Translate this page into:
Yanang water extract exhibits a protective effect against methomyl-induced cytotoxicity in RAW 264.7 cells via suppression of apoptosis and cell cycle arrest
⁎Corresponding author at: Department of Biochemistry, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand. nisaja@kku.ac.th (Nisachon Jangpromma)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract

Abstract
Methomyl, an extremely hazardous substance, is widely used for controlling insects and pests in agricultural production. This compound can cause several illnesses in humans. In this research, we determined the alleviation potential of Yanang water extract (YWE) against methomyl-induced cytotoxicity in RAW 264.7 macrophage cells and examined the underlying mechanisms of action. The YWE (2.5 – 10 μg/mL) exhibited no toxicity against RAW 264.7 cells, whereas methomyl significantly reduced RAW 264.7 cell growth, resulting in the progression of apoptosis and cell cycle arrest. Supplementation of YWE on methomyl treated RAW 264.7 cells revealed a pronounced protective effect via the suppression of caspase-9 and caspase-3 mRNA expression levels. Proteomics studies were used to assess the effect of methomyl on RAW 264.7 cells, revealing an increase of proteins associated with apoptosis and cell cycle arrest, whereas in the YWE co-treatment condition these proteins were suppressed. Moreover, an increase in proteins involved with cell cycle progression and anti-apoptosis was found in YWE co-treatment. Our findings suggest that YWE is potentially involved in mitigating methomyl-induced cytotoxicity, via the suppression of apoptosis and cell cycle arrest.
Keywords
Apoptosis
Bioactive compound
Cell cycle
Cytotoxicity
Insecticide
Tiliacora triandra
1 Introduction
Pesticides such as herbicides, insecticides, fungicides and rodenticides have been widely used in recent years to enhance agricultural yields in many countries. Although contributing to agricultural output improvement, pesticide contamination has negative impacts on human health and causes environmental imbalances (Tudi et al., 2021). Among the pesticides, insecticides are currently ranked as the top three pesticides used in Thailand, with a proportion of imports in the year 2021 of 10,294.3 metric tons (Sapbamrer et al., 2023).
Methomyl, a commonly used carbamate insecticide, is raising concerns due to its frequent detection in food and the environment, despite being less toxic than older alternatives (Trachantong et al., 2017). It induces acute cholinergic poisoning in mammals by inhibiting acetylcholinesterase, leading to the accumulation of acetylcholine and damage to the nervous system (Trachantong et al., 2017). Methomyl exposure triggers cellular damage (DNA, proteins, lipids) through reactive oxygen species (ROS)-induced oxidative stress and may activate MAPK pathways, leading to apoptosis or necrosis (Heikal et al., 2014; He et al., 2022). Studies highlight its significant oxidative stress in mammals (Djeffal et al., 2015). This necessitates the exploration of natural compounds to mitigate methomyl toxicity on immune cells.
Tiliacora triandra (Colebr.) Diels or Yanang is one medicinal plant that has been described for its detoxification ability in the human body and which might show the benefit of protecting against methomyl toxicity. Yanang belongs to the family of Menipermaceae and is an endemic plant of Southeast Asia (Singthong et al., 2009). In northeast Thailand, its leaf water extract is traditionally used in cooking to reduce bamboo shoot toxicity (Phunchago et al., 2015). Several studies have reported that Yanang leaf extract contains high levels of essential bioactive compounds including tannin, triterpene, saponin, phytol, α-tocopherol, flavonoids, phenolic compounds and minerals (Singthong et al., 2009; Phunchago et al., 2015; Thong-Asa et al., 2017). These bioactive compounds offer medicinal properties such as anti-fever, antibacterial, antimalarial, anticancer, anti-inflammation, antioxidant, and neuroprotective effects (Singthong et al., 2014; Thong-Asa et al., 2017). However, there is no research on the preventive effect of Yanang leaf extracts on methomyl-induced cytotoxicity in immune cell lines. Macrophages are one part of the innate immune response, having an important role in antigen presenting and the engulfing of antigens by phagocytosis (Nazimek et al., 2013). Accordingly, the roles of macrophages are in immune response initiation to regulate immunological function and are susceptible targets for chemical oxidants (Jang et al., 2015). Hence, macrophages may represent useful targets for developing new natural compounds to reduce toxicity from methomyl.
Therefore, this research aims to assess the protective effects of Yanang leaf extracts against methomyl-induced toxicity in RAW 264.7 murine macrophage cells. Various parameters were assessed, encompassing the percentage of cell cytotoxicity, apoptosis rate, and cell cycle arrest. Furthermore, alterations in mRNA expressions of apoptosis-related genes and proteomics profiling were examined to illuminate the mechanisms underlying the protective effects of Yanang leaf.
2 Materials and methods
2.1 Preparation of Yanang extract
The leaves of Yanang (Tiliacora triandra) were gathered from Khon Kaen province in the Northeastern region of Thailand. The leaves were washed under a tap, cut into small pieces, and ground with distilled water at a 1:8% w/v ratio. The extract was then filtered through cotton cloth. The filtrate was centrifuged at 6,068 ×g at 4 °C for 30 min. The pellet was discarded, and the supernatant was collected as Yanang water extract (YWE). The protein concentration and total phenolic content (TPC) were measured. The extract was kept cold (−20 °C) until further use in subsequent experiments.
2.2 Protein concentration measurement
The Bradford assay was employed to assess the concentration of protein (Bradford, 1976). Briefly, 1,000 µL of Bradford dye was mixed with 1 µL of YWE and incubated for 10 min. The absorbance was recorded at 595 nm (Microplate reader, Varioskan LUX, USA). The concentration of protein was subsequently determined using the BSA standard curve.
