Translate this page into:
ZnO-chlorogenic acid nanostructured complex inhibits Covid-19 pathogenesis and increases hydroxychloroquine efficacy
⁎Corresponding author at: Bio-screening and Preclinical Trial Lab, Biochemistry Department, Faculty of Science, Alexandria University, Alexandria, Egypt. d.ghareeb@alexu.edu.eg (Doaa A. Ghareeb)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
The study purpose was to compare the anti- novel coronavirus disease 2019 (COVID-19) property of chlorogenic acid (CGA) and Zinc oxide nanoparticles (ZnO-NP) with the new valid synthesized complex of ZnO /CGA-NPs.
Methods
The facile mixing method was utilized to prepare ZnO/CGA-NPs. The in vitro effect of different ZnO/CGA-NPs concentrations on papain-like protease (PLpro) and spike protein- receptor-binding domain (RBD) was measured by ELISA technique. The compounds effects on SARS-CoV2 were determined on viral entry, replication, and assembly by using plaque reduction assay, qPCR, and ELISA techniques. Their individual effects or mixed with hydroxychloroquine (HCQ) on erythrocytes (RBCs) and leukocytes (WBCs) were evaluated by routine cell culture technique. Finally, turbidity and agar well diffusion assays were done to evaluate their antimicrobial properties against Escherichia. coli, klebsila pneumonia, Streptococcus pyogenes, Staphylococcus aureus, and Candida albicans.
Results
The results confirmed that the uniformly dispersed ZnO-NPs were converted to aggregated form of ZnO/CGA-NPs upon the addition of CGA. The inhibitory concentration 50 (IC50) of ZnO /CGA-NPs against RBD, angiotensin-converting enzyme 2 (ACE2) and PLpro were 1647.7, 323.3 µg/mL and 38.7 µg/mL, respectively. Also, it inhibited E-gene, RdRp gene, E-protein, and spike protein with an IC50 of 0.11, 0.13, 0.48, and 0.37 µg/mL, respectively. It acted as an antimicrobial against all tested organisms with a minimum inhibitory concentration (MIC) of 26 µg/mL. Finally, ZnO/CGA-NPs Complex (0.1 IC50) prevented the cytotoxic effect of HCQ on RBCs and WBC by 92.3 and 90 %, respectively.
Conclusion
ZnO/CGA-NPs Complex can be considered as a new anti- severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) compound.
Keywords
VeroE6 toxicity
Plaque assay
Papain-like proteinase
RNA dependent RNA polymerase
- ACE2
-
angiotensin-converting enzyme 2
- CGA
-
chlorogenic acids
- COVID 19
-
coronavirus disease 2019
- HCQ
-
hydroxychloroquine
- PLpro
-
papain-like proteinase
- RBD
-
spike protein-receptor binding domain
- SARS-CoV-2
-
severe acute respiratory syndrome coronavirus 2
- ZnO
-
Zinc oxide
Abbreviations
1 Introduction
Till 30 July 2022 and according to the world health organization (WHO) statics, the pandemic disorder COVID-19 (Zhu et al. 2020) infected 581,304,308 cases from that there were 6,418,377 deaths worldwide. The viral proteins divided into structural protein which are envelop (E), membrane (M), spike (S) and nucleocapsid (N) protein as well as numerous non-structural proteins (NSP). Each one has a central role during virus entry, replication, and transcription. Each protein can be considered an effective drug target where N-protein is a drug target to stop viral replication (Jack et al. 2020; Boopathi et al. 2021). Both Spike and furin are targets to prevent the viral entry (Wan et al. 2020; Walls et al. 2020). Several studies reported that about one quarter of COVID-19 patients are co-infected with different microbes such as Staphylococcus aureus, Streptococcus pneumoniae, and Candida albicans (Contou et al. 2020; Ghareeb et al. 2021a), therefore the use of an empiric antibiotic is encouraged.
Despite, a few drugs as anti-COVID-19 was approved as anti-COVID 19, the new challenge for scientists is to find out new compounds derived from natural resources to be used as COVID-19 therapeutic agents. Because natural product can kill microbe, prevent the formation of biofilm, work on multitargets (Sharma et al., 2021), beside that it safer and more efficient than several synthetic drugs (Ghareeb et al. 2021b).
The in vitro and in silico reporters proved that synthetic and natural antioxidants compounds can be used for COVID-19 treatment (Chauhan et al. 2021), such as berberine, curcumin, vitamin C, and caffeic acid (CA). Chlorogenic acids (CGAs), which is CA derivatives, have a wide spectrum of biological activities, including antidiabetic, hypolipidemic, antioxidants, and anti-inflammatory (Prasad et al. 2011). This phenolic acid, that is esters of quinic acid with cinnamic acids, is richly found in green coffee beans as well as coffee (Matei et al. 2012). Recently, Wang et al. (2022) proved that CGA could be a good candidate for COVID-19 treatment as their molecular docking results revealed that CGA binds to several targets such as ACE and interleukin (IL) 6.
It is well known that respiratory infections are associated with zinc deficiency. Zinc oxide (ZnO) nanoparticles (NPs) show antimicrobial activity, inhabiting the replication of different life throttles viral infections and respiratory viral pathogens such as SARS-CoV1 (Liu et al. 2020) due to the ROS production (Sharma et al., 2021). ZnO-NPs can fight viral infections through several mechanisms, including modulating the viral particle entry and fusion, preventing the viral replication, which consequently interferes with translation of viral protein infection (Ghareeb et al. 2021a, Ghareeb et al., 2021b; Hamdi et al. 2021).
Despite, HCQ shows several side effects when used as anti-COVID-19 on clinical trials, it shows in vitro anti-COVID-19 properties (Ghareeb et al. 2021a; Infante et al. 2021).
Based on these findings, this work was carried out to investigate the efficacy of ZnO-NPs, CGA and ZnO/CGA-NPs complex as anti-COVID-19, antimicrobial, and hydroxychloroquine (HCQ) toxicity eliminators.
2 Material and methods
2.1 Materials
CGA (>99 purity), Milli-Q water, 3-(4, 5-dimethylthiazol −2-yl)-2, 5-diphenyltetrazolium bromide (MTT), Ethanol and methanol (HPLC grade) were purchased from Sigma Aldrich (Germany). Zinc acetate dihydrates and Sodium hydroxide pellets (Fisher Chemical) were used.
