negative ones (P = 0.011, P = 0.008), but the presence of esp, ace, cylA or gelE genes in isolates had no effect on biofilm formation. Isolates possessing hyl had much lower biofilm formation (P = 0.000). Present study showed that the esp, ace, gelE and cylA genes do not seem to be necessary nor sufficient for the production of biofilm in enterococci but the presence of efaA and asa1 correlates with increased biofilm formation of urinary tract isolates. Also the low prevalence of hyl among enterococci isolated from UTIS and its association with poor biofilm production indicate that the absence of this gene can be an advantage in the pathogenesis of UTIs.
Keywords
Enterococci
Biofilm
UTIs
Virulence genes
Colonization
Secretory factors
1 Introduction
Enterococci are Gram-positive member of the human gastrointestinal flora, and are also an important cause of opportunistic nosocomial infections (Marra et al., 2007). These organisms are capable of infecting numerous body sites, causing bacteremia, intra-abdominal infections, endocarditis, and urinary tract infections (Pillar and Gilmore, 2004). Enterococcus faecalis and Enterococcus faecium are the most common enterococci species, and they are responsible for up to 95% of human enterococcal infections (Hall et al., 1992; Jett et al., 1994; Jones et al., 2004).
Biofilm is a population of cells attached irreversibly on various biotic and abiotic surfaces and encased in a hydrated matrix of exopolymeric substances, proteins, polysaccharides and nucleic acids (Costerton, 2001). The ability of enterococci to form biofilms may confer an ecological advantage in certain situations. For example, clinical strains of E. faecalis isolated from infective endocarditis patients were significantly associated with the greater biofilm formation than nonendocarditis clinical isolates (Mohamed et al., 2004); this may be attributable in part to specific virulence factors in enterococci (Mohamed and Murray, 2005). Several enterococcal virulence factors have been identified, including adhesions and secreted virulence factors. The most important adhesion factors are Asa (aggregation substance), Esp (extracellular surface protein), EfaA (E. faecalis antigen A), Ace (adhesin of collagen from E. faecalis) and Ebp (endocarditis and biofilm-associated pili) (Fisher and Phillips, 2009) and secreted pathogenic factors of enterococci with a value in pathogenesis are CylA (cytolysin), GelE (gelatinase) and Hyl (hyaluronidase) (Kayaoglu and Ørstavik, 2004). Several studies investigated the role of these virulence factors in biofilm formation by enterococci (Shankar et al., 1999, 2001; Sandoe et al., 2003; Dupre et al., 2003). esp and gelE were the main factors investigated in strains from different origins (Shankar et al., 1999; Baldassarri et al., 2006). However, some studies claimed correlation among the presence of these factors and biofilm formation (Mohamed et al., 2003; Toledo-Arana et al., 2001) but others suggest that these genes do not seem to be necessary for the production of biofilm in enterococci (Baldassarri et al., 2006; Dupre et al., 2003). The purpose of this study was to investigate biofilm production by enterococcal strains isolated from UTIs and showing different virulence genes profiles, to establish a possible relationship between virulence profile and biofilm formation.
2 Materials and methods
2.1 Strains collection
One hundred and ninety six clinical isolates of enterococci from Urinary tract infections were collected from October 2009 till August 2010 from five university hospitals, including (Tehran) Baqiatalah, (Tehran) Kodakan, (Tehran) Milad, (Mashhad) Shariati and (Shiraz) Namazi. All isolates were identified by Mass Spectrophotometer (MALDI-TOF MS microflex, Bruker, Germany) and biochemical and PCR tests (Table 1) (Facklam, 1972; Kafil and Asgharzadeh, 2014; CLSI, 2012).