2.3 Total phenolic content (TPC) measurement
The TPC of YWE was assessed using the Folin-Ciocalteu (FC) reagent (Singthong et al., 2014). Briefly, 1,000 µL of YWE was mixed with 100 µL of FC reagent, 300 µL of 20 % sodium carbonate, and 500 µl of distilled water. The reaction mixture was incubated for 2 h at 50 °C, and a microplate reader (Varioskan LUX, USA) was used to record the absorbance at 765 nm. A calibration curve of standard gallic acid was used to convert the measured absorbance values into gallic acid equivalents (µg GAE/mL), which served as a measure of the total phenolic content.
2.4 Cell viability / cytotoxicity assay
RAW 264.7 cell viability (ATCC, Manassas, VA, USA) was evaluated using the MTT assay. In a 96-well plate, the cells (2.5 × 104 cells/well) were cultured for 24 h at 37 °C with 5 % CO2 in DMEM (Dulbecco's Modified Eagle Medium). Afterward, cells were exposed to YWE for an additional 24 h at doses ranging from 2.5 – 10 µg/mL. To assess the protective effect of YWE against methomyl-induced cytotoxicity in RAW 264.7, the cells were co-treated with 11,000 µM methomyl and 2.5 – 10 µg/mL of YWE for 24 h. Subsequently, the medium was substituted with 100 μL of MTT solution (0.5 mg/mL) and incubated for 30 min at 37 °C with 5 % CO2. The purple crystal (formazan) was then solubilized in 100 μL of DMSO, and the absorbance at 570 nm was quantified using a microplate reader (Varioskan LUX, USA). To calculate cell viability (%), the following formula was used: where A570test refers to the absorbance of cells treated with either YWE, methomyl, or co-treatment at 570 nm. A570control refers to the absorbance of untreated cells at 570 nm.
2.5 Dual fluorescent staining apoptosis assay
Apoptosis was studied using the dual staining approach with acridine orange/ethidium bromide (AO/EB). RAW 264.7 cells (1.0 × 105 cells/well) were plated into a 48-well plate and cultured for 24 h at 37 °C with 5 % CO2. After that, cells were incubated with 11,000 µM methomyl and supplemented with YWE (2.5 – 10 µg/mL) for an additional 24 h. After trypsinization, cells were resuspended in a fresh medium and centrifuged at 95 ×g for 5 min. Following this, a dual staining solution consisting of 10 mg/mL AO and 10 mg/mL EB in PBS was applied to stain the cell pellet in a 1:1 ratio. The cells were incubated in the dark for 15 min. A 10 μL aliquot of the stained cell suspension was transferred to microscope slides, coverslipped, and observed under a fluorescence microscope (Carl Zeiss Microscopy, USA) for analysis of apoptotic morphology.
2.6 Flow cytometry-based apoptosis assay
After being plated into a 12-well plate, RAW 264.7 cells (1.5 × 105 cells/well) were cultured for 24 h at 37 °C with 5 % CO2. After that, cells were exposed to 11,000 µM methomyl and supplemented with YWE (2.5 – 10 μg/mL) for 24 h. The cells were then trypsinized, suspended in fresh DMEM, and centrifuged at 95 ×g for 5 min. After rinsing the cell pellet with ice-cold PBS, cells were re-suspended in an appropriate volume of 1 × Annexin V binding buffer (BioLegend, USA) to achieve 1 × 106 cells/mL. Annexin V-FITC (5 μL) and propidium iodide (PI) (10 μL) (BioLegend, USA) were then added to 100 μL of this cell suspension. 400 μL of binding buffer was added after the mixture was left in the dark for 15 min. Apoptosis rates were analyzed using a flow cytometer (BD FACSCanto II, BD Biosciences, USA) with BD Accuri C6 software for data interpretation.
2.7 Flow cytometry-based cycle arrest assay
The cell cycle stage was assessed using PI staining. After treatment of RAW 264.7 cells with 11,000 µM methomyl and various concentrations of YWE, as described earlier, cell pellets were collected, rinsed with ice-cold PBS, and fixed with 400 μL of ice-cold 70 % ethanol at 4 °C overnight. The cells were then centrifuged at 95 ×g for 5 min and re-suspended in 200 μL of PBS. 100 μL of cell suspension was transferred into a flow tube and incubated in the dark with 2.5 μL of RNase A (20 mg/mL) and 2 μL of PI (1 mg/mL) for 30 min. After that, 400 μL of PBS was added to the cell suspension, and a Flow Cytometer (BD FACSCanto II, BD Biosciences, USA) was employed to assess the cell cycle.
2.8 Gene expressions study using real-time PCR
After RAW 264.7 cells were cultured and treated with 11,000 µM methomyl along with various concentrations of YWE, as described earlier, the cell pellets were harvested and processed for RNA extraction using Trizol reagent (Invitrogen, USA). The cDNA was synthesized according to the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc, USA). The PCR mixture included 2 μL of cDNA, 10 μL of LightCycle® SYBR® Green I Master mix (Roche, Switzerland), and 1 μL of each primer (10 μM) in a total volume of 20 μL. All PCR reactions were amplified using the LightCycler 480 instrument (Roche, Switzerland). The annealing temperatures for each gene-specific primer were determined individually as listed in Table 1. The comparative 2-ΔΔCT approach was employed to determine the relative alterations in gene expression levels. The GAPDH gene was used to normalize target gene expression levels.