2.2 ZnO nanoparticles and ZnO/CGA complex preparation and characterization
ZnO- NPs with 30 ± 5 nm hexagonal wurtzite structure was synthesized according to Aditya et al. (2018) and Ghareeb et al. (2021a). The facile mixing method was utilized to prepare the ZnO/CGA complex as reported by Belay et al. (2017). Firstly, the dispersed ZnO-NPs in Milli-Q water (225 µg/mL) by ultrasonication for 30 min was mixed with 2.6 mL of 100 µg/mL aqueous chlorogenic acid (CGA) solution and diluted under stirring to achieve a 1:0.5 ZnO:CGA mixture until the cloudy suspension of ZnO-NPs was instantly transformed to pale yellow. The solution was agitated at 25 °C for 2 h before using a freeze dryer to get the ZnO/CGA-dried yellow powder.
The ZnO-NPs and ZnO/CGA-complex spectra were performed at room temperature in the range of 200–500 nm using a UV–vis Spectrophotometer (Thermo Scientific™ Evolution™ 300). A Fourier transform infrared (FTIR, Shimadzu IRTracer-100 FT-IR) spectrophotometer was used to assess the interaction of ZnO and CGA confirmed using a wave number range of 4000–400 cm−1. Finally, the hydrodynamic size, morphology, and the crystalline nature of the ZnO-NPs and ZnO/CGA were measured using Zetasizer Nano ZS (Malvern, Worcestershire, UK), transmission electron microscopy (TEM) (JEOL, JEM 1400, Tokyo, Japan), and (Shimadzu XRD-6100 diffractometer) with a copper source (1.54 Å). Intensity (counts) values were detected in the 2θ range of 10-80°, with 30 mA (current) and 40 kV (tension).
2.3 In silico analysis
Chem Sketch program was used to draw the chemical structure of compounds in MOL format that converted to PDB file by Babel software. PDB of target proteins were obtained from www.rscb.org. Proteins’ co-ordinates energy minimization was done by SWISS-PDB VIEWER (SPDBV). Autodock.4 was used to analyze the ligand protein interaction (LPI) where the grid box size was (126 points in x, y and z dimensions), and spacing (0.375 Ã…) while the docking were used by lamarckian genetic algorithm (LGA) at 2,500,000 energy evaluations and the number of run (pose) was 10. The minimum binding energy was calculated by using Cygwin software. Finally, the LPI visualization was carried by the UCSF Chimera software.
2.4 Anti-COVID-19 assays
2.4.1 Determination of compounds inhibitory activity towards papain-like proteinase, spike protein
The inhibitory activities of ZnO-Nps, CGA, ZnO/CGA complex, and HCQ towards papain-like proteinase ((PLpro), Abcam, UK, cat# ab277615) and spike protein-receptor binding domain ((RBD), Abcam, UK, cat# ab273065) were determined according to Tai et al. (2020). While the compounds-ACE-2 binding efficiency was measured according to Ghareeb et al. (2021a).
2.4.2 In vitro VeroE6 toxicity
In a microwell-plate, tested compounds serial dilutions were added to 24 h cultured Vero E6 (30000 cells) then the plate was incubated for another 24 h in a calibrated CO2 incubator. The viable cells were visualized by MTT assay as described by Ghareeb et al. (2021a). The concentration that demonstrated 50 % cytotoxic effect (50 % cell death, IC50) was calculated from plot that contained % cytotoxicity (((Abs of cell without treatment-Abs of cells with treatment)/ Abs of cell without treatment) *100) on Y axis and sample concentration in X axis. The IC50 was used as the maximum concentration of the tested compound in plaque reduction assay.
2.4.3 Anti-COVID-19 activity (Plaque reduction assay)
The plaque reduction assay was carried out according to standardized published methods Xia et al. (2020), in which the plaques percentage inhibition was recorded as follows;
2.4.4 Antiviral activity
In cell culture plate (12wells), the seeded Vero E6 cell was infected with SARS-CoV-2 (provided from chemical warfare, Egyptian Army) at an 0.1 MOI and incubated for 2 h. After changing the media with fresh one containing serial dilutions of tested compounds or without (control), the plate was 3 days incubated at 37 °C in 5 % CO2. After centrifugation at 600 rpm for 10 min, the cells were scraped and added to 1 mL culture media. The virus was killed by exposing the supernatant and cells to routine UV disinfection protocol for 5 min, followed by cell lysis via sawing and freezing. The cellular RNA was isolated by using Qiagen viral RNA-isolation kit (#52906). cDNA was synthesized by using cDNA synthesis kit (Thermo Scientific, USA). Finally, the viral RdRp gene and E-gene amplification was carried out according to method of Xia et al. (2020). The gene expression was calculated by the -ΔΔCt method.
The spike protein and envelope protein levels were determined by ELISA technique (Xia et al., 2020).
2.5 In vitro bioscreening assays of the tested compounds
2.5.1 In vitro anti-hemolytic activity
The antihemolytic activities of ZnO-NPs, CGA, ZnO/CGA complex, and HCQ was determined as described by Ghareeb et al. (2021a). The effective concentration (EC50) that prevented a 50 % RBC hemolysis was measured in μg/mL by plotting % anti-hemolytic effect (((Abs in absences of treatment-Abs in presence of treatment)/ Abs in absences of treatment) *100) on Y axis and sample concentration in X axis.
2.5.2 In vitro peripheral blood monocytes (PBMCs) cytotoxicity
Rabbit PBMCs were isolated by adding Ficoll media. After centrifugation, the number of viable PBMCs was measured by trypan blue, and then resuspended at 1.0 × 105 cell/mL in RPMI culture medium. 100 µL of cells were cultured for 24 h in calibrated CO2 (5 % CO2, and 95 % humidity) then, serial dilutions of each compound (100 µL) were added to the cells, and the cells were re-incubated for an additional 24 h at the same conditions. MTT assay was used to determine the cell viability.
2.5.3 DPPH scavenging activity
The free radical scavenging activity of tested compounds was measured according to Brand-Williams et al. (1995).
2.5.4 Antimicrobial activity
Agar well diffusion and turbidity assays (Kadaikunnan et al. 2015) were used to detect the activity of the compounds against five microbial species Klebsila pneumonia (ATCC700603), E. coli (ATCC25922), Staphylococcus aureus (ATCC25923), Streptococcus pyogenes (EMCC1772), and Candida albicans (EMCC105). The minimum inhibitory concentration (MIC) was calculated for each compound tested.
2.6 Determination of in vitro HCQ toxicity elimination
For red blood cells, 100 µL of RBCs suspension (1 %) was mixed and incubated at 37 °C for 30 min with the concentration of tested compound (450 µL) that demonstrated the lowest hemolytic effect. Then, 450 μL of 27.5 μg/mL HCQ was added then this mixture was re-incubated for another 30 min. the absorptance of the supernatant was measured at 540 nm. Finally, the percentage of hemolysis was calculated.