Target gene
Primers (5′ → 3′)
Product (bp)
References
E. faecalis
ddlE1:ATCAAGTACAGTTAGTCTTTATTAG
941
Kariyama et al. (2000)
ddlE2: ACGATTCAAAGCTAACTGAATCAGT
E. faecium
ddlF1: TTGAGGCAGACCAGATTGACG
658
Cheng et al. (1997)
ddlF2: TATGACAGCGACTCCGATTCC
asa1
asa1: GCACGCTATTACGAACTATATGA
375
Vankerckhoven et al. (2004)
asa2: TAAGAAAGAACATCACCACGA
efaA
efaF: TGGGACAGACCCTCACGAATA
101
Lowe et al., (1995)
efaR: CGCCTGTTTCTAAGTTCAAGCC
gelE
gelF: TATGACAATGCTTTTTGGGAT
213
Vankerckhoven et al. (2004)
gelR: AGATGCACCCGAAATAATATA
ebpR
ebpA: AAAAATGATTCGGCTCCAGAA
101
Bourgogne et al. (2007)
ebpB: TGCCAGATTCGCTCTCAAAG
hyl
hylF: ACAGAAGAGCTGCAGGAAATG
276
Bourgogne et al. (2007)
hylR: GACTGACGTCCAAGTTTCCAA
esp
espA: GGAACGCCTTGGTATGCTAAC
95
Shankar et al. (1999)
espB: GCCACTTTATCAGCCTGAACC
ace
aceF: GGAGAGTCAAATCAAGTACGTTGGTT
101
Nallapareddy and Murray (2006)
aceR: TGTTGACCACTTCCTTGTCGAT
cylA
cylF: ACTCGGGGATTGATAGGC
688
Vankerckhoven et al. (2004)
cylR: GCTGCTAAAGCTGCGCTT
2.2 Genomic PCR
DNA extraction was done by the protocol described before (Asgharzadeh et al., 2008, 2011). PCR was performed in 25 μl volumes that contained 20–200 ng DNA, 0.5 μM of specific primers for each gene (Table 1), 1.5 mM MgCl2, and 200 μM of each dNTP, 1× PCR buffer and 2 U DNA polymerase (Cinnage, Iran). DNA was amplified by general PCR. An initial 10 min denaturation at 94 °C was followed by 35 cycles of 1 min denaturation at 94 °C, annealing at 58 °C (for ddlE, ddlF, esp, gelE, cylA, hyl, efaA and ace)/52 °C (for ebpR and asa1) for 1 min and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min. Positive controls for PCR were E. faecalis MMH594 (gelE, asa1, esp, cylA, ebpR positive), E. faecalis 29212 (gelE, asa1 positive), E. faecium C38 and C68 and E. faecalis 217 (Khan et al., 2005; Vankerckhoven et al., 2004; Kafil et al., 2013) PCR products were analyzed in agarose gels and visualized under UV after staining with 0.5 μg ml-1 ethidium bromide.
2.3 Biofilm assays
Biofilm assays were conducted based on a before described method (Hatt and Rather, 2008; Tenke et al., 2006). For each strain, few colonies suspended in physiological saline to 0.5 McFarland and Vortexes for 1 min. 96 well polystyrene Microtiter plates (Greiner CELLSTAR® flat-bottomed sterile cell-culture Nr. 655180) were filled with 180 μl Trypticase soy broth (TSB) + 0.5% glucose and 20 μl of bacteria suspension added to each well. 4 wells per strain were incubated and their mean considered as final absorbance. All plates were done in duplicate. Negative controls (Blank) were TSB + 0.5% glucose alone, which were dispensed into eight wells per tray. After stationary aerobic incubation for 24 h at 37 °C and 5% CO2, broth was carefully drawn off and the wells were washed three times with 300 μl of sterile phosphate buffered saline (PBS, room temperature). Biofilms were fixed with 150 μl methanol for 20 min, flick, and air dried in an inverted position in the warm room (about 30 min). Biofilms were stained with 150 μl of crystal violet solution in water (2%) for 15 min at room temperature and the wells were rinsed by placing the plate under running tap water. Microtiter plates were inverted on a paper towel and air dried. To quantify biofilm production, 150 μl of 33% acetic acid was added to each well to destain the biofilms and lidded plates were placed at room temperature for 30 min without shaking. Thereafter, the optical density of the resolubilized crystal violet was measured at 570 nm (OD570) by using a microtiter plate reader (Multiskan FC® Microplate Photometer, Thermo Scientific, Nr. 89087-320). The cut-off optical density (OD) for Biofilm formation by isolates was defined as the optical density higher than OD570 = 0.524 (absorbance of biofilm produced by E. faecalis ATCC 29212).