Gene
Primer sequences (5ʹ →3ʹ)
Annealing (°C)
Product size (bp)
Reference
Caspase-9
Forward
AGCCAGATGCTGTCCCATAC
55
124
AF262319
Reverse
CAGGAGACAAAACCTGGGAA
Caspase-3
Forward
CAGAGCTGGACTGCGGTATTGA
58
172
NM_012922
Reverse
AGCATGGCGCAAAGTGACTG
GAPDH
Forward
GAGAAACCTGCCAAGTATGATGAC
50
212
NM_008084.3
Reverse
TAGCCGTATTCATTGTCATACCAG
2.9 Proteomics analysis using liquid chromatography mass spectrometry
RAW 264.7 cells were cultured and treated with 11,000 µM methomyl and various concentrations of YWE as described earlier. Then, cells were harvested, and proteins were extracted using 50 mM Tris-HCl pH 7.0 and 0.5 % SDS, with sonication at amplitude levels of 80 % for 3 s. Centrifugation was performed at 9,481 ×g for 15 min and the supernatant was used for protein extraction by incubating with ice-cold acetone containing 0.1 mM DTT (1:5 v/v ratios) for 16 h at −20 °C. The protein was subsequently digested with trypsin at a ratio of 1:20 before being incubated at 37 °C for 6 h. Peptides were fractionated by ultra-performance liquid chromatography (UPLC) and C18-reverse phase chromatography. Quantitative data analysis was processed by Decyder Ms 2.0. Protein identification was conducted using Mascot. To visualize all possible intersections among proteome datasets, a Venn diagram was generated using the jvenn tool online.
2.10 Statistical methods
Data from three independent experiments are shown as means ± SD. SPSS 20 software was used to conduct the statistical analysis. Duncan's multiple range test was employed for post-hoc comparisons after one-way ANOVA was used to evaluate differences between the treatment and control groups. A 0.05 threshold for statistical significance was applied.
3 Results
3.1 Protective effect of YWE against methomyl-induced cytotoxicity in RAW 264.7 cells
The YWE was analyzed for total protein and total phenolic content using the Bradford assay and the Folin-Ciocalteu method, respectively. The results indicate that YWE showed protein and total phenolic content at 7.42 ± 0.24 µg/mL and 5.23 ± 0.41 µg GAE/mL, respectively. Rapid protein concentration determination with the Bradford method was used for estimating YWE concentration in all experiments. MTT assay confirmed that YWE (2.5–10 µg/mL) exhibited no cytotoxicity in RAW 264.7 cells (99.08 – 100.78 % cell viability; Fig. 1A). These non-toxic YWE concentrations were then tested for protection against methomyl-induced cytotoxicity. In Fig. 1B, it is evident that methomyl at a concentration of 11,000 μM significantly induced cytotoxicity in RAW 264.7 cells, resulting in a viability of 48.16 %. However, co-treatment with YWE at concentrations of 2.5, 5, and 10 µg/mL significantly reduced this effect, with cell viabilities of 57.13 %, 60.86 %, and 64.40 %, respectively (Fig. 1B).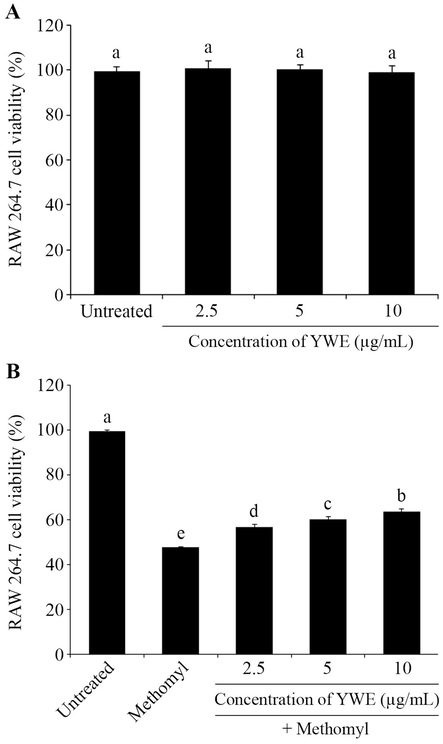
Toxicity and protective effect of Yanang water extract (YWE) on methomyl- induced cytotoxicity in RAW 264.7 cells. (A) RAW 264.7 cells were exposed to YWE at concentrations of 2.5 – 10 µg/mL for 24 h and then assessed using the MTT assay to determine their effect on cell viability. (B) The RAW 264.7 cells viability was measured using MTT assay after a 24 h incubation with 11,000 µM methomyl and supplementation with different concentrations of YWE (2.5 – 10 μg/mL) to evaluate its protective effect. The data are presented as mean ± SD. Different letters on the top of each bar are significantly different, p ≤ 0.05.
3.2 Protective effect of YWE on methomyl-induced apoptosis in RAW 264.7 cells
The morphological apoptotic changes of RAW 264.7 cells with AO/EB dual staining are shown in Fig. 2A. AO stains live cells green, while EB stains apoptotic cells orange or red. The untreated control group showed predominantly green live cells, whereas the methomyl-treated group exhibited more apoptotic cells. Co-treatment with YWE (2.5 – 10 μg/mL) led to a concentration-dependent reduction in apoptotic cells.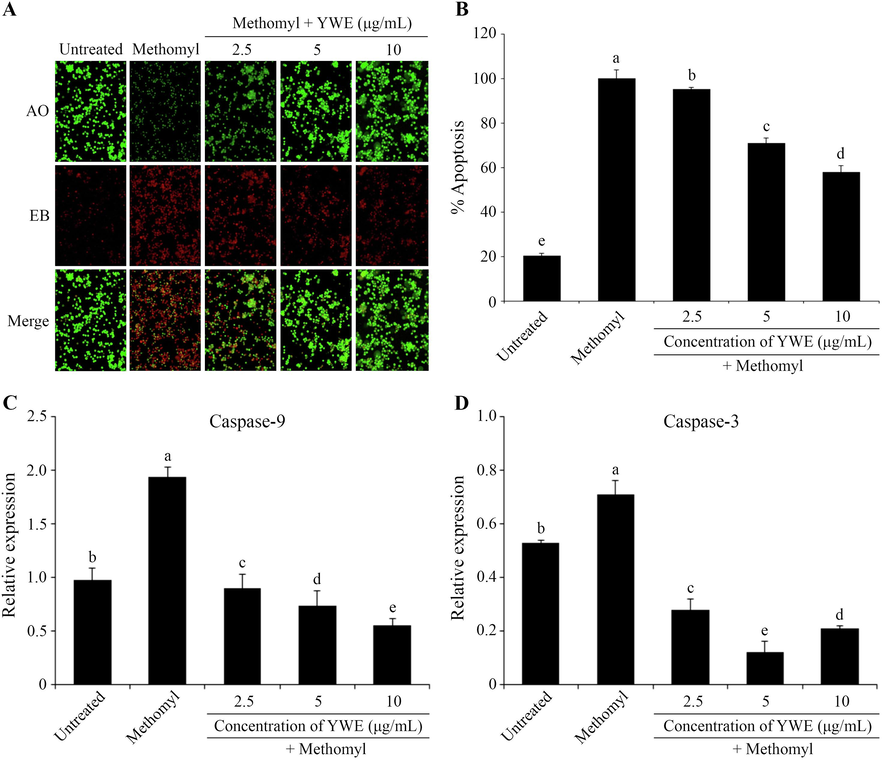
Protective effects of Yanang water extract (YWE) on methomyl-induced apoptosis in RAW 264.7 cells. After treatment with methomyl and different concentrations of YWE (2.5 – 10 μg/mL) for 24 h, RAW 264.7 cells were stained with AO/EB and observed under fluorescence microscopy (20×) (A). The cells undergoing apoptosis were further measured by annexin V-FITC/PI flow cytometry assay and the apoptosis rate was determined and represented as a bar graph (B). The protective effect of YWE on apoptosis related genes, Caspase-9 (C) and Caspase-3 (D) was assessed through real-time PCR. The data are presented as mean ± SD. Different letters on the top of each bar are significantly different, p ≤ 0.05.