For white blood cells (WBCs), the WBCs were isolated from 10 mL of blood, and then the palette was suspended in 10-fold resuspended in 1 mL culture RPMI (Dagur & McCoy 2015). The viable cells were then diluted to a concentration of 1 × 105 cell/mL in culture RPMI 1640 medium for 24 h in a CO2 incubator at standardized conditions. Following that, 100 µL of different concentrations of tested compounds were added alone or in the presence of HCQ concentration (that yield 50 % cell death) for 24 h in a CO2 incubator under standardized conditions. The cell proliferation was measured by MTT assay.
2.7 Statistical analyses
Data were expressed as means and standard deviations. The significant difference among groups was set at p < 0.05 using ANOVA, post hoc LSD test, and multiple comparison test in SPSS software package version 20.0 (Armonk, NY: IBM Corp).
3 Results
In the case of ZnO-NPs, only one absorption band at 372 nm was detected while four characteristic peaks for CGA at 217, 233, 299, and 324 nm (Fig. 1A) was found. The mixing of CGA with ZnO-Nps (Fig. 1B and 1C) showed hypochromic effects in the absorbance pattern which was associated with the appearance of the characteristic absorption peak of metal-phenolate at 372 nm. The addition of excess CGA to constant ZnO-Nps which calculated by
/
ratio (Fig. 1D) indicated that the addition of CGA after stoichiometry of ZnO/CGA = 2 was ineffective as the intensity of the CGA characteristic peak 324 nm was progressively increased while the metal-phenolate peak at 372 nm of ZnO/CGA remained constant.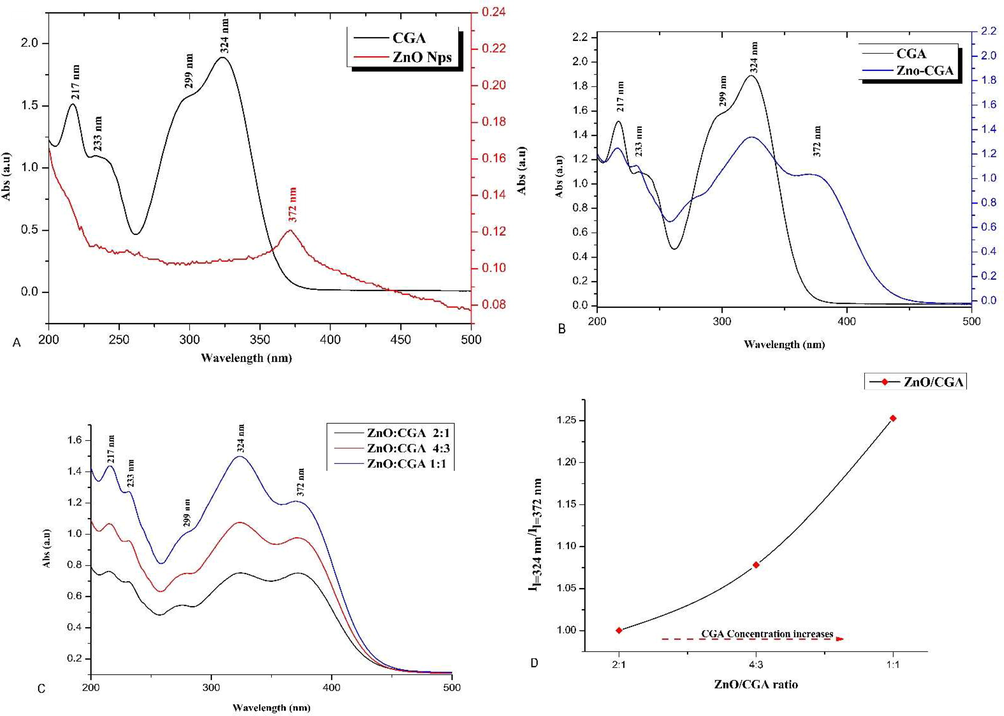
The UV–vis absorption spectrum of (A) CGA vs ZnO, (B) CGA vs ZnO/CGA, (C) ZnO vs CGA and ZnO/CGA, and (D) effect of excess CGA to constant ZnO-Nps.
The IR spectra (Fig. 2A) showed that in the ZnO/CGA spectrum, the characteristic Zn-O absorption bands of ZnO-Nps were observed at 428 cm−1, while the Zn-OH absorption band at 499 cm−1 was disappeared, moreover, almost all CGA characteristic absorption bands were presented. TEM images showed that the uniformly-highly dispersed particles of ZnO-NPs (30 ± 5 nm) (Fig. 2B) were converted into agglomerated clusters in ZnO/CGA-Nps (Fig. 2C) due to the presence of CGA on the surface of ZnO-NPs.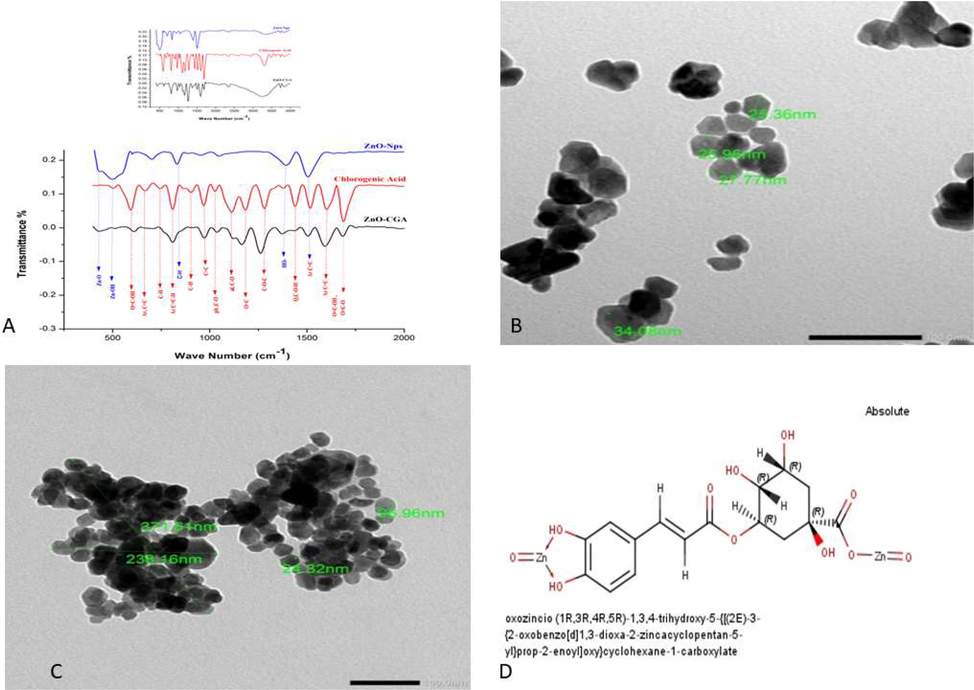
(A) The Fourier transform infrared (FTIR) spectrum of ZnO-Nps, CGA, and ZnO/CGA complex and (B and C, the magnification power was 100 nm) TEM and analysis of ZnO-NPs and ZnO/CGA and (D) The proposed structure of ZnO/CGA complex.