2.4 Statistical analysis
SPSS software version 16 (IBM SPSS Statistics) was used for statistic analysis. T test was performed for data analysis. P values below 0.05 were considered to be significant.
3 Results
From October 2009 till August 2010, one hundred ninety-six isolates of enterococci were collected from patients with urinary tract infections. One hundred and ten (56.12%) of isolates were E. faecalis and eighty-six (43.88%) were E. faecium. In biochemical analysis 2 E. faecium isolates were Arabinose negative and one E. faecalis was Tellurite negative. Also in PCR results 2 samples had no clear bonds. In mass spectroscopy analysis all strains were detected correctly and well defined. Based on the patients’ gender, 130 (66.32%) of the isolates were from female patients and 66 (33.67%) were from male patients. All isolates were from hospitalized patients with urinary tract infection. The samples were obtained from different wards, including internist, infectious disease, nephrology, pediatrics, intensive care units and women specialized wards. All isolates were investigated for the presence of virulence genes and 147 (75%) had esp gene, 76 (38.77%) had asa1 gene, 165 (84.18%) had ace gene, 160 (81.63%) had efaA gene, 183 (93.36) had ebpR gene, 67 (34.18%) had cylA gene, 160 (81.63%) had gelE gene and 34 (17.35%) had hyl gene. Biofilm experience was done in 4 wells per plate for every isolate (duplicates in two plates) with TSB+ 0.5% glucose (Mean of absorbance for all isolates is presented in Supplementary data). All means are presented according to the presence of