Efficacy of double-stranded RNA against white spot syndrome virus (WSSV) non-structural (orf89, wsv191) and structural (vp28, vp26) genes in the Pacific white shrimp Litopenaeus vannamei
*Corresponding author. Tel.: +52 687 8729626x87637 cesar_escobedomx@yahoo.com (César M. Escobedo-Bonilla)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Available online 5 December 2014
Peer review under responsibility of King Saud University.
Abstract
White spot syndrome virus (WSSV) is a major pathogen in shrimp aquaculture. RNA interference (RNAi) is a promising tool against viral infections. Previous works with RNAi showed different antiviral efficacies depending on the silenced gene. This work evaluated the antiviral efficacy of double-stranded (ds) RNA against two non-structural (orf89, wsv191) WSSV genes compared to structural (vp26, vp28) genes to inhibit an experimental WSSV infection. Gene orf89 encodes a putative regulatory protein and gene white spot virus (wsv)191 encodes a nonspecific nuclease; whereas genes vp26 and vp28 encode envelope proteins, respectively. Molecules of dsRNA against each of the WSSV genes were intramuscularly injected (4 μg per shrimp) into a group of shrimp 48 h before a WSSV challenge. The highest antiviral activity occurred with dsRNA against orf89, vp28 and vp26 (cumulative mortalities 10%, 10% and 21%, respectively). In contrast, the least effective treatment was wsv191 dsRNA (cumulative mortality 83%). All dead animals were WSSV-positive by one-step PCR, whereas reverse-transcription PCR of all surviving shrimp confirmed inhibition of virus replication. This study showed that dsRNA against WSSV genes orf89, vp28 and vp26 were highly effective to inhibit virus replication and suggest an essential role in WSSV infection. Non-structural WSSV genes such as orf89 can be used as novel targets to design therapeutic RNAi molecules against WSSV infection.
Keywords
WSSV
Aquatic diseases
Non-structural genes
dsRNA
Antiviral therapeutics
Litopenaeus vannamei
1 Introduction
White spot syndrome virus (WSSV) is a major pathogen affecting shrimp farming worldwide. Since its emergence in the mid 90s in various countries in America, it has caused major economic losses to shrimp farmers (Escobedo-Bonilla et al., 2008; Lightner, 2011). The WSSV genome consists of 531 putative genes (https://www.ncbi.nlm.nih.gov/nuccore/AF440570.1; accessed on 08/20/14), some of which may be essential for WSSV infection and/or replication. These include genes such as vp28, protein kinase, thymidine-thymidylate kinase, DNA polymerase and WSSV449 (Chen et al., 2002; Liu et al., 2001; Tsai et al., 2000; van Hulten et al., 2001a; Wang et al., 2011). Still, the role of many genes during virus infection is unknown. Silencing genes by specific double-stranded RNA represents a way to determine their putative roles (Fagutao et al., 2009; Labreuche et al., 2009; Nakayashiki et al., 2005).
Several studies have used RNA interference (RNAi) to inhibit viral infections in shrimp. Most of the genes used to inhibit WSSV infection encode structural proteins involved in virion architecture (Kim et al., 2007; Robalino et al., 2004, 2005; Tirasophon et al., 2005; Yodmuang et al., 2006). WSSV structural proteins such as vp28 and vp26 are involved in virus entry (Tang et al., 2007; van Hulten et al., 2001a; Youtong et al., 2011; Zhang et al., 2002). These proteins constitute targets against virus infection by recombinant proteins (Caipang et al., 2008; Witteveldt et al., 2006), neutralizing antibodies (Jiang et al., 2007; Natividad et al., 2007) and RNAi (Mejía-Ruíz et al., 2011; Robalino et al., 2004; Sarathi et al., 2008).
A few studies have assessed the antiviral efficacy of RNAi against non-structural WSSV proteins such as DNA polymerase, protein kinase, ribonucleotide reductase, thymidine-thymidylate kinase or protein WSSV449 (Attasart et al., 2009; Kim et al., 2007; Wang et al., 2011; Wu et al., 2007).
Since the publication of the complete WSSV genome (Chen et al., 2002; van Hulten et al., 2001b; Yang et al., 2001) many novel WSSV putative open reading frames (ORFs) have been described and some of these have been characterized (Hossain et al., 2004; Li et al., 2005), but the role of their products in virus infection is largely unknown. One of these genes is orf89, with 4436 nucleotides encoding a peptide 1437 amino acids long and apparent size of 165 kDa. This peptide localizes in the nucleus of transfected Sf9 insect cells. In vitro assays showed that the peptide had a negative regulation function of a WSSV protein kinase (orf61) and a thymidine-thymidylate kinase (Hossain et al., 2004). Other genes encoding putative enzymes related to nucleotide metabolism include white spot virus (wsv)067 (thymidylate synthase), wsv112 (dUTP pyrophosphatase), wsv172 (ribonucleotide reductase large subunit), wsv188 (ribonucleotide reductase small subunit [rr]), wsv191 (deoxyribonuclease I) and wsv395 (thymidylate kinase) (Yang et al., 2001). Of these, gene wsv191 has a sequence of 912 base pairs (bp), which encodes a non-specific nuclease of 304 residues with apparent size of 34.4 kDa. The assumed role of a non-specific nuclease in WSSV infection is degradation of nucleic acids of host cells, viral DNA repair and virus genome synthesis (Li et al., 2005).