The in silico study presented in Fig. 3 and Table 1 shows that ZnO/CGA had different ligand protein interaction (LPI) than both parents, where it binds with PLpro by two hydrogen bonds with Gln 270 and Tyr 274 and several hydrophobic interactions. This LPI showed the lowest binding energy (−17.8 kj) and estimated Ki (89.1 fM). Despite both CGA and ZnO-NPs bind with spike protein with hydrogen bonds, the ZnO/CGA complex binds with the protein through hydrophobic interactions only and it shows the lowest binding energy (−15.55 kj) and estimated Ki (4.01 nM). Finally, ZnO/CGA complex binds with RBD by five hydrogen bonds at LEU 91, LYS 94, ASP 206, ASN 210, and GLY 395 and it shows the lowest binding energy (-10.99 kj) and estimated Ki (8.82 nM). Table 2 shows that all tested compounds inhibited PLpro in a concentration-dependent manner where the highest inhibitory effect was shown in the case of ZnO/CGA complex followed by ZnO-NPs, then HCQ, and finally CGA where the relative IC50s were 38.67, 48.5, 270.3, and 3571.4 µg/mL, respectively. All tested compounds had ACE2 binding affinity and increased the formation of the RBD-ACE2 complex. The results are mean ± SD of three replicas., means in the same column with different superscripts letters are significantly different at p < 0.05, where a is the lowest means and d is the highest one.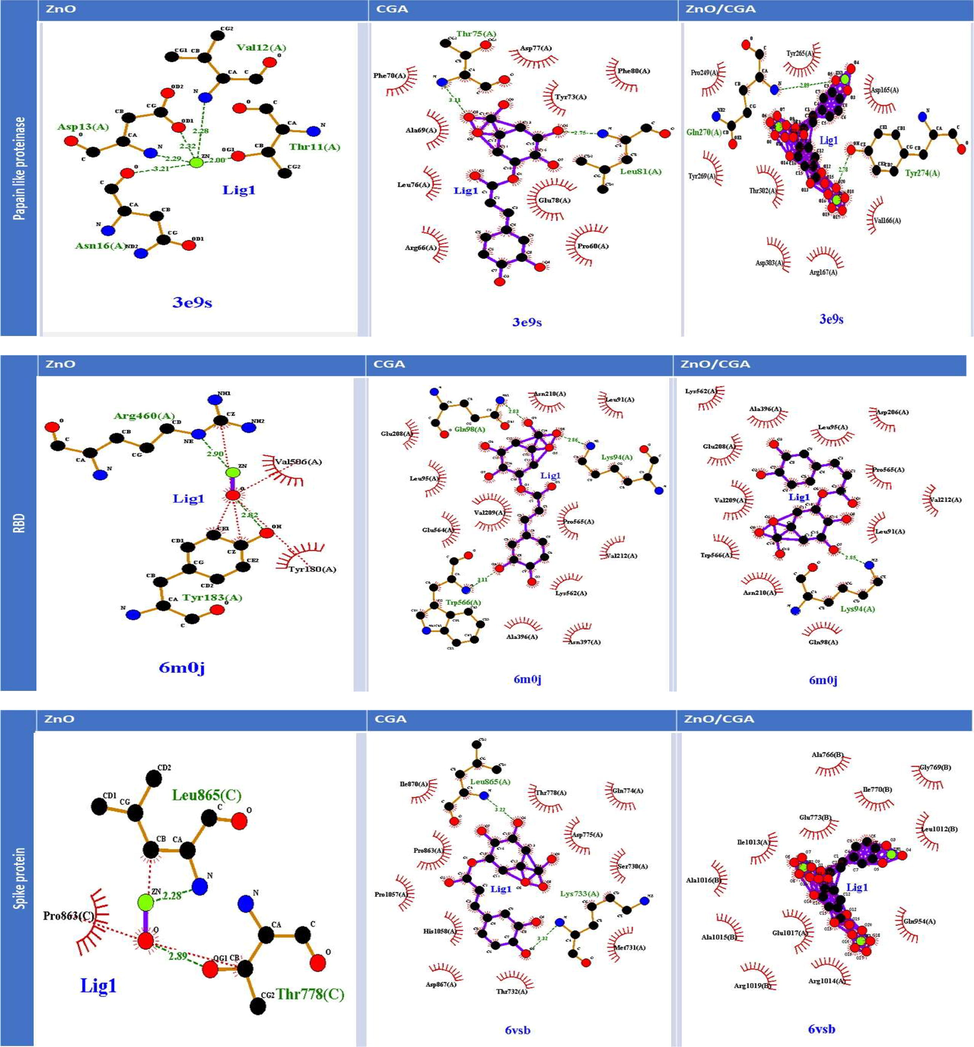
Molecular docking and LigPlus analysis illustrating the interaction between ZnO-NPs, CGA, and ZnO/CGA and the studied proteins (papain-like proteinase, spike protein, and spike protein receptor-binding domain).