virulence genes in isolates. No biofilm were detected in negative control wells. Biofilm formation of isolates with different colonization genes profile is presented in Fig. 1. By comparing isolate absorbance, asa1 positive isolates had significantly higher biofilm formation than asa1 negative isolates (P = 0.011) as well as efaA positive isolates had higher biofilm formation than efaA negative isolates (P = 0.008). No significant differences were found when comparing ace-positive and -negative isolates (P > 0.05). Also esp positive and negative isolates had no significant difference in biofilm formation (P > 0.05). ebpR gene was found almost in all isolates of UTI, therefore we were not able to investigate difference in ebpR positive and negative isolates. On comparing biofilm formation in esp positive isolates possess secretory factors, differences were not significant with gelE and cylA positive isolates (P > 0.05; P > 0.05, respectively). hyl positive isolates had lower biofilm formation tendency (P = 0.000). The mean of absorbance of esp related isolates is presented in Fig. 2. Absorbance of isolates with different secretory genes profiles is presented in Fig. 3. gelE and cylA positive isolates had higher biofilm formation than isolates with gelE and hyl (P = 0.011). Isolates with all colonization factors had no difference in biofilm formation with isolates having all secretory factors (P > 0.05) (Figs. 2 and 3).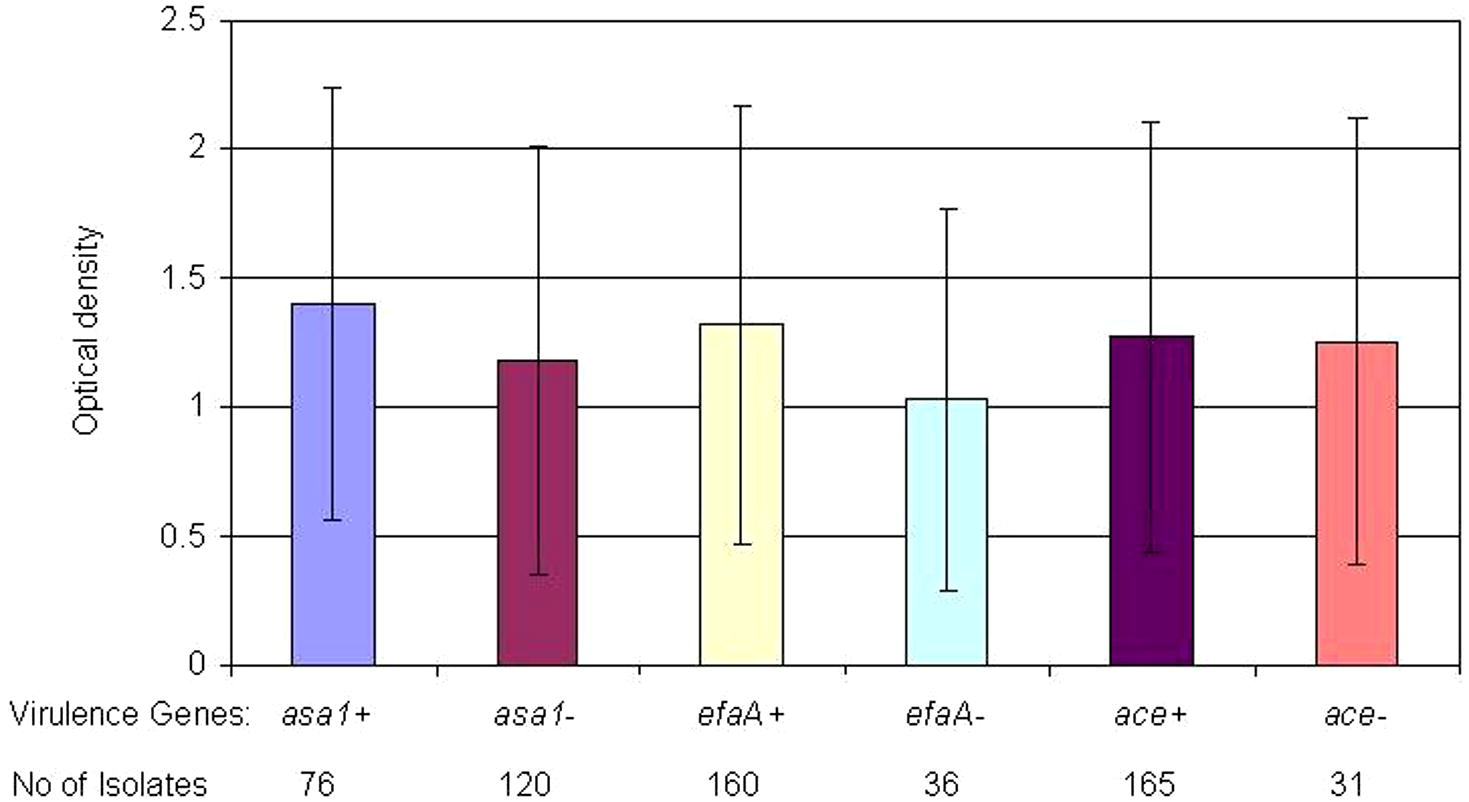
Biofilm formation (OD570) by the enterococcal isolates according to the presence or absence of colonization factors.