Annexin V-FITC/PI flow cytometry confirmed methomyl-induced apoptosis in RAW 264.7 cells, which was significantly reduced with YWE treatment. Apoptotic cell percentages decreased in a concentration-dependent manner to 95.54 %, 71.26 %, and 57.87 % with 2.5, 5, and 10 μg/mL of YWE, respectively (Fig. 2B).
Quantitative real-time PCR analysis revealed that methomyl treatment increased the expression of Caspase-9 and Caspase-3, key regulators of apoptosis, compared to the untreated group. Conversely, YWE co-treatment (2.5 – 10 µg/mL) downregulated these genes, suggesting that YWE may exert its protective effect against methomyl-induced apoptosis by regulating caspase expression (Fig. 2C, 2D).
3.3 Protective effects of YWE on cell cycle arrest induced by methomyl toward RAW 264.7 cells
DNA flow cytometric analysis indicated that methomyl significantly induced G2/M cell cycle arrest by increasing the G2/M phase from untreated control to 7.30 % ± 0.28. However, the percentages of RAW 264.7 cells in the G2/M phase were significantly reduced by co-treatment with YWE (2.5, 5, and 10 μg/mL) to 6.45 % ± 0.49, 5.85 % ± 0.30, and 3.30 % ± 0.18, respectively. These findings suggest that YWE exerts protective effects by preventing methomyl-induced cell cycle arrest in RAW 264.7 cells, as evidenced by the reduced G2/M phase population (Fig. 3).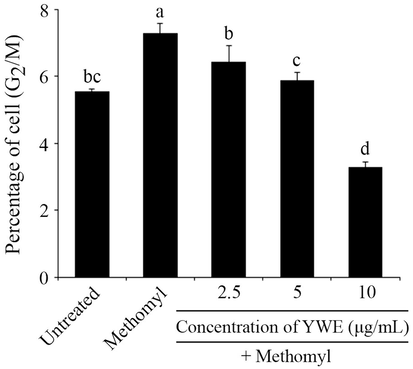
Protective effect of Yanang water extract (YWE) on methomyl-induced cell cycle arrest in RAW 264.7 cells. After treatment with methomyl and different concentrations of YWE (2.5 – 10 μg/mL) for 24 h, RAW 264.7 cells were stained with PI and measured by flow cytometry. The percentage of RAW 264.7 cells in G2/M peak is presented as mean ± SD. Different letters on the top of each bar are significantly different, p ≤ 0.05.
3.4 Protective effects of YWE on the protein patterns in methomyl-induced toxicity in RAW 264.7 cells
The proteomic analysis revealed 681 proteins expressed in RAW 264.7 cells under 5 different conditions, including untreated control, methomyl alone, and co-treatment of methomyl with 2.5, 5, and 10 µg/mL of YWE. The number of expressed proteins in each group was 550, 536, 575, 565, and 561, respectively (Fig. 4). A Venn diagram of differentially expressed proteins (DEPs) indicated that 423 proteins were shared commonly among all 5 conditions, while 7 proteins were only observed in the untreated control and methomyl treated groups, 5, 8 and 1 proteins were only observed in co-treatment of methomyl with 2.5, 5, and 10 μg/mL of YWE, respectively (Table S1). The functional proteins related to cell death and cell cycle pathway are indicated in Table 2-4. Table 2 lists the proteins exclusively observed in each group, which have known functions in apoptosis and cell cycle pathways. Of these, 4 proteins were only found in the methomyl treated group, including RAC-alpha serine/threonine-protein kinas (AKT1), Gamma-aminobutyric acid receptor subunit alpha-3 (GBRA3), LAG1 longevity assurance homolog 1 (LASS1) and programmed cell death protein 2-like (PDD2L). Meanwhile, co-treatment with 2.5 – 10 μg/mL of YWE altered functional proteins by promoting an expression of CUB and sushi domain-containing protein 1 (CSMD1) at a dose of 2.5 μg/mL, Homeobox protein DLX-6 (DLX6), Spectrin alpha chain (SPTA1) and E3 ubiquitin-protein ligase TRIP12 (TRIPC) at a dose of 5 μg/mL, and RING finger protein 113A (R113A) at a dose of 10 μg/mL.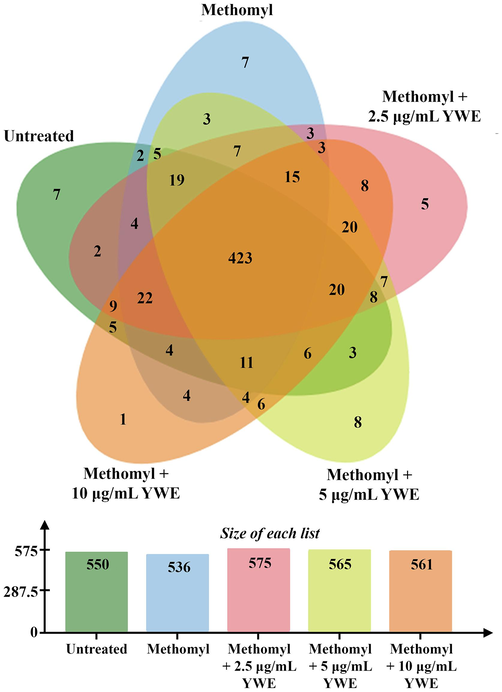
Venn diagram presenting the expressed proteins of 5 treatments, including untreated control (550 proteins), methomyl treatment (536 proteins), and supplementation of 2.5 μg/mL Yanang water extract (YWE) (575 proteins), 5 μg/mL YWE (565 proteins), and 10 μg/mL YWE (561 proteins) to methomyl treated RAW 264.7 cells.