Parameters
CGA
ZnO
ZnO/ CGA
Reference inhibitor
pH
6.75
PAPAIN-LIKE PROTEINASE
Name of Reference inhibitor
5-Amino-2-Methyl-N-[(1r)-1-Naphthalen-1-Ylethyl]benzamide
Hydrogen bond
LIG 1O-THR 75.A OG1
LIG 1O-LEU 76.A O
LIG 1O-ASP 77.A O
LIG 1O-GLU 78.A O
LEU 81.A N-LIG 1ONo hydrogen bond
LIG 1O- Gln 270. A N
LIG 1O- Tyr 274. A OH
Hydrophobic interaction
LIG 1 N /A GLU 168 OE1 no hydrogen 2.647 N/A
LIG 1 N /A GLU 180 OE2 no hydrogen 3.064 N/A
Binding energy
−10.58
−2.77
−17.8
−3.98
Estimated Ki
17.49 nM
9.28 mM
89.10 fM
1.21 mM
Reference RMSD
36.54
32.56
44.24
62.53
SPIKE PROTEIN
Name of Reference inhibitor
Geraniin
Hydrogen bond
LIG 1O-MET 731.A O
LIG 1O-ASP 775.A OD1
LIG 1O-LEU 865.A O
LYS 733.A N-LIG 1OLIG1O-ASP775.C OD1
LIG1O-THR778.C-OG1No hydrogen bond
Hydrophobic interactionLIG O/Gln 314 N
Binding energy
−9.77
−2.74
−15.55
−16.68
Estimated Ki
69.39 nM
9880 uM
4.01 pM
595.11 fM
Reference RMSD
385.4
385.03
378.33
263.02
SPIKE RECEPTOR-BINDING DOMAIN
Name of Reference inhibitor
Amentoflavone
Hydrogen bond
LIG 1O-GLU 208.A O
LYS 94.A NZ-LIG 1O
GLN 98.A NE2-LIG 1O
TRP 566.A N- LIG 1OLIGO-TYR183.A OH
LIGO-VAL506.A OLIG 1O-LEU 91.A O
LIG 1O-ASP 206.A O
LIG 1O-GLY 395.A O
LYS 94.A NZ-LIG 1O
ASN 210.A ND2-LIG 1OASN 210 N---LIG O
Binding energy
−10.34
−2.31
−10.99
−12.17
Estimated Ki
26.30 nM
20,180 uM
8.82 nM
1.19 nM
Reference RMSD
50.17
66.63
55.6
60.26
Groups
Concentration µg/mL
PLpro inhibition %
SP- RBD-ACE2 complex formation fold
ACE-2 Binding %
ZnO
104
71.06 ± 1.3e
20.25 ± 0.5e
16.4 ± 0.9d
52
62.2 ± 0.90d
13.375 ± 0.3d
15.6 ± 0.7d
26
45.6 ± 1.1c
6.625 ± 0.2c
13.1 ± 1.3c
13
35.31 ± 0.77b
3.5 ± 0.3b
10.7 ± 0.8b
6.5
22.5 ± 0.12a
0.5 ± 0.02a
8.2 ± 0.08a
IC50 (µg/mL)
48.5
256.8
317.1
CGA
1000
14.5 ± 0.9c
14.25 ± 0.5d
18.3 ± 1.2b
500
13.6 ± 1.1c
13.875 ± 0.3d
18.1 ± 2.1b
250
11.2 ± 0.7b
4.375 ± 0.1c
17.2 ± 1.1ab
125
10.6 ± 1.01b
1 ± 0.01b
16.4 ± 1.1a
62.5
6.9 ± 0.4a
0.75 ± 0.002a
14.8 ± 1.2a
IC50 (µg/mL)
3571.4
3508.8
2732.2
ZnO/CGA
156.5
80.3 ± 1.9d
4.75 ± 0.2e
24.2 ± 0.1e
78.25
65.1 ± 2.8c
1.625 ± 0.07d
17.1 ± 0.2d
39.215
50.7 ± 3.4b
0.625 ± 0.003c
15.6 ± 0.1c
19.563
45.6 ± 2.1b
0.375 ± 0.007b
11.5 ± 0.07b
9.782
38.6 ± 1.2a
0.01 ± 0.001a
9.2 ± 0.05a
IC50 (µg/mL)
38.67
1647.4
323.3
HCQ
200
37 ± 0.42e
0.5 ± 0.001d
15.6 ± 1.4b
100
30 ± 0.23d
0.5 ± 0.001d
14.8 ± 0.9b
50
22.5 ± 0.15c
0.375 ± 0.003c
14.5 ± 0.8b
25
11.2 ± 0.39b
0.25 ± 0.001b
13.8 ± 1.1b
12.5
8.1 ± 0.74a
0.125 ± 0.007a
9.02 ± 0.5a
IC50 (µg/mL)
270.3
20,000
641.03
In accordance with the previous data, Table 3 shows that all tested compounds acted as anti-SARS-CoV2 where the highest antiviral effect was shown in the case of ZnO/CGA complex followed by CGA and HCQ and finally ZnO-Nps. The obtained data confirmed that these compounds could be considered potent antiviral candidates because the obtained EC50s (effective antiviral concentration 50) for all tested compounds were lower than their IC50s (Vero E6 inhibitory concentration 50). The results are mean ± SD of three replicas., means in the same column with different superscripts letters are significantly different at p < 0.05, where a is the lowest means and d is the highest one.
Vero E6 IC50 (µg/mL)
Reduction %
Dosage (µg/mL)
Anti-SARS CoV2 EC50 (µg/mL)
EC50/IC50 %
CGA
23.6 ± 1.2d
76 ± 1.9b
15.4 ± 0.21c
4.9 ± 0.021d
21 ± 0.9b
ZnO
12.6 ± 0.42c
67.5 ± 1.2a
5.5 ± 0.41b
4.11 ± 0.012c
33 ± 1.2d
ZnO/CGA
1.65 ± 0.003a
80 ± 3.4c
1.56 ± 0.0002a
0.312 ± 0.002a
19 ± 0.78a
HCQ
1.4 ± 0.1b
76 ± 2.5b
1.1 ± 0.13a
0.385 ± 0.01b
28 ± 1.7c
Interestingly, our data which is represented in Table 4 confirmed that ZnO-NPs, CGA, and ZnO/CGA complex inhibited the ORF, E- and RdRp-gene expression in a concentration-dependent manner where the ZnO/CGA complex showed the maximum gene down-regulation patterns. In accordance with gene expression data, the ZnO/CGA complex showed the maximum inhibition E-protein and spike protein pattern. The results are mean ± SD of three replicas., means in the same column with different superscripts letters are significantly different at p < 0.05, where a is the lowest means and e is the highest one.