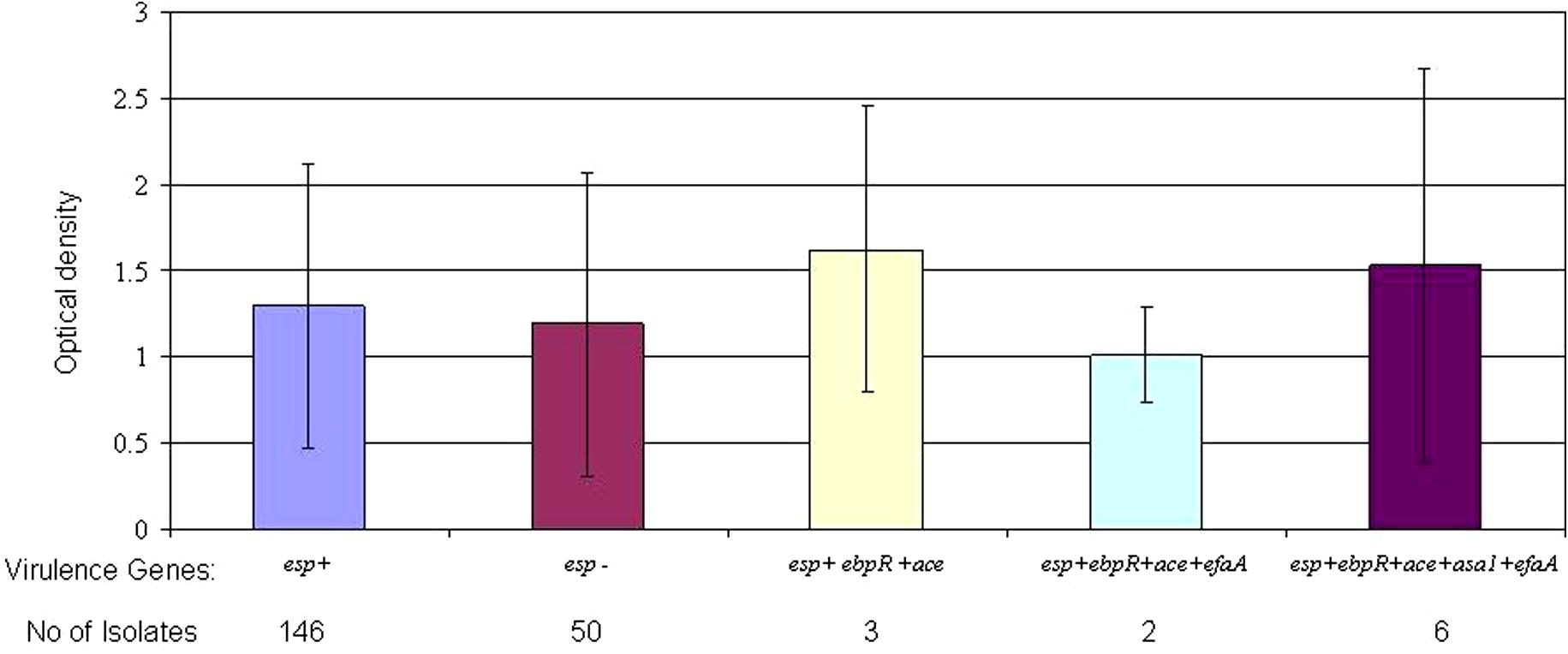
Biofilm formation (OD570) by the enterococcal isolates according to association of esp gene with other colonization factors.
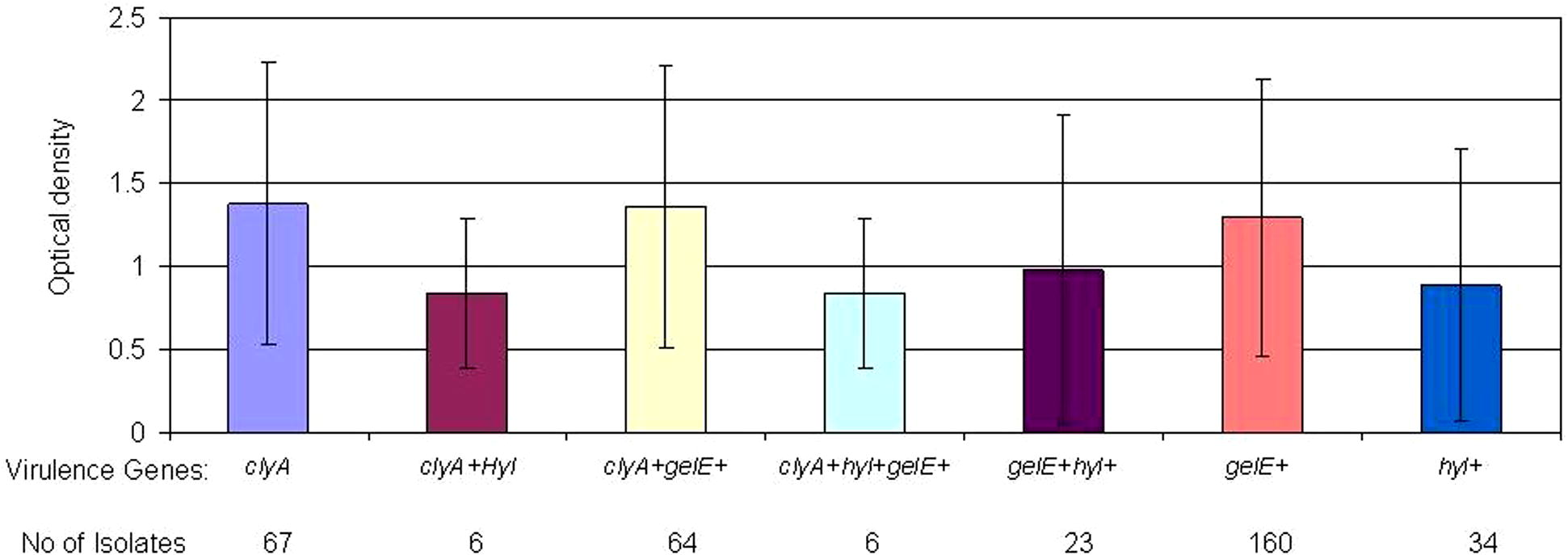
Biofilm formation (OD570) by the enterococcal isolates according to the presence of secretory factors.
4 Discussion
Bacterial urinary tract infections represent the most common type of nosocomial infections. Often, the ability of bacteria to both establish and maintain these infections is directly related to biofilm formation on indwelling devices or within the urinary tract itself (Hatt and Rather, 2008). Enterococci (especially E. faecalis) are one of the main causative agents of urinary tract infection and Catheter-associated urinary tract infections (CAUTIs) besides Gram-negative pathogens (Tenke et al., 2006; Guiton et al., 2010). In these infections, biofilm provides a favorable milieu for microbial survival within the host as the organisms are shielded from the host immune response, as well as antibiotics and antimicrobial agents (Yasuda et al., 1994; Lewis, 2001). Several studies were conducted to introduce main virulence genes of enterococci that are associated with biofilm formation in these bacteria (Shankar et al., 1999; Mohamed et al., 2003; Baldassarri et al., 2006), but virulence mechanism and related genes in biofilm formation are not well understood (Duggan and Sedgley, 2007). In this study, we investigated biofilm formation of clinical enterococci isolates isolated from UTIs. These strains were characterized for the presence of adhesions and secretory virulence factors. Our investigated isolates had diverse presence of virulence factors from lack to high amount of virulence genes. Several earlier studies investigated the relation of the presence of virulence genes and biofilm formation, especially the presence of esp and gelE. esp has been implicated as a contributing factor in the colonization and