Previous works have shown different antiviral efficacies of RNAi molecules depending on the targeted genes (Robalino et al., 2005; Wu et al., 2007). It is possible that antiviral efficacy depends on the role of the silenced gene in virus infection/replication (Kim et al., 2007). Then, RNAi silencing of WSSV genes with a critical role in virus replication might show stronger virus inhibition and lower shrimp mortality. Hence, the aim of this study was to evaluate the antiviral efficacy of double-stranded (ds)RNA against sequences of the non-structural WSSV genes orf89 and wsv191 compared to dsRNA against two WSSV structural proteins (vp28 and vp26) under experimental conditions.
2 Material and methods
2.1 Shrimp
Healthy shrimp Litopenaeus vannamei (n = 147; mean body weight [MBW] = 23.3 ± 4.2 g) from a hatchery in Sonora, Mexico were used. Shrimp were determined to be WSSV-negative by PCR using DNA from hemolymph and primers amplifying a segment of the vp28 gene (Table 1).
| Primer name | Sequence 5′ → 3′ | PCR-product size (bp) |
|---|---|---|
| orf89 F1 | AGGACCCGATCGCTTACTTTGA | 482 |
| orf89 R1 | CTCCCTCCCTTGCGAACTTG | |
| wsv191 F1 | AAGTGGGTGCGCAACAAAATA | 490 |
| wsv191 R1 | TGTAGAGGGCATGAGGGATAG | |
| vp28 F3 | AACTGCAGATGGATCTTTCTTTC | 610 |
| vp28 R3 | AACTGCAGTTACTCGGTCCTCAG | |
| vp26 F4 | ATCCAACCAACACGTAAAGG | 580 |
| vp26 R4 | CAATTGCCACTTTACTTCTTCCTG | |
| LacZ F2 | ACCAGAAGCGGTGCCGGAAA | 1012 |
| LacZ R2 | CCACAGCGGATGGTTCGGAT | |
| β-Actin F2 | GAAGTAGCCGCCCTGGTTG | 416 |
| β-Actin R2 | CGGTTAGCCTTGGGGTTGAG | |
| BLOCK-iT. T7 | GATGACTCGTAATACGACTCACTA |
2.2 Shrimp maintenance
Before the experiments, shrimp were maintained in tanks with artificial seawater, which was prepared using drinking water added with artificial marine salt (Instant Ocean, Blacksburg VA, USA) to 25 g/l salinity, at temperature of 27 ± 2 °C and continuous aeration. Water exchange (50% of total volume) was done every third day. Monitoring of other water quality parameters such as ammonia, nitrate or nitrite was not done.
2.3 WSSV inoculum
A WSSV inoculum was prepared from naturally-infected shrimp from Sinaloa following the method of Escobedo-Bonilla et al. (2005). Briefly, carcasses without exoskeleton and hepatopancreas were minced and suspended in 10 volumes of phosphate buffered saline (PBS) [NaCl 137 mM, KCl 2.69 mM, Na2HPO4 10.16 mM, KH2PO4 1.37 mM, pH 7.4]. Tissue suspension was homogenized with a grinder (Braun Multiquick, Spain) and the suspension was clarified by two consecutive centrifugation steps at 3000g and 16,600g for 20 min at 4 °C, respectively. The resulting suspension was filtered through a 0.45 μm membrane, aliquoted in cryovials (0.8 ml) and stored at −80 °C until used.
2.4 In vivo titration
A batch of shrimp L. vannamei (n = 30, MBW = 13.1 ± 1.53 g) from the same hatchery were used for the in vivo titration of WSSV inoculum as described earlier (Escobedo-Bonilla et al., 2005). Briefly, tenfold serial dilutions (10−2 − 10−7) of the WSSV stock were done with cold PBS. Per dilution, five shrimp were intramuscularly inoculated (50 μl) and individually kept in 12 L artificial seawater (Instant Ocean, Blacksburg VA, USA) at salinity of 25 g/l and temperature 27 ± 2 °C. Shrimp were monitored for 120 h post inoculation (hpi) every 12 h for clinical signs of WSSV infection (anorexia, lethargy) and mortality. The shrimp infectious dose 50% endpoint (SID50/ml) (the dose that causes infection to 50% of the inoculated animals) was 105.6 SID50/ml according to infection data by PCR. Likewise, the lethal dose 50% endpoint (LD50/ml) (the dose that kills 50% of the inoculated animals) was 105.6 LD50/ml, determined with mortality data at the end of the experiments (120 hpi). A dose of 2500 SID50 in 100 μl was selected to perform the WSSV challenge bioassays. This dose was selected because we wanted to make sure that every shrimp inoculated with such a dose would become infected and die.
2.5 Synthesis of dsRNA molecules
DNA from the four WSSV genes (vp26, vp28, vp191 and orf89) were amplified using specific primers (Table 1). PCR amplifications were done with the kit pure Ready-To-Go PCR beads (Illustra, GE Health Life Science, USA) following manufacturer’s instructions. PCR amplifications were done in a thermocycler (Perkin-Elmer Gene Amp PCR System 97000, USA) with an initial denaturation step (94 °C) for 4 min; followed by 30 cycles of denaturation (94 °C) 1 min; annealing (58–62 °C, depending on the gene) for 1 min and extension (72 °C) 1 min. A final extension step (72 °C) for 10 min was done. PCR products were stored at 4 °C. Quantity and integrity of PCR products were analyzed through 1% agarose gel electrophoresis stained with 2 μl (10 mg/ml) ethidium bromide.