Gene name
Matched protein
t-Test (p)
Function
Reference
Appears only in methomyl treated group
AKT1
RAC-alpha serine/threonine-protein kinase
0.96
Relates to cell proliferation, apoptosis and survival
Hou et al., 2022
GBRA3
Gamma-aminobutyric acid receptor subunit alpha-3
0.91
Promotes apoptosis and differentiation in T-helper cell type-2
Meng et al., 2016
LASS1
LAG1 longevity assurance homolog 1
0.75
Inhibits proliferation, induces apoptosis in HepG2 HB cells through mitochondrial apoptotic, NF–κB and cell cycle signaling pathways (LASS2)
Yang et al., 2019
Related to ceramide synthesis which might promote cell cycle arrest and apoptosis (LASS5)
Xu et al., 2005; Gomez-Larrauri et al., 2020
PDD2L
Programmed cell death protein 2-like
1.00
Deletes lymphocytes by reducing lymphocyte proliferation and cytokine production (PD-1)
Berntsson et al., 2018
Promotes apoptosis, suppresses cell transformation by inhibiting protein translation (PDCD-4)
Lankat-Buttgereit and Goke, 2003
Appears only in co-treatment of methomyl and 2.5 μg/mL YWE group
CSMD1
CUB and sushi domain-containing protein 1
0.64
Suppresses tumor progression
Blom, 2017
Appears only in co-treatment of methomyl and 5 μg/mL YWE group
DLX6
Homeobox protein DLX-6
0.68
Anti-necrotic effects and promotes cell proliferation (DLX-2 and family)
Lee et al., 2011
SPTA1
Spectrin alpha chain, erythrocyte
0.62
Target of caspases
Karanam et al., 2010
TRIPC
E3 ubiquitin-protein ligase TRIP12
0.98
Regulates cell cycle progression
Brunet et al., 2020
Appears only in co-treatment of methomyl and 10 μg/mL YWE group
R113A
RING finger protein 113A
0.57
Controls stability, trafficking and activity of proteins, cell proliferation and differentiation, apoptosis, immune regulation, signaling and mitochondrial dynamics
Nakamura, 2011
Gene name
Matched protein
t-Test (p)
Log2 expression value
Function
Reference
Methomyl
Methomyl + YWE (μg/mL)
2.5
5
10
DIDO1
Death-inducer obliterator 1
0.04
7.79 ± 0.55
7.62 ± 0.46
–
–
Pro-apoptotic protein and is induced by apoptosis
Guerfali et al., 2008; Kumar et al., 2013
CUL5
Cullin-5
0.14
7.92 ± 1.60
8.71 ± 1.87
6.03 ± 0.18
–
Inhibits cell growth through ubiquitylating and degrading adenosine 3′,5′-monophosphate-responsive element binding protein 1 (CREB1)
Chen et al., 2023
Gene name
Matched protein
t-Test (p)
Log2 expression value
Function
Reference
Untreated
Methomyl
Methomyl + YWE (μg/mL)
2.5
5
10
MCM5
DNA replication licensing factor MCM5
0.67
8.10 ± 0.27
7.74 ± 0.21
8.21 ± 0.11
8.18 ± 0.14
8.08 ± 0.13
Improves growth and survival via inactivation of p53
Agarwal et al., 2007
CCNB2
G2/mitotic-specific cyclin-B2
0.44
8.69 ± 0.33
8.83 ± 0.08
8.98 ± 0.33
9.19 ± 0.11
9.01 ± 0.39
Positively regulates cell proliferation
Wu et al., 2021
IQGA1
Ras GTPase-activating-like protein IQGAP1
0.12
8.93 ± 0.33
7.37 ± 1.08
8.97 ± 0.02
8.09 ± 0.19
8.79 ± 0.10
Promotes cell proliferation and cell cycle progression, inhibits apoptosis
Sato et al., 2010; Zeng et al., 2018
RBBP6
Retinoblastoma-binding protein 6
0.01
8.45 ± 0.46
7.16 ± 0.38
7.92 ± 0.33
7.27 ± 0.06
8.20 ± 0.08
Promotes cell proliferation and inhibits apoptosis via p53 degradation
Motadi et al., 2011
JAK2
Tyrosine-protein kinase JAK2
0.77
5.22 ± 0.05
4.26 ± 0.35
4.42 ± 0.36
5.48 ± 1.76
5.07 ± 0.48
Enhances cell growth and survival
Kovanen et al., 2000
STOX2
Storkhead-box protein 2
0.19
9.98 ± 2.25
9.59 ± 2.10
9.42 ± 0.01
10.10 ± 0.11
9.73 ± 1.49
Suppresses apoptosis and promotes growth
Sasahira et al., 2016
PAWR
PRKC apoptosis WT1 regulator protein
0.99
4.77 ± 0.32
5.26 ± 0.84
6.01 ± 0.40
5.82 ± 0.05
4.99 ± 0.37
Promotes apoptosis
Shukla et al., 2016
Moreover, 39 proteins were differentially expressed across the groups of methomyl treatment, and co-treatments of methomyl with YWE were also evaluated (Table S2). Of these, the expression levels of 2 apoptosis-related proteins, Death-inducer obliterator 1 (DIDO1) and Cullin-5 (CUL5) were found to have a marked decrease in a dose-dependent manner of YWE (Table 3).