persistence of infection within the urinary tract (Shankar et al., 2001). In the present study, no association between the presence of esp and biofilm-forming ability was observed among isolates collected from urinary tract infections from different hospitals (P > 0.05) (Fig. 2). Conflicting outcomes have been published regarding the role of the esp gene in biofilm formation. Some authors have suggested that esp promotes biofilm formation; however, additional determinants may contribute to biofilm formation in enterococci (Upadhyaya et al., 2011; Moniri et al., 2013). Other studies suggest that the esp gene does not seem to be necessary nor sufficient for the production of biofilm in enterococci (Maestre et al., 2012; Dworniczek et al., 2005; Ramadhan and Hegedus, 2005). Heikens et al. also, did not find esp as an essential factor for adherence and intestinal colonization of Enterococcus in mice (Heikens et al., 2009). These findings add more contrary to the role of esp on biofilm formation by enterococci. Also we could not find any difference among esp positive isolates and gelE or cylA positive isolates. These results can show that the presence or absence of esp gene had no effect on biofilm formation by urinary tract isolates of enterococci. Also cytolysin operon that is in close association with the esp gene on the chromosome of enterococci, had no significant association with biofilm formation of isolates. On looking at other colonization factors, findings of this study showed that isolates with asa1 and efaA genes produced more biofilms than negative ones and it seems that these proteins have the highest contribution in biofilm formation in the urinary tract isolates (P > 0.011, P > 0.008; respectively). efaA has been shown to have an important role in pathogenesis of enterococci in infective endocarditis (Mohamed et al., 2004; Preethee et al., 2012) but its importance in urinary tract infections is not well described.