Groups
Concentration (µg/mL)
ORF gene expression
ORF gene downregulation %
E-gene expression
E-gene downregulation %
RdRp expression
RdRp down regulation %
E protein inhibition %
Spike protein inhibition %
ZnO
5.48
0.15 ± 0.001a
85.29 ± 1.1c
0.73E-05 ± 0.1 E-06a
99.99 ± 3.6b
0.001 ± 0.0001a
99.9 ± 3.8a
85.71 ± 7.1c
83.33 ± 3.1d
2.74
0.18 ± 0.002b
81.96 ± 1.6c
3.36 E-05 ± 0.1 E-06b
99.99 ± 1.2b
0.018 ± 0.005b
98.2 ± 4.1a
78.57 ± 3.6b
88.89 ± 2.1c
1.37
0.37 ± 0.001c
62.63 ± 1.4b
5.36 E-05 ± 0.1 E-06c
99.99 ± 3.1b
0.019 ± 0.0002b
98.1 ± 3.1a
71.43 ± 1.8b
76.67 ± 3.6b
0.685
0.47 ± 0.01d
53.35 ± 0.9a
7.36E-05 ± 0.1 E-06d
89.78 ± 2.1a
0.028 ± 0.0001c
97.2 ± 2.3a
64.29 ± 2.4a
67.78 ± 1.8a
IC50
µg/mL
0.641
0.38
0.24
0.365
0.51
CGA
10
0.04 ± 0.0001a
95.99 ± 2.8d
1 E-04 ± 0.1 E-04a
99.99 ± 1.8a
0.001 ± 0.1 E-05b
99.89 ± 3.9a
92.5 ± 7.9d
91.66 ± 3.2d
5
0.096 ± 0.0001b
90.33 ± 3.1c
2 E-04 ± 0.1 E-04b
99.97 ± 1.6a
0.0011 ± 1.1 E-05b
99.89 ± 5.2a
87.5 ± 6.4c
83.33 ± 1.3c
2.5
0.11 ± 0.002c
88.88 ± 2.1b
2 E-04 ± 0.1 E-04b
99.97 ± 1.7a
0.0029 ± 1.1 E-06c
99.715 ± 1.2a
75 ± 4.8b
50 ± 0.9b
1.25
0.13 ± 0.001d
87.7 ± 1.1a
2 E-03 ± 0.1 E-04d
99.8 ± 0.9a
0.0033 ± 0.1 E-05d
99.67 ± 1.4a
50 ± 3.6a
25 ± 0.8a
IC 50
µg/mL
0.71
0.63
0.63
1.25
2.5
ZnO/CGA
1.565
2.5E-05 ± 0.00001a
99.99 ± 1.9d
0.003 ± 0.0001a
99.7 ± 1.4d
1.52E-05 ± 0.1E-06a
99.99 ± 1.8b
96 ± 7.8c
93.3 ± 3.6d
0.783
0.015 ± 0.0001b
98.5 ± 2.8c
0.078 ± 0.0001b
92.2 ± 0.1c
0.001 ± 2.3 E-06b
99.89 ± 7.8b
80 ± 6.3b
80 ± 2.7c
0.391
0.88 ± 0.0001c
11.7 ± 0.03b
0.1 ± 0.001c
90 ± 0.2b
0.001 ± 1.1 E-06b
99.89 ± 3.7b
40 ± 1.5a
53.3 ± 3.1b
0.195
0.93 ± 0.001d
6.7 ± 0.01a
0.14 ± 0.001d
86.03 ± 1.8a
0.234 ± 0.001b
76.67 ± 6.1a
40 ± 2.1a
40 ± 0.9a
IC 50
µg/mL
0.53
0.11
0.13
0.48
0.37
Table 5 proves that CGA was the safest compound toward RBC, PBMCs and WBCs where it showed the highest IC50 followed by ZnO/CGA complex, ZnO-NPs and finally HCQ. In addition, the ZnO/CGA complex presented the highest antioxidants capacity followed by CGA, HCQ, and finally ZnO-NPs. The results are mean ± SD of three replicas., means in the same column with different superscripts letters are significantly different at p < 0.05, where a is the lowest means and d is the highest one.
RBC
PBMC
WBC
Antioxidants
IC50 (µg/mL)
CGA
370.83 ± 12.3c
235 ± 17.8c
655 ± 12.5d
250 ± 10.8b
ZnO
41.67 ± 2.31b
64 ± 0.5b
25 ± 1.1b
439 ± 18.9d
ZnO/CGA
372.15 + 9.4c
230 + 2.01c
395 ± 3.5c
78 ± 1.3a
HCQ
24.55 ± 0.8a
53.2 ± 0.05a
20 ± 1.23a
352 ± 17.8c
Table 6 shows that all tested compounds acting as anti- Klebsila pneumonia, Staphylococcus aureus, Streptococcus pyogenes, and Candida albicans where the highest antimicrobial effect was shown in the case of ZnO/CGA complex. *Diameter includes 5 mm well diameter. - ND; Not detected. - MIC; Minimum inhibition concentration.
Agar well diffusion assay
Turbidity assay
Concentrations of the tested compounds
Inhibition zone diameter (mm)**
MIC (µg/mL)
Percent of inhibition (%)
MIC (µg/mL)
100 %
50 %
25 %
100 %
50 %
25 %
Gram negative Bacteria
klebsila pneumonia ATCC700603
ZnO-NPs
11 ± 0.0
ND
ND
104
65.87 ± 0.0
61.84 ± 0.0
44.13 ± 0.0
26
CGA
ND
ND
ND
ND
87.38 ± 0.0
71.32 ± 0.0
58.21 ± 0.0
13
ZnO/CGA
35 ± 0.0
ND
ND
104
88.23 ± 0.0
77.66 ± 0.0
65.11 ± 0.0
26
Positive control (Amoxicillin 0.1 %)
35 ± 0.0
92.48 ± 0.0
Gram positive Bacteria
Staphylococcus aureus ATCC25923
ZnO-NPs
17 ± 0.0
ND
ND
104
87.77 ± 0.0
33.24 ± 0.0
8.09 ± 0.0
26
CGA
ND
ND
ND
ND
61.34 ± 0.0
59.81 ± 0.0
40.37 ± 0.0
13
ZnO/CGA
ND
ND
ND
ND
74.39 ± 0.0
61.88 ± 0.0
50.95 ± 0.0
26
Positive control (Amoxicillin 0.1 %)
25 ± 0.0
95.51 ± 0.0
Streptococcus pyogenes EMCC1772
ZnO-NPs
12 ± 0.0
ND
ND
104
68.31 ± 0.0
50.85 ± 0.0
41.49 ± 0.0
26
CGA
ND
ND
ND
ND
68.85 ± 0.0
60.12 ± 0.0
41.53 ± 0.0
13
ZnO/CGA
32 ± 0.0
20 ± 0.0
ND
52
69.95 ± 0.0
63.93 ± 0.0
59.65 ± 0.0
26
Positive control (Amoxicillin 0.1 %)
35 ± 0.0
80.98 ± 0.0
Yeast
Candida albicans EMCC105
ZnO-NPs
30 ± 0.0
25 ± 0.0
20 ± 0.0
26
25.00 ± 0.0
23.44 ± 0.0
20.31 ± 0.0
26
CGA
ND
ND
ND
ND
34.26 ± 0.0
33.04 ± 0.0
17.52 ± 0.0
13
ZnO/CGA
ND
ND
ND
ND
83.15 ± 0.0
65.08 ± 0.0
63.30 ± 0.0
26
Positive control (Fluconazole 0.1 %)
40 ± 0.0
94.14 ± 0.0
Table 7 showed that ZnO-NPs prevented the HCQ hemolytic effect by 82.9 % and increased the WBC viability by 5.6fold while CGA decreased hemolysis by 93 % and increased WBC viability by 9.2fold and finally the ZnO/CGA complex decreased RBCs hemolysis by 92.3 % and increased the WBC viability by 9.7fold. The results are mean ± SD of three replicas., means in the same column with different superscripts letters are significantly different at p < 0.05, where a is the lowest means and g is the highest one. Note, the concentration of ZnO-NPs, CGA and ZnO/CGA were 1/10 of IC50s while HCQ concentration was IC50.