PCR products were used for the TOPO linking reaction. This assay introduced a T7 promoter to the WSSV PCR products. Reactions were done following the manufacturer instructions (Block-iT RNAi TOPO Transcription kit, K3500-01, Invitrogen, USA). Briefly, 1 μl of a WSSV PCR product (20 ng/μl), 1 μl saline solution, 3 μl sterile water and 1 μl of Block-iT T7-TOPO Linker were mixed. The reaction was incubated for 15 min at 37 °C and afterwards it was chilled on ice.
Products were used in two separate PCR assays to get sense and antisense WSSV gene strands containing the T7 promoter. For the WSSV-T7 TOPO sense strand, each of the WSSV reverse primers was used along with the Block-iT T7 primer. For the WSSV-T7 TOPO antisense strand, each of the WSSV forward primers was used along with the Block-iT T7 primer. The amount and quality of the resulting products were verified by 1% agarose gel electrophoresis stained with ethidium bromide.
Then, transcription of each sense or antisense RNA strand was done separately in a 1.5 ml microtube. The reaction contained RNAse-free water (21 μl), 75 mM NTPs (8 μl), sense or antisense secondary PCR amplification (1–10 μl), 10× transcription buffer (4 μl) and Block-iT T7 enzyme mix (6 μl). The reaction was mixed and incubated at 37 °C for 6 h. Afterwards, each reaction was added 2 μl DNAse I and it was incubated for 15 min at 37 °C.
Each RNA strand transcript was purified with 160 μl of RNA binding buffer with 1% (v/v) β-mercaptoethanol and 100 μl absolute ethanol. The sample was placed in a RNA spin cartridge following manufacturer instructions (Invitrogen USA). A final volume of 40 μl of each single-stranded RNA was obtained. Then, the purified transcripts were aligned to form dsRNA. Equimolar concentrations of the respective sense and antisense RNAs were mixed in a sterile microtube floating in boiling water. The mix was left to cool down for 90 min. Afterwards, dsRNA was quantified by nanospectrophotometer (nanodrop 2000c, Thermo Scientific, Wilmington DE, USA) and stored at −80 °C until used. In addition, a LacZ dsRNA was synthesized and used as a control.
2.6 WSSV challenge bioassays
WSSV-free shrimp L. vannamei were used. Animals were acclimatized to experimental conditions (80 l tanks containing artificial seawater at 25 g/l salinity continuous aeration and temperature of 25 ± 2 °C) for 24 h. Afterwards, groups of 10 shrimp were intramuscularly injected (4 μg dsRNA in 40 μl cold PBS) with one of the dsRNAs (vp26, vp28, vp191, orf89). Another group of animals was treated with dsRNA against bacterial LacZ gene and used as a control. A group of 10 shrimp was mock-treated with 40 μl PBS and used as a positive control. Three replicates were done per treatment.
Later, (48 h) all the treated animals were intramuscularly challenged with a high dose (2500 SID50 in 50 μl) of WSSV. Shrimp were monitored for 10 d after WSSV challenge for clinical signs of disease and mortality twice daily. Moribund and dead shrimp were recorded for cumulative mortality and removed from the tanks. Tissue samples were taken for WSSV detection. At the end of the experiment, surviving shrimp were sacrificed and gills were sampled for RNA extraction and WSSV PCR analyses. During the experiments, water exchange (50% total volume) was done every fourth day to maintain water quality.
2.7 Nucleic acid extractions
WSSV-infected gills from shrimp were ground using DNAzol reagent (Molecular Research Center, Cincinnati, OH, USA) following the manufacturer’s instructions. The resulting DNA pellet was dissolved in ultra pure water and stored at −20 °C until used for PCR analyses.
RNA extraction was done only in surviving shrimp using Trizol reagent (Molecular Research Center, Cincinnati, OH, USA). Briefly, gill tissues were homogenized in microtubes containing 1 ml Trizol. Samples were added 0.2 ml chloroform, mixed and incubated at room temperature for 3 min. Samples were centrifuged at 12,000g for 15 min at 4 °C. The upper aqueous phase was recovered and RNA was precipitated by adding 0.5 ml isopropanol. After incubation for 10 min at room temperature and centrifugation at 12,000g for 10 min, RNA was washed with 75% isopropanol. The pellet was dried and resuspended in RNAse-free water at 55–60 °C for 10 min. Samples were treated with 1 μl DNAse I at 37 °C for 15 min. Extracted RNA was directly used for WSSV detection using the reverse-transcriptase (RT)-PCR assay with the kit Ready-To-Go RT-PCR beads (Illustra, GE Health Life Science, USA) following manufacturer’s instructions.
2.8 Diagnosis of WSSV infection
WSSV detection by PCR was done using primers vp28 F3/vp28 R3, which amplify a segment of the WSSV gene vp28 (Table 1). Amplification conditions were: initial denaturation at 95 °C for 5 min; then, 30 cycles with denaturation at 95 °C for 45 s; alignment at 58 °C for 45 s and extension at 72 °C for 45 s. A final extension step was done at 72 °C for 10 min. An internal control (β-actin, 416 bp) using the same amplification conditions as WSSV vp28 was included. PCR products were resolved in 1% TAE-agarose gel electrophoresis stained with ethidium bromide.
2.9 Evaluation of antiviral efficacy
The antiviral efficacy of the different dsRNAs against WSSV genes was assessed using cumulative mortality (mean ± standard error, expressed as percentage) and number of WSSV-infected shrimp (determined by PCR and/or RT-PCR).