Among the 423 proteins commonly identified in all 5 conditions (Table S3), 7 proteins are involved in apoptosis and cell cycle arrest. These include DNA replication licensing factor MCM5 (MCM5), G2/mitotic-specific cyclin-B2 (CCNB2), Ras GTPase-activating-like protein IQGAP1 (IQGA1), Retinoblastoma-binding protein 6 (RBBP6), Tyrosine-protein kinase JAK2 (JAK2), Storkhead-box protein 2 (STOX2), and PRKC apoptosis WT1 regulator protein (PAWR). Their expression levels exhibited slight changes, as shown in Table 4.
4 Discussion
Methomyl induces ROS formation, leading to oxidative stress (Jang et al., 2015), which can result in cell cycle arrest, apoptosis, or necrosis (He et al., 2022). Our findings consistently demonstrate methomyl induced apoptosis and cell cycle arrest in RAW 264.7 cells, as shown by AO/EB double staining and flow cytometry. However, supplementation with YWE significantly reduced these effects. One of the mechanisms in apoptosis induction involves the caspases. These enzymes are a conserved protein family activated after a cell receives pro-apoptotic signals (Heikal et al., 2014). Caspases associated with apoptosis have been classified into two groups based on their mechanisms: the initiator caspases, represented by caspase-9, and the effector caspases, represented by caspase-3 (Shi, 2004). The literature indicates that exposure to methomyl, both caspase-9 and caspase-3, results in increased mRNA expression levels in experimental animals (Heikal et al., 2014; He et al., 2022). Our study confirms methomyl increases caspase-9 and −3 expression in RAW 267.4 cells, and YWE treatment reduces these levels.
Proteomic analysis suggests methomyl disrupts RAW 264.7 cell functions like apoptosis, cell cycle, and survival. RAW 264.7 cells exposed to methomyl displayed 4 unique proteins, which disappeared upon co-treatment with YWE. This suggests potential mitigation of methomyl cytotoxicity by YWE across all doses. RAC-alpha serine/threonine-protein kinase (AKT1) is a well-known protein kinase that plays a critical role in various vital biological processes, including cell survival, cell proliferation, and apoptosis (Hou et al., 2022). Gamma-aminobutyric acid receptor subunit alpha-3 (GBRA3), found exclusively in the methomyl treatment group, is a component of the Gamma-aminobutyric acid receptor (GABA). Despite its main function in the central nervous system, GABA has been linked to immune suppression, including promoting apoptosis and differentiation in T-helper cells (Meng et al., 2016). LAG1 longevity assurance homolog 1 (LASS1) and Programmed cell death protein 2-like (PDD2L or PD-2) functions are less known, but their family regulates apoptosis and proliferation. Yang et al. (2019) found LASS2 suppresses cell proliferation and induces apoptosis. LASS5 has been reported as a regulator in ceramide synthesis. An increase in ceramide has commonly been assumed to induce apoptosis and cell cycle arrest (Xu et al., 2005; Gomez-Larrauri et al., 2020). Similarly, the interaction of PD-1 with its ligand triggers lymphocyte deletion by downregulating proliferation and cytokine production (Berntsson et al., 2018). Meanwhile, PD-4 can initiate apoptosis by inhibiting protein translation (Lankat-Buttgereit and Goke, 2003). The data reported here appear to support the assumption that 2.5 – 10 μg/mL YWE shows a protective effect against apoptosis and cell cycle arrest via down-regulation of these proteins.
On the other hand, anti-apoptotic proteins were observed to be exclusively expressed only in the co-treatment group with YWE and methomyl. The function of CUB and sushi domain-containing protein 1 (CSMD1) remains limited due to a lack of thorough investigation. One explanation of its function is tumor suppressive activity in human breast cancer cells (Blom, 2017). Hemeobox protein DLX-6 (DLX6) belongs to the DLX family in humans, which includes DLX-1, DLX-2, DLX-3, DLX-5, and DLX-7. Interestingly, DLX-2 has been reported to hold anti-necrotic activity and could promote cell proliferation (Lee et al., 2011). Spectrin alpha chain (SPTA1), is one of the caspase targets, as well as myosin and vimentin. Karanam et al. (2010) suggest that SPTA1 levels depend on specific regulation and may indicate caspase activity linked to the mitochondrial apoptotic pathway. This supports our finding that SPTA1 presence might suggest an anti-apoptotic effect. E3 ubiquitin-protein ligase TRIP12 (TRIPC) plays crucial roles in various vital biological processes, including cell cycle progression, DNA/chromatin repair, and cell differentiation. Inactivation of TRIPC leads to cell cycle arrest and apoptosis (Brunet et al., 2020). The last outstanding protein in this group is RING finger (RNF) protein 113A (R113A). Common functions of RNF protein impact many routes of cellular and physiological processes, including cell stability, integrity, trafficking, proliferation, differentiation, apoptosis, signaling, and mitochondrial dynamics (Nakamura, 2011). The evidence from this study suggests that YWE dose influences protein expression through different cellular mechanisms. Notably, the higher dose appeared to have a significantly greater impact on inhibiting apoptosis and cell cycle arrest, while also inducing cell cycle progression.