Among secretory factors, however Arciola et al. showed importance of gelatinase in Biofilm formation of Enterococci in implant infections (Arciola et al., 2008), the presence or absence of cytolysin and gelatinase in our set had no significant effect on biofilm formation, but isolates possessing hyl had a significantly lower biofilm formation (P = 0.000) which indicates that isolates carrying this gene prefer a planktonic to a biofilm lifestyle. Also only 17.35% of urinary tract isolates carried this gene. Low prevalence of hyl positive isolates in UTI and the low biofilm formation tendency can indicate that the absence of this gene can be an advantage for pathogenesis of enterococci in UTI. Comparing isolates carrying all colonization factors with the ones carrying all secreted factors showed no significant difference.
In conclusion, results of the present study showed that the presence of esp, ace, gelE and cylA genes did not seem to be necessary nor sufficient for the production of biofilm in enterococci, but the presence of efaA and asa1 in isolates was associated with the higher biofilm formation of urinary tract isolates. Also low prevalence of hyl positive isolates in UTI and low biofilm formation tendency of these isolates can indicate that the absence of this gene can be an advantage for pathogenesis of enterococci in urinary tract infections.
Acknowledgments
We would like thank all staff of labs for cooperation in collecting samples. Also we thank Dr. Hossein Navidinia and Dr. Mohammad Momenian for their helpful comments in manuscript preparation. This study was financially supported by Special grant from National Elite foundation of Iran for Hossein Samadi Kafil with Grant No. 3.12.118.
References
- Strong biofilm production, antibiotic multi-resistance and high gelE expression in epidemic clones of Enterococcus faecalis from orthopaedic implant infections. Biomaterials. 2008;29:580-586.
- [Google Scholar]
- Comparison of mycobacterial interspersed repetitive unit-variable number tandem repeat and IS6110–RFLP methods in identifying epidemiological links in patients with tuberculosis in Northwest of Iran. Ann. Microbiol.. 2008;58:333-339.
- [Google Scholar]
- Tuberculosis transmission in Northwest of Iran: using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect. Genet. Evol.. 2011;11:124-131.
- [Google Scholar]
- Virulence factors in enterococcal infections of orthopedic devices. Int. J. Artif. Organs.. 2006;29:402-406.
- [Google Scholar]
- EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J. Bacteriol.. 2007;189:6490-6493.
- [Google Scholar]
- A PCR assay for identification of Enterococcus faecium. J. Clin. Microbiol.. 1997;35:1248-1250.
- [Google Scholar]
- M100-S22. Performance Standards for Antimicrobial Susceptibility Testing: 22nd Informational Supplement. Wayne, PA: CLSI; 2012.
- Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol.. 2001;9:50-52.
- [Google Scholar]
- Biofilm formation of oral and endodontic Enterococcus faecalis. J. Endod.. 2007;33:815-818.
- [Google Scholar]
- Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy) J. Med. Microbiol.. 2003;52:491-498.
- [Google Scholar]
- Virulence of Enterococcus isolates collected in Lower Silesia (Poland) Scand. J. Infect. Dis.. 2005;37:630-636.
- [Google Scholar]
- Recognition of group D streptococcal species of human origin by biochemical and physiological tests. Appl. Microbiol.. 1972;23:1131-1139.
- [Google Scholar]
- The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749-1757.
- [Google Scholar]
- Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect. Immun.. 2010;78(10):4166-4175.
- [Google Scholar]
- Epidemiology of Enterococcus faecalis urinary tract infection in a teaching hospital in London, United Kingdom. J. Clin. Microbiol.. 1992;30(8):1953-1957.