3 Results
Shrimp from the mock-treated group and those treated with dsRNA against LacZ first showed clinical signs of WSSV infection such as reduced feeding, erratic swimming and/or lethargy at 24 hpi. The first mortalities were recorded at 36 hpi. The positive control group and the group treated with LacZ dsRNA both had 100% mortality at 108 hpi (Fig. 1). All dead animals in both groups were WSSV-positive by PCR (Fig. 2).
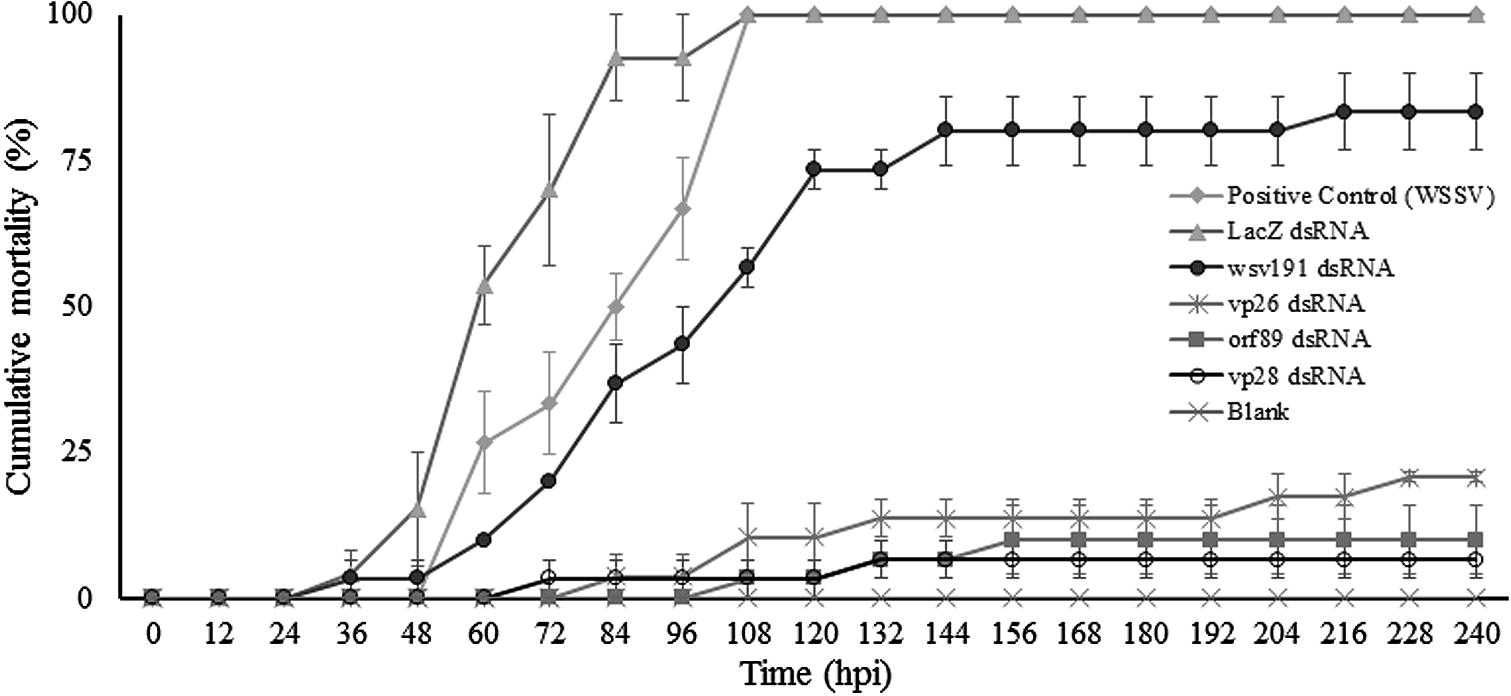
- Cumulative mortality (mean ± standard error) of shrimp treated with dsRNA against WSSV genes vp28, vp26, orf89 and wsv191. Groups of mock-treated shrimp, LacZ dsRNA and a blank were included. The blank is a group of shrimp which was not challenged with WSSV nor treated with any dsRNAs, and it was only kept in the same conditions of density and water quality as the treatments and control groups.