Death-inducer obliterator 1 (DIDO1) and Cullin-5 (CUL5) were interestingly observed to be down-regulated in the co-treatment with YWE and methomyl. DIDO1, a pro-apoptotic protein, becomes overexpressed upon activation of the apoptotic signal, ultimately leading to cell death (Guerfali et al., 2008; Kumar et al., 2013). Meanwhile, CUL5 acts as a tumor suppressor. Its overexpression leads to inhibition of cell growth via ubiquitylating and degrading adenosine 3′,5′-monophosphate-responsive element binding protein 1 (CREB1) (Chen et al., 2023). It is crucial to emphasize that suppression of these proteins was controlled in a dose-dependent manner by YWE. One possible implication of this is that the bioactive compound of YWE might be able to disrupt apoptotic signaling and promote cell cycle progression, by governing the critical functions of these proteins.
A total of 423 shared common datasets across 5 experimental groups were screened, of which 7 proteins were associated with apoptotic proteins. Among these 7 proteins, 6 proteins were up-regulated, and 1 protein was down-regulated. The first up-regulated protein was the DNA replication licensing factor MCM5 (MCM5). MCM5 is relevant to inactivation of p53 signaling, resulting in enhanced cell growth and survival (Agarwal et al., 2007). G2/mitotic-specific cyclin-B2 (CCNB2) is one of the most important proteins involved in cell cycle regulation. Its defect leads to the failure of the G2/M checkpoint during the cell cycle progression (Wu et al., 2021). Ras GTPase-activating-like protein IQGAP1 (IQGA1) is upregulated in breast cancer and promotes cell proliferation while regulating the cell cycle and suppressing apoptosis (Sato et al., 2010; Zeng et al., 2018). Retinoblastoma-binding protein 6 (RBBP6) interacts with p53 and pRb proteins, promoting cell proliferation by facilitating p53 protein degradation through its RNF domain (Motadi et al., 2011). Meanwhile, Tyrosine-protein kinase JAK2 (JAK2) and Storkhead-box protein 2 (STOX2) were similarly reported to promote cell growth, cell survival, and prevent apoptosis (Kovanen et al., 2000; Sasahira et al., 2016). In contrast, PRKC apoptosis WT1 regulator protein (PAWR or Par-4) was observed to be down-regulated in YWE supplemented groups. Par-4 is one of the most well-known pro-apoptotic proteins and was identified in cancer cells. Secretion of Par-4 promotes apoptosis in cancer cells with specific activation through cell surface receptor GRP78 (Shukla et al., 2016). Moreover, their expression levels were gradually agreeable with doses of YWE. Taken together, the observed up- and down-regulation of proteins by YWE suggests its potential to mitigate methomyl-induced toxicity, possibly by interfering with apoptotic and cell cycle pathways.
5 Conclusions
YWE protects RAW 264.7 cells from methomyl toxicity by reducing apoptosis and cell cycle arrest via suppressing caspase-9 and caspase-3 mRNA expression. Proteomic analysis reveals methomyl induces proteins associated with immune suppression and cell cycle arrest. Co-treatment with YWE and methomyl upregulates anti-apoptotic and cell cycle progression proteins, while suppressing pro-apoptotic proteins. This suggests that YWE disrupts apoptotic signaling and promotes cell cycle progression, preventing methomyl-induced cytotoxicity.
CRediT authorship contribution statement
Boonyarit Kukaew: Writing – original draft, Visualization, Investigation, Formal analysis. Wanna Sirisangtragul: Writing – original draft, Supervision, Methodology. Sittiruk Roytrakul: Writing – review & editing, Methodology, Investigation, Formal analysis. Anupong Joompang: Writing – original draft, Formal analysis. Napaporn Roamcharern: Writing – original draft, Validation, Formal analysis. Anupong Tankrathok: Writing – review & editing, Formal analysis. Pattralak Songserm: Writing – original draft, Visualization. Sakda Daduang: Writing – review & editing, Supervision. Sompong Klaynongsruang: Supervision, Funding acquisition. Nisachon Jangpromma: Writing – review & editing, Visualization, Supervision, Methodology, Investigation, Funding acquisition.
Acknowledgements
This work was supported by the Fundamental Fund of Khon Kaen University, the National Science, Research and Innovation Fund (NSRF), Thailand, and the Protein and Proteomics Research Center for Commercial and Industrial Purposes (ProCCI), Faculty of Science, Khon Kaen University, Thailand.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- DNA replication licensing factor minichromosome maintenance deficient 5 rescues p53-mediated growth arrest. Cancer Res.. 2007;67(1):116-121.
- [Google Scholar]
- Expression of programmed cell death protein 1 (PD-1) and its ligand PD-L1 in colorectal cancer: Relationship with sidedness and prognosis. Oncoimmunology. 2018;7(8):e1465165.
- [Google Scholar]
- The role of complement inhibitors beyond controlling inflammation. J. Intern. Med.. 2017;282(2):116-128.
- [Google Scholar]
- A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem.. 1976;72:248-254.
- [Google Scholar]
- E3 Ubiquitin Ligase TRIP12: Regulation, structure, and physiopathological functions. Int. J. Mol. Sci.. 2020;21(22):8515.
- [Google Scholar]
- Cullin-5 deficiency orchestrates the tumor microenvironment to promote mammary tumor development through CREB1-CCL2 signaling. Sci. Adv.. 2023;9(3):eabq1395.
- [Google Scholar]
- Role of bioactive sphingolipids in physiology and pathology. Essays Biochem.. 2020;64(3):579-589.
- [Google Scholar]
- Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genom.. 2008;9:238.
- [Google Scholar]
- Oxidative stress induced by methomyl exposure reduces the quality of early embryo development in mice. Zygote. 2022;30(1):57-64.