- [Google Scholar]
- Role of bacterial biofilms in urinary tract infections. Curr. Top. Microbiol. Immunol. 2008;322:163-192.
- [Google Scholar]
- Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BMC Microbiol.. 2009;9:19.
- [Google Scholar]
- Emerging resistance among bacterial pathogens in the intensive care unit – a European and North American Surveillance study (2000–2002) Ann. Clin. Microbiol. Antimicrob.. 2004;3:14.
- [Google Scholar]
- Adhesion and virulence factor properties of enterococci isolated from clinical samples in Iran. Indian J. Pathol. Microbiol.. 2013;56:238-242.
- [Google Scholar]
- Vancomycin-resistant Enterococcus faecium and Enterococcus faecalis Isolated from Education Hospital of Iran. Mædica J. Clin. Med.. 2014;9:323-327.
- [Google Scholar]
- Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol.. 2000;38:3092-3095.
- [Google Scholar]
- Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit. Rev. Oral. Biol. Med.. 2004;15:308-320.
- [Google Scholar]
- Molecular characterization of multidrug-resistant Enterococcus spp. from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Mol. Cell. Probes. 2005;19:27-34.
- [Google Scholar]
- Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect. Immun.. 1995;63:703-706.
- [Google Scholar]
- In vitro interference of tigecycline at subinhibitory concentrations on biofilm development by Enterococcus faecalis. J. Antimicrob. Chemother.. 2012;67(5):1155-1158.
- [Google Scholar]
- Enterococcal virulence determinants may be involved in resistance to clinical therapy. Diagn. Microbiol. Infect. Dis.. 2007;58:59-65.
- [Google Scholar]
- Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun.. 2004;72:3658-3663.
- [Google Scholar]
- Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. J. Clin. Microbiol.. 2005;43:5405-5407.
- [Google Scholar]
- Influence of clinical origin and of various genes on biofilm formation by Enterococcus faecalis.Program Abstracts of the 43rd Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, abstract B-821. Washington, DC: American Society for Microbiology; 2003. p. :52.
- Virulence gene’s relationship with biofilm formation and detection of aac (6′)/aph (2″) in Enterococcus faecalis isolated from patients with urinary tract infection. Jundishapur J. Microbiol.. 2013;6(5):e6244.
- [Google Scholar]
- Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host E. faecalis interaction. Infect. Immun.. 2006;74:4982-4989.
- [Google Scholar]
- Enterococcal virulence pathogenicity island of E. faecalis. Front. Biosci.. 2004;9:2335-2346.
- [Google Scholar]
- Molecular identification of an Enterococcus faecalis endocarditis antigen efaA in root canals of therapy-resistant endodontic infections. J. Conserv. Dent.. 2012;15(4):319-322.
- [Google Scholar]
- Biofilm formation and esp gene carriage in enterococci. J. Clin. Pathol.. 2005;58:685-686.
- [Google Scholar]
- Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol.. 2003;52:547-550.
- [Google Scholar]
- Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun.. 1999;67:193-200.
- [Google Scholar]
- Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun.. 2001;69:4366-4372.
- [Google Scholar]
- The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol.. 2001;67:4538-4545.
- [Google Scholar]
- Comparative study among clinical and commensal isolates of Enterococcus faecalis for the presence of esp gene and biofilm production. J. Infect. Dev. Ctries.. 2011;5(5):365-369.
- [Google Scholar]
- Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol.. 2004;42:4473-4479.
- [Google Scholar]
- Interaction between human polymorphonuclear leucocytes and bacteria released from in-vitro bacterial biofilm models. J. Med. Microbiol.. 1994;41:359-367.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jksus.2014.12.007.
Appendix A
Supplementary data
Supplementary Table 1
Supplementary Table 1
Biofilm formation (OD570) by the enterococcal isolates collected from UTIs testing positive for cly, gel or hyl, cly and hyl, cly and gel, cyl, hyl and gel, gel and hyl.