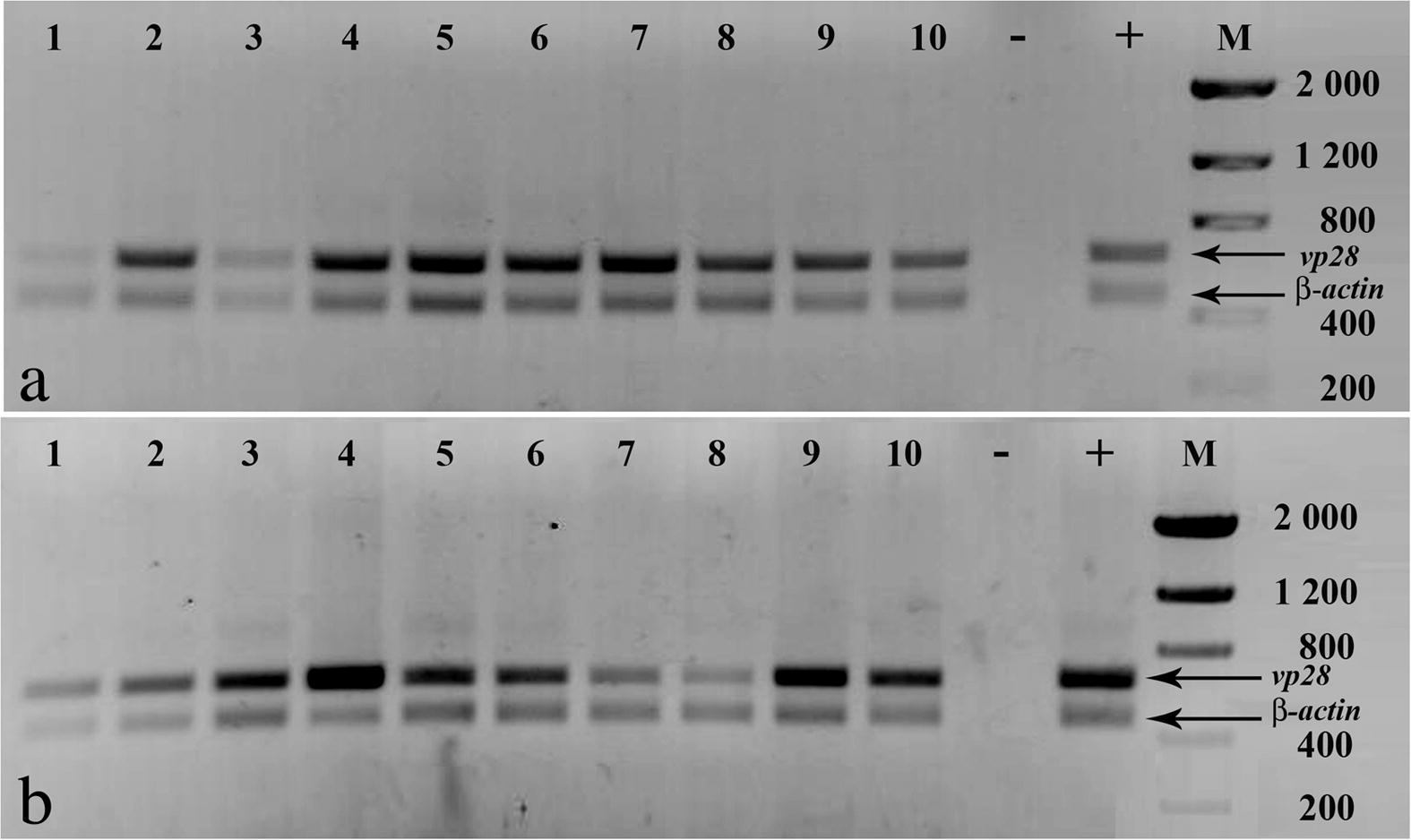
- PCR analyses of moribund and dead shrimp in (a) mock-treated, and (b) LacZ dsRNA groups. Amplification of shrimp β-actin was done as internal control. Negative control (−), WSSV positive control (+), molecular weight marker (M) size in base pairs.
Shrimp treated with dsRNA against four WSSV genes showed different mortality rates. Shrimp treated with vp28 dsRNA showed first mortalities at 72 hpi and the final cumulative mortality was 7%. Animals treated with orf89 dsRNA displayed first mortalities at 108 hpi and cumulative mortality at the end of the experiments was 10%. Shrimp treated with vp26 dsRNA showed first mortalities at 84 hpi and cumulative mortality was 21%. Animals treated with wsv191 dsRNA showed first mortalities at 24 hpi and cumulative mortality was 83% at the end of the experiments (Fig. 1).
All dead animals in these treatments were WSSV-positive by PCR (Fig. 3). Surviving shrimp of these four treatments did not display clinical signs of WSSV infection and all were WSSV-negative by RT-PCR assays (Fig. 4).
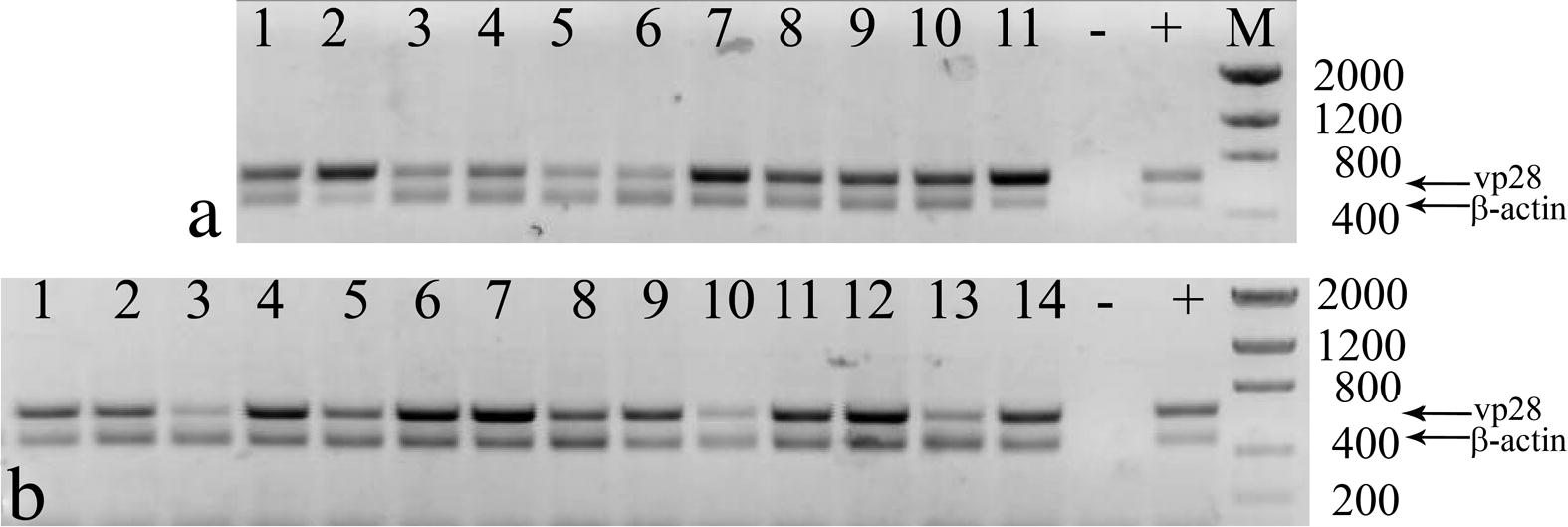
- PCR analyses of moribund and dead shrimp treated with dsRNA against WSSV genes. (a) WSSV-positive shrimp treated with dsRNA against vp28 (lanes 1 and 2), orf89 (lanes 3–5) and vp26 (lanes 6–11). (b) WSSV-positive shrimp treated with wsv191 dsRNA. Amplification of shrimp β-actin was done as internal control. Negative control (−), WSSV positive control (+), molecular weight marker (M) size in base pairs.