Compounds
RBC hemolysis
WBC cytotoxicity
Concentration (µg/mL)
Hemolytic (%)
Concentration (µg/mL)
Viability (%)
ZnO
4.1
5.88 ± 0.002e
2.5
53 ± 1.3c
CGA
37
3.52 ± 0.01a
65.5
57 ± 1.3c
ZnO/CGA
37
4.08 ± 0.03c
19.5
57 ± 2.8c
+ve Control (HCQ)
24.5
56 ± 1.3 g
20
6 ± 0.04a
HCQ + ZnO
24.5 + 4.1
9.6 ± 0.002f
200 + 2.5
34 ± 2.3b
HCQ + CGA
24.5 + 37
3.76 ± 0.05b
200 + 62.5
55 ± 3.7c
HCQ + ZnO/CGA
24.5 + 37
4.32 ± 0.07d
200 + 19.5
58 ± 2.9c
4 Discussion
This study was aimed to synthesize a ZnO/CGA complex then investigate its efficacy as anti-COVID-19, antimicrobial, and eliminator of hydroxychloroquine (HCQ) toxicity in comparing to its parents’ efficiencies.
The characterization data prove the synthesis of a new ZnO/CGA complex that can block the viral entry, replication, and assembly. Besides that, it is considered a potent antioxidant and antimicrobial and it could be used in combination with HCQ as adjuvant therapy. ZnO-NPs had one while CGA UV-spectrum showed four characteristic peaks producing from the Pi transitions that occurred in double bond and the aromatic ring frame (Kalinowska et al. 2020). Moreover, the addition of CGA to ZnO-NPs produced hypochromic effect which indicated the adsorption of CGA onto the ZnO-Nps surface by forming interaction between phenolic catecholate and carboxylate group of CGA with Zn (Kalinowska et al. 2020). The IR absorption spectra indicated the adsorption of CGA on the ZnO-Nps surface. The characteristic CGA absorption bands (667, 809, 1516, and 1600 cm) were attributed to the aromatic ring (Kalinowska et al. 2020). Several alterations had been noticed in ZnO/CGA complex where the redshift intensity was reduced to lower wave numbers (1506 and 1592 cm−1) and the vibration mode of the quinic ring C-OH peak (1438 cm−1) was lost. This could be due to the enrollment quinic ring C-OH in the formation of hydrogen bonds with ZnO surface –OH which also indicated by the band boarding at 3304 cm−1 which is specific for the OH group. There were a forceful reduction with the redshift in both the C⚌O functional group stretching of ester bond band (1687 cm−1) and the C⚌O carboxyl functional groups stretching band (1183 cm−1) to 1682 and 1165 cm−1 which may be suggested that the carboxylate anion may be involved in coordination with zinc (Kalinowska et al. 2020). The the C—O—C stretching vibration in ester bond band (1282 cm−1) has appeared with slightly higher intensity and redshift to 1261 cm−1, the bands of the C—O phenolic and alcoholic stretching (1113 and 1027 cm−1, respectively) were forcibly reduced with the blueshift to 1120 and 1037 cm−1, respectively, indicating that CGA interacted with the surface of ZnO-Nps and ZnO/CGA ground state complex was formed. Altogether, these spectra indicate that CGA interacted with ZnO-NPs by noncovalent bonds. In accordance, the proposed structure of the ZnO/CGA complex was sketched and checked by Chemaxon Marvin sketch software (Fig. 2D).
It is reported that CGA prevents the early stages of the viral infection during virus growth, moreover, it acts as anti-Herpes Simplex Virus I and HCoV-NL63 (Yu et al. 2020; Weng et al. 2019). The obtained results confirmed that the parents and the newly formed complex ZnO/CGA are acting as anti-SARS-CoV2 activity as they increased the cellular pH which leads to endosomal pathway (Xia et al. 2020). The parents and their complex uncompetitively inhibited the binding between RBD and ACE2 through the formation of hydrogen bonds and hydrophobic interaction with RBD that alters the ACE2 active site-specificity to the substrate (Chandel et al. 2020). In this type of inhibition ZnO/CGA binds with enzyme allosteric site and with active site leading to the formation of inactive enzyme-substrate-inhibitor complex. In agreement with these results, the docking study of Yu et al. (2020) proved that CGA interacts with ACE2 at position Gln42 andAsp38 which prevented the ACE2 attachment with S protein. Also, Adem et al. (2021) in silico results indicated that CGA interacts with main-proteinase, S proteins subunits and endoribonuclease. Furthermore, ZnO/CGA complex downregulated and inhibited several viral proteins PLpro, RdRp, E and spike proteins which lead to viral replication inhibition. Moreover, CGA binds to cellular heat shock protein A5 which is receptor for COVID-19 (Elfiky, 2021).
Our results indicate that the ZnO/CGA complex binds PLpro active site Gln270 and Tyr274 (Báez-Santos et al. 2015) also it binds to the ubiquitin-like domine 2 (UL2) (amino acids 75–88) N-terminal of PLpro which is linked to the catalytic site. UL2 blocks the formation of the interferon pathway (Shin et al. 2020) therefore our tested compounds could be reduced the viral replication through stimulation of the antiviral interferon pathway.
The toxicity of a new compound must be detected in vitro on different normal cells such as WBC, RBC, and fibroblast (Deore et al. 2019). It is known that compounds that acting as antioxidants could be used as adjuvant therapy during COVID-19 infection to decrease the oxidative stress and cytokines storm (Soto et al. 2020). ZnO/CGA complex had the highest antioxidants property. Despite, ZnO-NPs increased cellular oxidative stress, CGA acts as a potent antioxidant, anticancer (Moon et al. 2017and anti-inflammatory (Al-Hatamleh et al. 2020). We must notice that the concentration of ZnO-NPs in this complex is lower than the recommended doses about 15folds, which indicated the elimination of cytotoxic effect of ZnO-NPs (Sharma et al., 2021).
Some COVID-19 infected people had bacterial infection, and this is the reason for the administration of antibiotics for these patients (Contou et al. 2020). All tested compounds had inhibitory ability toward Klebsila pneumonia, Staphylococcus aureus, Streptococcus pyogenes, and Candida albicans. CGA inhibits bacterial arginase and agmatinase and viral neuraminidase (Moon et al. 2017). CGA is also acting as anti- Streptococcus pneumoniae by inhibiting neuraminidase (Guan et al. 2020).
Despite, the presence of HCQ in COVID-19 therapeutic protocol is controversial (Risch 2020) due to associated side effects (Barnabas et al. 2021), it has anti-inflammatory and anti-SARSCoV2 properties. Therefore, we measured the protective effect of tested compounds on RBC hemolysis and WBC destruction by HCQ, and the results proved that ZnO /CGA protects the cells that could be due to the antioxidant and anti-inflammatory properties of CGA (Moon et al. 2017).