- [Google Scholar]
- Protective effects of vitamin C against methomyl-induced injuries on the testicular antioxidant status and apoptosis-related gene expression in rat. J. Environ. Anal. Toxicol.. 2014;5(2):1-7.
- [Google Scholar]
- A RAC-alpha serine/threonine-protein kinase (CgAKT1) involved in the synthesis of CgIFNLP in oyster Crassostrea gigas. Fish. Shellfish. Immunol.. 2022;127:129-139.
- [Google Scholar]
- Paraquat induces apoptosis through a mitochondria-dependent pathway in RAW264.7 cells. Biomol. Ther. (Seoul). 2015;23(5):407-413.
- [Google Scholar]
- Proteome analysis reveals new mechanisms of Bcl11b-loss driven apoptosis. J. Proteome Res.. 2010;9(8):3799-3811.
- [Google Scholar]
- Regulation of Jak2 tyrosine kinase by protein kinase C during macrophage differentiation of IL-3-dependent myeloid progenitor cells. Blood. 2000;95(5):1626-1632.
- [Google Scholar]
- Lipoic acid prevents Cr(6+) induced cell transformation and the associated genomic dysregulation. Environ. Toxicol. Pharmacol.. 2013;36(1):182-193.
- [Google Scholar]
- Programmed cell death protein 4 (pdcd4): A novel target for antineoplastic therapy? Biol. Cell. 2003;95(8):515-519.
- [Google Scholar]
- Homeobox gene Dlx-2 is implicated in metabolic stress-induced necrosis. Mol. Cancer. 2011;10:113.
- [Google Scholar]
- Propofol inhibits T-helper cell type-2 differentiation by inducing apoptosis via activating gamma-aminobutyric acid receptor. J. Surg. Res.. 2016;206(2):442-450.
- [Google Scholar]
- Expression and function of retinoblastoma binding protein 6 (RBBP6) in human lung cancer. Immunobiology. 2011;216(10):1065-1073.
- [Google Scholar]
- The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes (Basel). 2011;1(4):354-393.
- [Google Scholar]
- Macrophage function in allergic and autoimmune responses. J. Phys. Ther. Health Promot.. 2013;1(1):36-45.
- [Google Scholar]
- Tiliacora triandra, an anti-intoxication plant, improves memory impairment, neurodegeneration, cholinergic function, and oxidative stress in hippocampus of ethanol dependence rats. Oxid. Med. Cell. Longev.. 2015;2015:918426
- [Google Scholar]
- Important role of the government in reducing pesticide use and risk sustainably in Thailand: Current situation and recommendations. Front. Public Health. 2023;11:1141142.
- [Google Scholar]
- Storkhead box 2 and melanoma inhibitory activity promote oral squamous cell carcinoma progression. Oncotarget. 2016;7(18):26751-26764.
- [Google Scholar]
- Association of RNase L with a Ras GTPase-activating-like protein IQGAP1 in mediating the apoptosis of a human cancer cell-line. FEBS J.. 2010;277(21):4464-4473.
- [Google Scholar]
- Caspase activation, inhibition, and reactivation: A mechanistic view. Protein Sci.. 2004;13(8):1979-1987.
- [Google Scholar]
- Shukla, N., Zhao, Y., Rangnekar, V., 2016. PAWR (PRKC apoptosis WT1 regulator protein; Prostate apoptosis response-4, Par-4), Atlas Genet Cytogenet Oncol Haematol.
- Extraction and physicochemical characterisation of polysaccharide gum from Yanang (Tiliacora triandra) leaves. Food Chem.. 2009;114(4):1301-1307.
- [Google Scholar]
- Bioactive compounds and encapsulation of Yanang (Tiliacora triandra) leaves. Afr. J. Tradit. Complement. Altern. Med.. 2014;11(3):76-84.
- [Google Scholar]
- Tiliacora triandra (Colebr.) Diels leaf extract enhances spatial learning and learning flexibility, and prevents dentate gyrus neuronal damage induced by cerebral ischemia/reperfusion injury in mice. Avicenna. J. Phytomed.. 2017;7(5):389-400.
- [Google Scholar]
- Lethal and sublethal effects of a methomyl-based insecticide in Hoplobatrachus rugulosus. J. Toxicol. Pathol.. 2017;30(1):15-24.
- [Google Scholar]
- Agriculture development, pesticide application and its impact on the environment. Int. J. Environ. Res. Public Health. 2021;18(3):1112.
- [Google Scholar]
- Cyclin B2 (CCNB2) stimulates the proliferation of triple-negative breast cancer (TNBC) cells in vitro and in vivo. Dis. Markers. 2021;2021:5511041.
- [Google Scholar]
- LASS5 is the predominant ceramide synthase isoform involved in de novo sphingolipid synthesis in lung epithelia. J. Lipid Res.. 2005;46(6):1229-1238.
- [Google Scholar]
- LASS2 inhibits proliferation and induces apoptosis in HepG2 cells by affecting mitochondrial dynamics, the cell cycle and the nuclear factor-kappaB pathways. Oncol. Rep.. 2019;41(5):3005-3014.
- [Google Scholar]
- Ras GTPase-activating-like protein IQGAP1 (IQGAP1) promotes breast cancer proliferation and invasion and correlates with poor clinical outcomes. Med. Sci. Monit.. 2018;24:3315-3323.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103229.
Appendix A
Supplementary material
The following are the Supplementary data to this article:Supplementary Data 1
Supplementary Data 1
The list of functional proteins that noticeably appear only in each treatment
Supplementary Data 2
Supplementary Data 2
The list of functional proteins expressed by the protective effect of Yanang water extract (YWE). The relative quantitation ratios of each sample are presented as log2
Supplementary Data 3
Supplementary Data 3
The list of the 423 differentially expressed functional proteins that shared common datasets across all treatment groups. The relative quantitation ratios of each sample are presented as log2