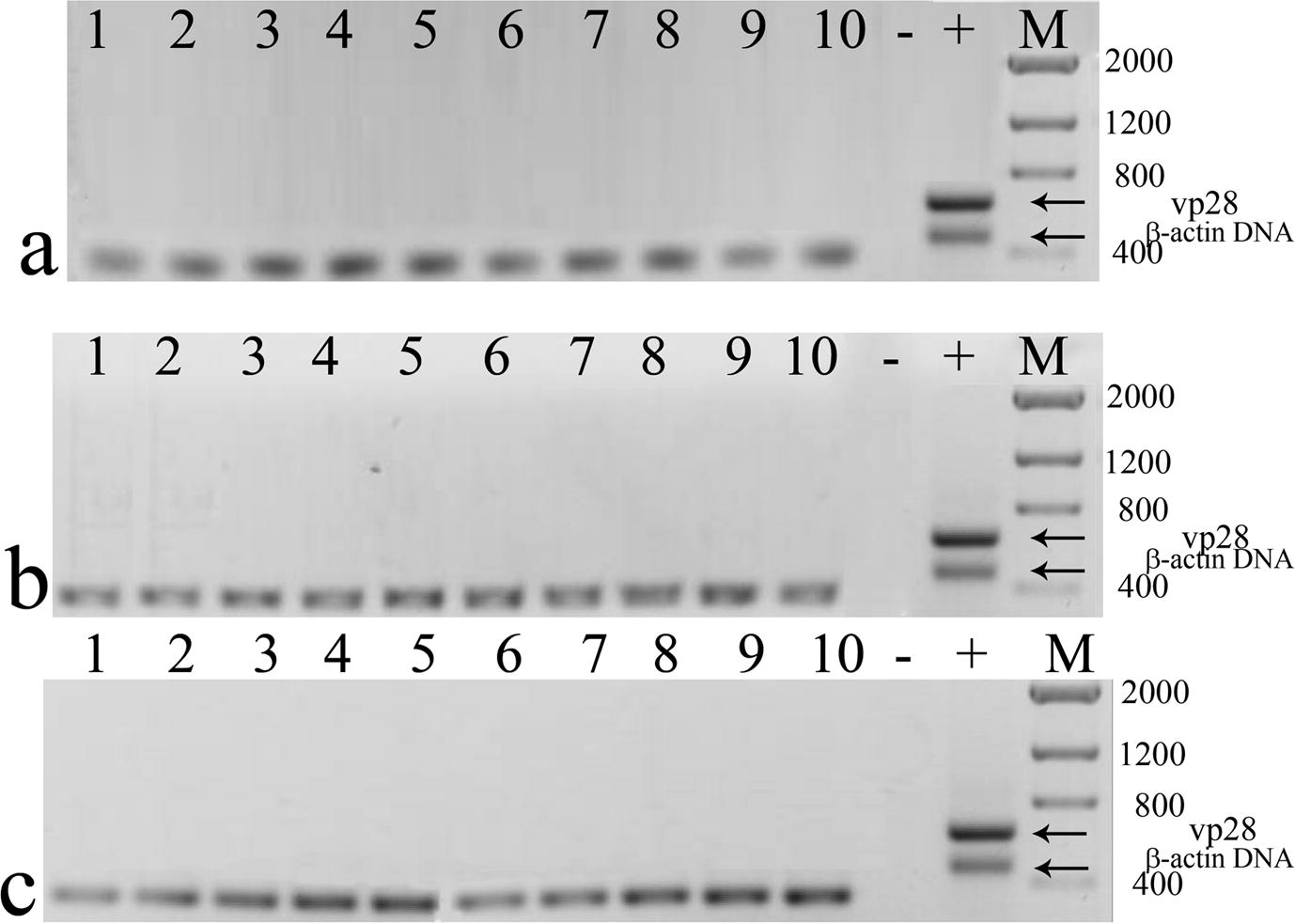
- RT-PCR from surviving shrimp treated with dsRNA against (a) vp28, (b) orf89, (c) vp26. No vp28 cDNA amplification was observed. Amplification of shrimp β-actin cDNA was done as internal control. Size of β-actin cDNA was slightly lower than β-actin DNA (≈330 bp). Negative control (−), WSSV positive control (+), molecular weight marker (M) size in base pairs.