5 Conclusion
ZnO/CGA-NPs Complex was successfully prepared by facile mixing method. Where, CGA molecules are interacted by non-covalent interaction on ZnO-Nps surface that formed the ground-state complex. Nano-ZnO /CGA complex exhibited anti-COVID-19 properties as it inhibited several steps in virus lifecycle where it blocked RBD and bind with ACE2 (viral entry targets) Also, it blocked virus replication by inhibiting PLpro and RdRp. This comples is a very potent antimicrobial agent against respiratory tract pathogens. Finally, ZnO/CGA-NPs Complex could be used to eliminate the HCQ associated toxicity.
Acknowledgments
The Deanship of Scientific Research, College of Applied medical sciences, University of Bisha, Kingdom of Saudi Arabia has funded this work through the COVID-19 initiative project under the grant (UB-COVID-14-1441).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Caffeic acid derivatives (CAFDs) as inhibitors of SARS-CoV-2: CAFDs-based functional foods as a potential alternative approach to combat COVID-19. Phytomedicine.. 2021;85:153310-153326.
- [Google Scholar]
- ZnO nanoparticles modified with an amphipathic peptide show improved photoprotection in skin. ACS Appl. Mater. Interfaces.. 2018;11(1):56-72.
- [Google Scholar]
- Antiviral and immunomodulatory effects of phytochemicals from honey against COVID-19: Potential mechanisms of action and future directions. Molecules.. 2020;25:5017-5039.
- [Google Scholar]
- The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res.. 2015;115:21-38.
- [Google Scholar]
- Hydroxychloroquine as postexposure prophylaxis to prevent severe acute respiratory syndrome coronavirus 2 infection: a randomized trial. Ann. Intern. Med.. 2021;174(3):344-352.
- [Google Scholar]
- Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn.. 2021;39:3409-3418.
- [Google Scholar]
- Use of a free radical method to evaluate antioxidant activity. LWT.. 1995;28(1):25-30.
- [Google Scholar]
- Structure-based drug repurposing for targeting Nsp9 replicase and spike proteins of severe acute respiratory syndrome coronavirus 2. J. Biomol. Struct. Dyn.. 2020;24:1-4.
- [Google Scholar]
- Natural products as environmentally safe and green approach to combat Covid-19. Curr. Res. Green Sustain. Chem.. 2021;4:100114.
- [Google Scholar]
- Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care.. 2020;10:1-9.
- [Google Scholar]
- Collection, storage, and preparation of human blood cells. Curr. Protoc. Cytom.. 2015;73:5-11.
- [Google Scholar]
- The stages of drug discovery and development process. Asian J. Pharm. Res. Dev.. 2019;7:62-67.
- [Google Scholar]
- Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn.. 2021;39:3194-3203.
- [Google Scholar]
- Nanoparticles of ZnO/Berberine complex contract COVID-19 and respiratory co-bacterial infection in addition to elimination of hydroxychloroquine toxicity. J. Pharm. Investig.. 2021;51(6):735-757.
- [Google Scholar]
- Potential therapeutic and pharmacological strategies for SARS-CoV2. J. Pharm. Investig.. 2021;51(3):281-296.
- [Google Scholar]
- Exploration of binding mechanism of a potential streptococcus pneumoniae neuraminidase inhibitor from herbaceous plants by molecular simulation. Int. J. Mol. Sci.. 2020;21:1003-1015.
- [Google Scholar]
- Investigating the internalization and COVID-19 antiviral computational analysis of optimized nanoscale zinc oxide. ACS Omega.. 2021;6(10):6848-6860.
- [Google Scholar]
- Hydroxychloroquine in the COVID-19 pandemic era: in pursuit of a rational use for prophylaxis of SARS-CoV-2 infection. Expert Rev. Anti Infect. Ther.. 2021;19(1):5-16.
- [Google Scholar]
- Jack A, Ferro L, Trnka M, Wehri E, Nadgir A, Nguyenla X, et al. 2020. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. BioRxiv. doi:10.1101/2020.09, 14, v3.
- In-vitro antibacterial, antifungal, antioxidant and functional properties of Bacillus amyloliquefaciens. Ann. Clin. Microbiol. Antimicrob.. 2015;14(1):9-19.
- [Google Scholar]
- Zn (II) complex of plant phenolic chlorogenic acid: Antioxidant, Antimicrobial and structural studies. Materials.. 2020;13:3745-3762.
- [Google Scholar]
- Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol.. 2020;92(5):491-494.
- [Google Scholar]
- Investigating the chemical changes of chlorogenic acids during coffee brewing: Conjugate addition of water to the olefinic moiety of chlorogenic acids and their quinides. J. Agric. Food Chem.. 2012;60(49):12105-12115.
- [Google Scholar]
- Anti-cancer therapy: Chlorogenic acid, gallic acid and ellagic acid in synergism. IOSR J. Pharm. Biol. Sci.. 2017;12:48-52.
- [Google Scholar]
- Inhibitory effect of caffeic acid on cancer cell proliferation by oxidative mechanism in human HT-1080 fibrosarcoma cell line. Mol. Cel. Biochem.. 2011;349(1):11-19.
- [Google Scholar]
- Risch responds to “how to consider low reported death rates in COVID-19”. Am. J. Epidemiol.. 2020;189:1230-1231.
- [Google Scholar]
- Insights into the synthesis and mechanism of green synthesized antimicrobial nanoparticles, answer to the multidrug resistance. Mater. Today Chem.. 2021;19
- [Google Scholar]
- Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature.. 2020;587(7835):657-662.
- [Google Scholar]
- Is antioxidant therapy a useful complementary measure for Covid-19 treatment? An algorithm for its application. Medicina.. 2020;56:386-414.
- [Google Scholar]
- Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol.. 2020;17(6):613-620.
- [Google Scholar]
- Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell.. 2020;181(2):281-292.
- [Google Scholar]
- Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol.. 2020;94(7)
- [Google Scholar]
- Chlorogenic acid, a natural product as potential inhibitor of COVID-19: virtual screening experiment based on network pharmacology and molecular docking. Nat. Prod. Res.. 2022;36(10):2580-2584.
- [Google Scholar]
- Antiviral activity of Sambucus Formosana Nakai ethanol extract and related phenolic acid constituents against human coronavirus NL63. Virus Res.. 2019;273:197767-197774.
- [Google Scholar]
- Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res.. 2020;30(4):343-355.
- [Google Scholar]
- Exploring the active compounds of traditional Mongolian medicine in intervention of novel coronavirus (COVID-19) based on molecular docking method. J. Funct. Foods.. 2020;71:104016-104025.
- [Google Scholar]
- A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med.. 2020;382(8):727-733.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102296.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