4 Discussion
The present study showed the antiviral efficacy of dsRNA against four WSSV genes using a standardized challenge procedure with a high virus dose. A single dsRNA dose of 4 μg per shrimp was enough to trigger an antiviral response against a lethal WSSV challenge. Sequence-specific dsRNA effectively inhibited virus replication and reduced mortality in treated shrimp. At the end of the experiments (10 dpi), most of the animals treated with sequence-specific dsRNA survived (Fig. 1) and had the lowest rates of WSSV infection (Figs. 3 and 4). In contrast, shrimp treated with sequence-independent dsRNA (bacterial LacZ gene) became infected (Fig. 2) and only underwent a delay in time to mortality (Fig. 1). Previous studies have reported a mild antiviral effect of sequence-independent dsRNA treatment using either a low amount of infectious virus (Robalino et al., 2007) or even with the same high infectious dose (Mejía-Ruíz et al., 2011). This finding confirms that in shrimp, only sequence-specific dsRNA triggers a protective RNAi response upon a high WSSV dose.
Shrimp treated with dsRNA against vp28 and vp26 were protected against a highly lethal WSSV challenge. This result endorses previous findings where dsRNA targeting any of these two genes gave a similar protection against an experimental WSSV infection (Mejía-Ruíz et al., 2011). Genes vp28 and vp26 encode structural proteins vp28 and vp26, respectively. These proteins have been shown to play important roles in virion structure, virus attachment and infection (van Hulten et al., 2001a; Xie and Yang, 2005). Protein vp28 is located in the viral envelope, which is the outermost layer of the WSSV virion. This protein has been determined to play a key role in the infection process (van Hulten et al., 2000) such as attachment to cell membrane and cytoplasm entry (Yi et al., 2004), virus morphogenesis and virion assembly (Wang et al., 1999). These functions make gene vp28 an important target against WSSV infection and the inhibition of this protein clearly affects virus infection/replication.
Protein vp26 is another major component of the virus envelope (Zhang et al., 2002; Tang et al., 2007). This peptide probably anchors with other proteins such as VP51 in the nucleocapsid to give structure to the virion, as vp26 might help keep attached the envelope to the nucleocapsid (Xie et al., 2005; Wan et al., 2008). It was shown that protein vp26 may be involved in the cytoplasmic transport of the nucleocapsid, since it is able to interact with actin fibers from the cellular cytoskeleton (Xie et al., 2005). Therefore, silencing vp26 may also impair infection and virus morphogenesis thus preventing virus replication.
The efficacy of two new dsRNAs against WSSV genes orf89 and wsv191 was evaluated and compared to the antiviral efficacy of the two previously tested dsRNAs against WSSV genes vp28 and vp26. Results showed that silencing orf89 gene had a strong antiviral effect since reduction of shrimp mortality was very similar to that observed in the group treated with vp28 dsRNA and it even surpassed the antiviral effect in shrimp treated with vp26 dsRNA. In contrast, dsRNA against wsv191 had the least antiviral effect of all the sequence-specific dsRNAs tested.
The function of the two non-structural proteins encoded by these WSSV genes may help explain differences in antiviral activity. WSSV gene orf89 is expressed early during virus replication and encodes a viral protein that localizes in the nucleus of transfected cells and has a negative regulatory function (Hossain et al., 2004). In contrast, gene wsv191 probably is not expressed early, since it lacks a TATA box and a putative early transcription signal (Witteveldt et al., 2001). This gene encodes a putative non-specific endonuclease with both DNAse and RNAse activities (Li et al., 2005; Witteveldt et al., 2001). Previous works indicated that the antiviral efficacy of silenced genes is related to their role in virus infection/replication (Kim et al., 2007). Even at low levels, some essential genes that are expressed early during infection can be effective antiviral targets (Shekhar and Lu, 2009; Yodmuang et al., 2006). This seems to be the case for WSSV gene orf89.
In conclusion, the present study showed that dsRNA against a novel WSSV gene orf89 was highly effective to inhibit virus replication and reduced shrimp mortality upon a high-dose WSSV challenge. Such an efficacy was as good as that observed in dsRNA against the WSSV gene vp28 encoding an essential structural protein. Therefore, WSSV orf89 represents a novel RNAi target against WSSV infection.
References
- Inhibition of white spot syndrome virus replication in Penaeus monodon by combined silencing of viral rr2 and shrimp PmRab7. Virus Res.. 2009;145:127-133.
- [Google Scholar]
- Enhanced survival of shrimp, Penaeus (Marsupenaeus) japonicus from white spot syndrome disease after oral administration of recombinant vp28 expressed in Brevibacillus brevis. Fish Shellfish Immunol.. 2008;23:315-320.
- [Google Scholar]
- Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology. 2002;301:136-147.
- [Google Scholar]
- A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis.. 2008;31:1-18.
- [Google Scholar]
- In vivo titration of white spot syndrome virus (WSSV) in SPF Litopenaeus vannamei by intramuscular and oral routes. Dis. Aquat. Organ.. 2005;66:163-170.
- [Google Scholar]
- Increased bacterial load in shrimp hemolymph in the absence of prophenoloxidase. FEBS J.. 2009;276:5298-5306.
- [Google Scholar]
- Characterization of orf89 – A latency-related gene of white spot syndrome virus. Virology. 2004;325:106-115.
- [Google Scholar]
- Neutralizing assay of monoclonal antibodies against white spot syndrome virus (WSSV) Aquaculture. 2007;272:216-222.
- [Google Scholar]
- Protection of shrimp (Penaeus chinensis) against white spot syndrome virus (WSSV) challenge by double-stranded RNA. Fish Shellfish Immunol.. 2007;23:242-246.
- [Google Scholar]
- Lack of evidence for Litopenaeus vannamei Toll receptor (lToll) involvement in activation of sequence-independent antiviral immunity in shrimp. Dev. Comp. Immunol.. 2009;33:806-810.
- [Google Scholar]
- Functional identification of the non-specific nuclease from white spot syndrome virus. Virology. 2005;337:399-406.
- [Google Scholar]
- Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): a review. J. Invertebr. Pathol.. 2011;106:110-130.
- [Google Scholar]
- Cloning, characterization, and phylogenetic analysis of a shrimp white spot syndrome virus gene that encodes a protein kinase. Virology. 2001;289:362-377.
- [Google Scholar]
- Double-stranded RNA against white spot syndrome virus (WSSV) vp28 or vp26 reduced susceptibility of Litopenaeus vannamei to WSSV, and survivors exhibited decreased susceptibility in subsequent re-infections. J. Invertebr. Pathol.. 2011;107:65-68.
- [Google Scholar]
- RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol.. 2005;42:275-283.
- [Google Scholar]
- White spot syndrome virus (WSSV) inactivation in Penaeus japonicus using purified monoclonal antibody targeting viral envelope protein. Aquaculture. 2007;269:54-62.
- [Google Scholar]
- Double-stranded RNA and antiviral immunity in marine shrimp: inducible host mechanisms and evidence for the evolution of viral counter-responses. Dev. Comp. Immunol.. 2007;31:539-547.
- [Google Scholar]
- Double-stranded RNA induces sequence-specific antiviral silencing in addition to non-specific immunity in marine shrimp: convergence of RNA interference and innate immunity in the invertebrate antiviral response? J. Virol.. 2005;79:13561-13571.
- [Google Scholar]
- Induction of antiviral immunity of double-stranded RNA in a marine invertebrate. J. Virol.. 2004;78:10442-10448.
- [Google Scholar]
- Silencing vp28 gene of white spot syndrome virus of shrimp by bacterially expressed dsRNA. Mar. Biotechnol.. 2008;10:198-206.
- [Google Scholar]
- Application of nucleic-acid-based therapeutics for viral infections in shrimp aquaculture. Mar. Biotechnol.. 2009;11:1-9.
- [Google Scholar]
- Crystal structures of major envelope proteins vp26 and vp28 from white spot syndrome virus shed light on their evolutionary relationship. J. Virol.. 2007;81:6709-6717.
- [Google Scholar]
- Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem. Biophys. Res. Commun.. 2005;334:102-107.
- [Google Scholar]
- Identification and characterization of a shrimp white spot syndrome virus (WSSV) gene that encodes a novel chimeric polypeptide of cellular-type thymidine kinase and thymidylate kinase. Virology. 2000;277:100-110.
- [Google Scholar]
- Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology. 2000;266:227-236.
- [Google Scholar]
- White spot syndrome virus envelope protein vp28 is involved in the systemic infection of shrimp. Virology. 2001;285:228-233.
- [Google Scholar]
- vp26 of white spot syndrome virus functions as a linker protein between the envelope and nucleocapsid of virions by binding with vp51. J. Virol.. 2008;82:12598-12601.
- [Google Scholar]
- The Shrimp NF-kB pathway is activated by white spot syndrome virus (WSSV) 449 to facilitate the expression of WSSV069 (ie1), WSSV303 and WSSV371. PLoS One. 2011;6:e24773.
- [Google Scholar]
- Histopathology and cytopathology of white spot syndrome virus (WSSV) in cultured Penaeus monodon from peninsular Malaysia with emphasis on pathogenesis and the mechanism of white spot formation. Dis. Aquat. Organ.. 1999;39:1-11.
- [Google Scholar]
- Identification and phylogeny of a non-specific endonuclease gene of white spot syndrome virus of shrimp. Virus Genes. 2001;23:331-337.
- [Google Scholar]
- Increased tolerance of Litopenaeus vannamei to white spot syndrome virus (WSSV) infection after oral application of the viral envelope protein vp28. Dis. Aquat. Organ.. 2006;70:167-170.
- [Google Scholar]
- Inhibition of white spot syndrome virus in Litopenaeus vannamei shrimp by sequence-specific siRNA. Aquaculture. 2007;271:21-30.
- [Google Scholar]
- Interaction of white spot syndrome virus vp26 protein with actin. Virology. 2005;336:93-99.
- [Google Scholar]
- Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol.. 2001;75:11811-11820.
- [Google Scholar]
- vp28 of shrimp white spot syndrome virus is involved in the attachment and penetration into shrimp cells. J. Biochem. Mol. Biol.. 2004;37:726-734.
- [Google Scholar]
- YHV-protease dsRNA inhibits YHV replication in Penaeus monodon and prevents mortality. Biochem. Biophys. Res. Commun.. 2006;341:351-356.
- [Google Scholar]
- WSSV: vp26 binding protein and its biological activity. Fish Shellfish Immunol.. 2011;30:77-83.
- [Google Scholar]
- Transcription and identification of an envelope protein gene (p22) from shrimp white spot syndrome virus. J. Gen. Virol.. 2002;83:471-477.
- [Google Scholar]







